Abstract
Little is known about how genetic variation at the nucleotide level contributes to competitive fitness within species. During a 6,000-generation study of Bacillus subtilis evolved under relaxed selection for sporulation, a new strain, designated WN716, emerged with significantly different colony and cell morphologies; loss of sporulation, competence, acetoin production, and motility; multiple auxotrophies; and increased competitive fitness (H. Maughan and W. L. Nicholson, Appl. Environ. Microbiol. 77:4105–4118, 2011). The genome of WN716 was analyzed by OpGen optical mapping, whole-genome 454 pyrosequencing, and the CLC Genomics Workbench. No large chromosomal rearrangements were found; however, 34 single-nucleotide polymorphisms (SNPs) and +1 frameshifts were identified in WN716 that resulted in amino acid changes in coding sequences of annotated genes, and 11 SNPs were located in intergenic regions. Several classes of genes were affected, including biosynthetic pathways, sporulation, competence, and DNA repair. In several cases, attempts were made to link observed phenotypes of WN716 with the discovered mutations, with various degrees of success. For example, a +1 frameshift was identified at codon 13 of sigW, the product of which (SigW) controls a regulon of genes involved in resistance to bacteriocins and membrane-damaging antibiotics. Consistent with this finding, WN716 exhibited sensitivity to fosfomycin and to a bacteriocin produced by B. subtilis subsp. spizizenii and exhibited downregulation of SigW-dependent genes on a transcriptional microarray, consistent with WN716 carrying a knockout of sigW. The results suggest that propagation of B. subtilis for less than 2,000 generations in a nutrient-rich environment where sporulation is suppressed led to rapid initiation of genomic erosion.
INTRODUCTION
The phenotypic plasticity of bacterial genomes enables bacteria to alter their gene expression in response to environmental stresses (18). A well-studied example of one such stress is the transition of Bacillus subtilis cells from exponential growth into the stationary phase upon nutrient limitation (49). Transition state genes in B. subtilis include those involved in such global responses as chemotaxis and motility; production of extracellular enzymes; production of, and resistance to, antibiotics; genetic competence; and initiation of sporulation (reviewed in reference 46). Sporulation itself is a complex developmental process requiring over 125 genes and taking about 6 to 8 h (7, 13, 48). Of these genes, about 21% are pleiotropic, i.e., also involved in housekeeping and other cellular processes (20). Thus, relaxing selective pressure for sporulation would be predicted to reduce the genetic interference from sporulation in pleiotropic genes and to allow greater potential for their variation in a population (14). Understanding how sporulating bacteria evolve under constant sporulation-repressing conditions can help us further elucidate how genes interact at the whole-organism level.
Laboratory evolution experiments have become important tools for exploring the underlying processes of evolution in microorganisms (6, 33). In previous communications, we have reported on evolution of five B. subtilis populations, WN624A through WN624E, that were cultivated for 6,000 generations in a sporulation-repressing, high-nutrient medium (R medium) (19–21). Populations 624A and 624C had reduced ability to sporulate, and three populations (624B, 624D, and 624E) lost this trait entirely (20). During the course of this evolution experiment, we noted a novel small-colony variant that appeared in, swept through, and became fixed in three out of the five evolving populations (624A, 624B, and 624E) (21). We showed that these small-colony variants had gained increased fitness in R medium and displayed a number of new phenotypic traits related to growth (a higher exponential growth rate and cell filamentation) and to the transition state (continued growth in postexponential phase; loss of motility, sporulation, and genetic competence; and altered fermentation profiles) (21).
To better understand the genetic causes of increased fitness in the small-colony variants, the WN624A population was chosen for further studies. In this population, the small-colony variant appeared at generation 1,330 and completely swept through the culture in ∼450 generations. Representative colonies from before (WN715) and after (WN716) the population sweep were isolated. The small-colony variant strain WN716 exhibited an unusual long, filamentous cell morphology that was distinct from the single rod-shaped cells characteristic of traditional B. subtilis, and the motility of WN716 was lost or greatly reduced. In competition experiments using R medium, even when strains WN715 and WN716 were inoculated at a 1,000:1 ratio, respectively, strain WN716 became the sole inhabitant and strain WN715 disappeared within ∼40 generations (21).
Transcriptional microarrays were performed on RNA extracted from early-stationary-phase cells of WN715 versus WN716 to identify expression changes greater than 8-fold. The microarrays revealed genes involved in purine and pyrimidine biosynthesis and stress responses to be preferentially upregulated during early stationary phase in WN716. Genes for major autolysins, transport proteins, cytochromes, early sporulation functions, competence, antibiotic production, acetoin fermentation, and motility were preferentially downregulated in WN716 (21). These results suggested that the increased fitness of WN716 was not due to a single mutation but that a number of changes must have occurred during its evolution in order to result in its complex alteration of multiple phenotypic traits.
In this communication, we describe experiments undertaken with the purpose of identifying genomic changes leading to the observed phenotypes of evolved strain WN716, using two approaches. First, optical mapping was used to search for potential large-scale genetic changes (e.g., large deletions, insertions, or inversions). Second, whole-genome 454 pyrosequencing was used to identify small-scale changes (e.g., single-nucleotide polymorphisms [SNPs] or small insertions/deletions [indels]). Three strains from the 624A evolution experiment were subjected to analysis; ancestral strain WN624, presweep strain WN715, and postsweep strain WN716. Predicted phenotypes resulting from mutations identified in WN716 were tested.
MATERIALS AND METHODS
B. subtilis strains, media, and growth conditions.
The construction of the ancestral B. subtilis strain WN624 (trpC2 amyE::spc) and the evolution of strains WN715 (trpC2 amyE::cat) and WN716 (multiply auxotrophic; amyE::spc) have been described in detail previously (21). B. subtilis subsp. spizizenii strains NRRL B-23049T and NRRL B14821 were generously donated by Mike Roberts, Team Qinetiq North America (31). The media used included R medium (21), LB medium (23), Schaeffer sporulation medium (SSM) (40), and Spizizen minimal medium (SMM) (47). When required, media were supplemented with the following antibiotics (final concentrations): chloramphenicol (5 μg/ml), spectinomycin (100 μg/ml), and rifampin (50 μg/ml). Fosfomycin and mitomycin C sensitivities were tested in liquid LB medium by 2-fold serial dilutions, giving final concentrations ranging from 0 to 800 μg/ml and 1 to 128 ng/ml, respectively, and cells were grown in 96-well microtiter plates at 37°C for 8 h before growth was measured. Where appropriate, SMM was supplemented with the following auxotrophic requirements (final concentrations): arginine (25 μg/ml), biotin (5 ng/ml), glutamate (25 μg/ml), histidine (25 μg/ml), and tryptophan (25 μg/ml). All growth was at 37°C with aeration. Testing of the resistance of strains WN624, WN715, and WN716 to the antibiotics produced by B. subtilis subsp. spizizenii was performed as described previously (2).
Optical mapping.
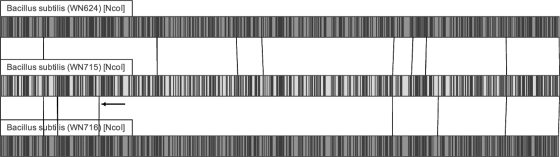
Frozen cells of strains WN624, WN715, and WN716 were sent to OpGen (Madison, WI) for optical mapping. Each whole genome was digested with NcoI and scanned using their Argus optical-mapping technology. The optical maps of WN624, WN715, and WN716 were compared to each other and to an in silico map of the reference strain 168 to check for large-scale deletions, inversions, and translocations in their genomes. Palindromic regions were ignored or, often, misidentified by the program as inversions (Fig. 1), and changes smaller than 5 kbp were too small for the technology to resolve.
Fig. 1.
Optical maps of ancestral strain WN624 (top), presweep strain WN715 (middle), and postsweep strain WN716 (bottom). Whole genomes of ancestral strain WN624 and evolved strains WN715 and WN716 were digested with NcoI, and optical maps of each strain were created by OpGen technology. The ∼4-kb deletion detected in strain WN716 by optical mapping is indicated by the arrow. All other lines connecting maps are due to small inversions in palindromic regions generated by the mapping program (see Materials and Methods).
DNA isolation, 454 pyrosequencing, and mutation identification.
DNA from B. subtilis strains was isolated as described previously (37). Whole-genome sequences of strains WN624, WN715, and WN716 were determined by 454 pyrosequencing. Sequencing libraries were prepared using the standard protocol and kit provided by 454 Life Sciences (Roche Diagnostics). NextGen (University of Florida) sequenced each genome on a 454 GS-FLXTM Standard 454 pyrosequencing platform. Each strain was then subjected to a genome-wide comparison to the reference genome, 168, using the CLC Genomics Workbench (CLC GW) alignment and annotation tools. Mutations (SNPs and indels) that were consistent among WN624, WN715, and WN716 (even if all three strains differed from the 168 reference sequence at the position in question) were discarded. We compiled a list of the remaining mutations that were the same in WN624 and WN715 but different in WN716 and verified each mutation by direct visual inspection of the sequence in CLC GW. Mutations that had fewer than 4 reads or less than 100% agreement were discarded.
Since homopolymeric regions are hot spots for misreads, mutations that occurred in these regions (fliR, alsR, clpE, and lytD), were verified by PCR amplification of the region and automated Sanger sequencing using an ABI 3130 DNA sequencer. Mutations that occurred within open reading frames of genes with known annotated functions (as predicted on NCBI for the reference genome) were further analyzed to determine their possible contributions to the phenotype of strain WN716. Mutations within intergenic regions were analyzed as follows. The intergenic regions surrounding these mutations were identified using the Subtilist database (http://genolist.pasteur.fr/SubtiList/) (28, 29) and further examined using DBTBS (http://dbtbs.hgc.jp/; 44) to identify their locations within putative promoters, transcription factor binding sites, or transcriptional terminators. Mutations located within terminators were further analyzed using MFOLD (http://mfold.rna.albany.edu/; 54) to determine changes in stability possibly caused by the mutation in question.
RESULTS AND DISCUSSION
Optical maps.
Long-term evolution of microbes under constant conditions often leads to irreversible loss of genomic material, a phenomenon known as genomic erosion (27). Previously, we had noted that a strain evolved from WN624 after ∼6,000 generations of relaxed selective pressure for sporulation had sustained a deletion of ∼10 kbp in the nonessential ppsABCDE operon (18). Therefore, in order to search for possible large-scale deletions or other gross alterations in genome architecture, an OpGen optical map was created for strains WN624, WN715, and WN716, as described previously (11). The genome maps were aligned using the Map Solver software supplied by OpGen. Comparison of the optical maps showed a single large deletion of about 4 kb in postsweep strainWN716 that was not present in ancestral strain WN624 or presweep strain WN715 (Fig. 1). However, subsequent 454 sequencing of the three strains revealed that this deletion was not present in strain WN716. Thus, this putative deletion was likely incorrectly identified due to the resolution limits of optical mapping (approximately 5 kb). Aside from the above observation, the overall optical maps of the three genomes aligned well with each other, indicating that the phenotypic changes observed in WN716 were not the result of major chromosomal rearrangements occurring before 1,800 generations (Fig. 1).
454 sequencing and alignment.
To identify individual mutations that might account for the increased fitness of WN716, we used 454 pyrosequencing to sequence the genomes of strains WN715 and WN716. The resulting sequencing statistics are shown in Table 1. Before processing, our preliminary data revealed a total of 196 SNPs, 105 of which were verified according to our stringency qualifications as described in Materials and Methods. Of these 105 total SNPs, 11 were located in intergenic regions. Of the remaining 94 SNPs, located in coding regions, 27 were determined to result in silent mutations. Of the 67 remaining SNPs, 34 were located in genes of known function and 33 in genes of unknown function. Four mutations were found in homopolymeric regions, which are known to be susceptible to misreading by 454 pyrosequencing. In these cases, the regions in question were resequenced by Sanger sequencing, and the Sanger sequencing results agreed with the 454 data in each case (data not shown).
Table 1.
Genome assembly statisticsa
| Parameter | WN715 |
WN716 |
||||
|---|---|---|---|---|---|---|
| Count | Avg length | Total no. of bases | Count | Avg length | Total no. of bases | |
| Reads | 150,530 | 224.6 | 33,808,075.0 | 89,868 | 224.8 | 20,204,273.0 |
| Matched | 148,387 | 224.9 | 33,369,451.0 | 88,738 | 225.1 | 19,973,927.0 |
| Not matched | 2,143 | 204.7 | 438,624.0 | 1,130 | 203.9 | 230,346.0 |
| Reference | 1 | 4,215,606.0 | 4,215,606.0 | 1 | 4,215,606.0 | 4,215,606.0 |
The genomes of strains WN715 and WN716 were sequenced using 454 pyrosequencing and aligned to the reference B. subtilis subsp. subtilis strain 168 genome (NCBI reference NC 000964.3).
Of the 34 coding-sequence SNPs in genes of known function, we deduced 29 missense, 1 nonsense, and four +1 frameshift mutations (Table 2). These mutations were clustered in several categories of pathways, especially motility (fliI, fliL, fliR, and motA), sporulation (cotX, spsI, minD, citB, and phrE), stress response (clpE and clpC), biosynthesis of amino acids (argH and carA), cofactors (bioF), and antibiotics (pksL, ppsE, srfAA, and sigW) (Table 2). SNPs in some genes suspected to induce mutator phenotypes, including enzymes for DNA repair (uvrB) and recombination (addA), were also identified. Several mutations corresponded to genes previously found by microarray analysis to be up- or downregulated in WN716 compared to WN715 (21) (Table 3).
Table 2.
Identified mutations that altered the amino acid sequence encoded by genes of known function in strain WN716a
| Gene or operon | Function | WN716 coverage at 100% | Amino acid change |
|---|---|---|---|
| Category 1. Cell envelope and cellular processes | |||
| 1.1 Cell wall | |||
| dacC | Peptidoglycan biosynthesis, early stationary phase | 4 | G300S |
| lytD | Involved in cell separation, cell wall turnover, antibiotic-induced lysis | 4b | S185L |
| wapA | Cell wall-associated protein precursor | 8 | P1578S |
| 1.2 Transport/binding proteins and lipoproteins | |||
| gltP | Glutamate uptake symport protein | 4 | +1 frameshift at codon 9 |
| 1.4 Membrane bioenergetics (electron transport chain, ATP synthase) | |||
| qoxB | Cytochrome aa3 quinol oxidase | 4 | A238V |
| 1.5 Motility and chemotaxis: | |||
| fliI | Flagellar ATP synthase | 5 | R338G |
| fliL | Required for flagellar formation | 4 | T54A |
| fliR | Required for flagellar formation | 6b | +1 frameshift at codon 66 |
| motA | Flagellar motor rotation | 4 | V221L |
| 1.7 Cell division | |||
| minD | Cell division inhibition (septum placement) | 6 | R33C |
| 1.8 Sporulation | |||
| cotX | Spore coat protein | 6 | A87V |
| spsI | Spore coat polysaccharide synthesis | 5 | R95H |
| phrE | Negative control of RapE activity | 4 | A37V |
| Category 2. Intermediary metabolism | |||
| 2.1 Metabolism of carbohydrates and related molecules | |||
| 2.1.1 Specific pathways | |||
| nagB | N-Acetylglucosamine utilization | 7 | Nonsense at codon 150 |
| 2.1.2 Main glycolytic pathways | |||
| pykA | Pyruvate kinase | 5 | E78G |
| tpiA | Triose phosphate isomerase | 6 | E249G |
| 2.1.3 TCAc cycle | |||
| citB | Aconitase | 7 | T616I |
| 2.2 Metabolism of amino acids and related molecules | |||
| argH | Arginine biosynthesis | 6 | A143V |
| carA | Arginine biosynthesis | 4 | A354V |
| hutU | Histidine utilization | 4 | V225A |
| 2.5 Metabolism of coenzymes and prosthetic groups | |||
| bioF | Biotin biosynthesis | 4 | A247S |
| Category 3. Information pathways | |||
| 3.2 DNA restriction/modification and repair | |||
| uvrB | Excision of UV light-induced pyrimidine dimers in DNA | 4 | V498A |
| 3.3 DNA recombination | |||
| addA | Initiation stage of recombination | 4 | E522K |
| 3.5 RNA synthesis | |||
| 3.5.1 Transcription initiation | |||
| sigW | RNA polymerase ECF-type sigma factor | 4 | +1 frameshift at codon 13 |
| 3.5.2 Transcription regulation | |||
| alsR | Regulation of the acetoin operon (alsSD) | 3b | A21D |
| 3.5.3 Transcription elongation | |||
| rpoB | RNA polymerase | 4 | N269D |
| 3.6 RNA modification | |||
| miaA | Translation | 8 | S36N |
| Category 4. Other functions | |||
| 4.1 Adaptation to atypical conditions | |||
| clpC | Class III stress response ATPase | 10 | S589P |
| clpE | ATP-dependent Clp protease-like (class III stress gene) | 3b | +1 frameshift at codon 70 |
| rsbS | Negative regulation of σB activity | 7 | L29S |
| 4.3 Antibiotic production | |||
| pksS | Polyketide synthase of type I | 5 | H2900R |
| ppsE | Plipastatin synthetase | 5 | Q1064R |
| srfAA | Surfactin production and competence | 4 | A782V |
| 4.4 Phage-related function | |||
| xkdT | PBSX prophage gene | 5 | A175V |
SNPs and indels were identified by aligning the whole genomes of WN624, WN715, and WN716 with the reference genome of B. subtilis 168 and identifying mutations present in WN716 and absent in the other strains. Silent mutations and mutations in genes of unknown function are not included in the table.
Mutation was confirmed by Sanger sequencing.
TCA, tricarboxylic acid.
Table 3.
Mutations found between WN715 and WN716 that correlate with microarray dataa
| Gene | Function | Fold expression change in WN716 | Amino acid change |
|---|---|---|---|
| pksS | Polyketide synthase type I | −8 | H2900R |
| srfAA | Surfactin synthetase subunit 1 | −17 | A782V |
| fliI | Flagellum-specific ATP synthase | −12 | R338G |
| fliR | Required for flagellum formation | −24 | +1 frameshift at codon 66 |
| lytD | N-Acetylglucosaminidase, major autolysin | −7 | S185L |
| bioF | 8-Amino-7-oxononanoate synthase | −4 | A247S |
| alsR | Transcriptional regulator of alsSD operon | −122 (alsS), −87 (alsD) | A21D |
Microarray data from reference 21.
As can be seen in Table 2, most SNPs found within coding sequences were missense mutations, which might be predicted to affect the activity, stability, etc., of the target gene itself. However, some SNPs were either nonsense mutations or +1 frameshift mutations, which might be expected to also exert polar effects on downstream genes in the same operon. An exception to this supposition would be the monocistronic gltP and clpE genes (Table 2); +1 frameshifts in these two genes would not be expected to exert any polar effect. A nonsense mutation at codon 150 in nagB (Table 2) might affect the expression of the downstream yvoA gene, the function of which is unknown; however, microarray analysis detected only 0.58-fold less yvoA mRNA in WN716 than in WN715 (21). Downstream from the sigW cistron is located the rsiW cistron encoding an antisigma factor, RsiW, that regulates SigW activity (41). A +1 frameshift at codon 13 in sigW could result in loss of expression of the RsiW protein. However, the likely phenotype of a SigW− RsiW− double mutant would be identical to that of a SigW− single mutant, because (i) the target of RsiW regulation, SigW, is itself missing and (ii) expression of the sigW-rsiW operon is under SigW control (4). An interesting finding was a +1 frameshift located at codon 66 in the fliR gene, which is the 20th cistron in a large (31-gene) flagellar-chemotaxis (fla-che) operon (32). A frameshift in fliR might be expected to cause premature transcription termination, affecting the expression of the 11 downstream fla-che genes. It is interesting that the 30th cistron in the fla-che operon is sigD, which encodes the SigD sigma factor responsible for transcription of all flagellar, motility, and chemotaxis genes, including the fla-che operon (and sigD) itself. We previously noted that the mRNA levels of all motility genes, including the fla-che operon, were severely downregulated in strain WN716, but DNA sequencing failed to reveal any mutations in or around sigD (21). A polar effect of a frameshift in fliR could explain these observations.
Linking the WN716 phenotype with mutation in sigW.
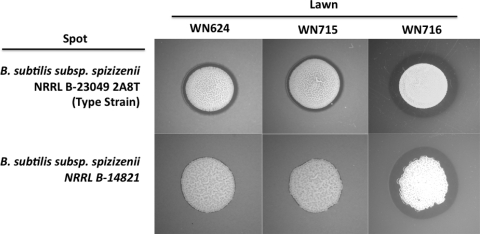
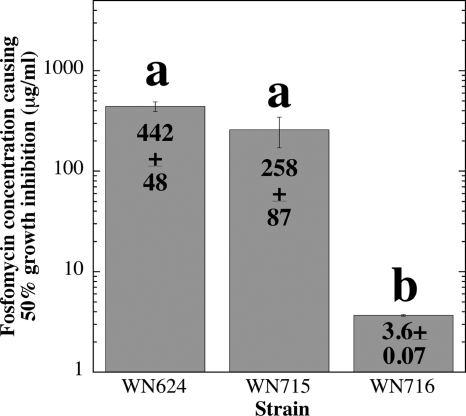
Several mutations identified in WN716 would be predicted to result in specific altered phenotypes. Where possible, these predictions were tested directly. For example, 454 sequencing identified a +1 frameshift mutation at codon 13 of the sigW gene, which encodes the alternative sigma factor σW (Table 2). The σW regulon in B. subtilis includes target genes involved in resistance to a number of bacteriocins and antibiotics produced by various microbes, including other Bacillus species (2–4). Therefore, loss of σW would be predicted to result in sensitivity of strain WN716 to antibiotics produced by other Bacillus spp. This prediction was tested by spotting cells of two strains of B. subtilis subsp. spizizenii onto lawns of B. subtilis strains WN624, WN715, and WN716 (Fig. 2). Strain WN716 was indeed more sensitive to the antibiotic(s) produced by the two B. subtilis subsp. spizizenii strains, as demonstrated by its larger zone of growth inhibition (Fig. 2). This observation is in excellent agreement with results obtained in a sigW knockout mutant (2). In addition, fosfomycin resistance, encoded by the sigW-dependent fosB gene (3), was tested by growing strains WN624, WN715, and WN716 in liquid LB medium containing various concentrations of fosfomycin and comparing their sensitivity profiles, presented here as the concentration of fosfomycin that inhibited the growth rate of cells by 50% (Fig. 3). Strain WN716 was observed to be dramatically more sensitive to fosfomycin than either ancestral strain WN624 or presweep strain WN715 (Fig. 3). Taken together, these results strongly suggest that the observed phenotypes are indeed caused by the sigW mutation in postsweep strain WN716.
Fig. 2.
Bacteriocin sensitivity assay. Lawns of strains WN624, WN715, and WN716 were plated onto LB medium, and aliquots of overnight liquid cultures of B. subtilis subsp. spizizenii were spotted onto the lawns. Zones of growth inhibition of the lawns can be seen as dark rings surrounding the colony spots.
Fig. 3.
Sensitivities of strains WN624, WN715, and WN716 to fosfomycin. The concentration of fosfomycin causing a 50% reduction in the growth rate was determined for each strain. The data are shown as averages ± standard deviations (n = 3). The lowercase letters above the bars denote significant differences (analysis of variance [ANOVA]; P < 0.05).
Previously, we reported on the results of cDNA microarray experiments comparing the transcriptomes of WN715 and WN716 in early stationary-phase of growth in liquid R medium (21). The discovery of a frameshift mutation inactivating sigW in WN716 would be predicted to affect the transcription of genes belonging to the SigW regulon, which has been delineated previously (4). These observations prompted us to compare our microarray data with the microarray data published by Cao et al. (4), the results of which are presented in Table 4. We observed that several known members of the SigW regulon were downregulated in both microarrays, for example, the yjoB, yknW, yoaF, and yvlA genes and the yqeZ-yqfAB, pspA-ydjGHI, and sppA-yteJ operons, as well as the sigW-rsiW operon itself (Table 4). Thus, the microarray data support the prediction that WN716 has sustained a mutation inactivating sigW. However, it should be noted that comparison of the two sets of microarray data also revealed a number of discrepancies. For example, a set of genes (abh, divIC, yfhKLM, yjbC-spx, yqjL, ywoA, and yxzE) was observed to be downregulated in WN716 but not in the data of Cao et al. (4) (Table 4). Conversely, a number of genes (pbpE-racX, ybfO, ydbS, yobJ, ysdB, ythPQ, yuaF, ywrE, and yxjJI) were observed to be downregulated in the data of Cao et al. (4) but not in WN716 (Table 4). Finally, a substantial number of known SigW-dependent genes were not significantly downregulated in either set of microarray data (Fig. 4). These apparent discrepancies are likely due to at least two factors. First, the two microarray experiments were performed on different pairs of strains, grown in different media, and harvested at different growth phases (4, 21). Second, the SigW regulon has been defined by a combination of several different approaches, including microarray analyses, bioinformatics techniques, and runoff macroarray (ROMA) analyses, none of which, applied by itself, was able to completely define the regulon (4). Thus, the lack of complete concordance between the two data sets is not surprising.
Table 4.
SigW-dependent genes downregulated in the transcriptome of strain WN716 vs. that of WN715a
| Gene or operonb | Annotated functionc | Fold downregulation in WN716d |
|---|---|---|
| abh | Transcriptional regulator of transition state genes (AbrB-like) | 7.0 |
| divIC | Cell division initiation protein (septum formation) | 5.4 |
| fosB (yndN) | Cysteine-dependent fosfomycin resistance protein | <2.0 (<2.0) |
| pbpE | Penicillin-binding protein 4 | <2.0 (7.7) |
| racX | Amino acid racemase | <2.0 (11) |
| pbpX | Penicillin-binding protein | <2.0 |
| pspA (ydjF) | Alkaline shock-induced protein | 2.1 (3.4) |
| ydjG | Conserved protein | 3.7 |
| ydjH | Conserved membrane protein | 2.3 |
| ydjI | Conserved protein | 10.9 |
| sigW | RNA polymerase ECF-type sigma factor | 5.1 (10) |
| rsiW (ybbM) | Possible anti-SigW protein | 9.4 (17) |
| sppA (yteI) | Signal peptide peptidase | 2.2 (2.5) |
| yteJ | Conserved membrane protein | 12.5 (2.2) |
| xpaC | 5-Bromo-4-chloroindolyl phosphate hydrolysis protein | <2.0 (<2.0) |
| yaaN | Similar to toxic cation resistance protein | <2.0 |
| ybfO | Similar to erythromycin esterase | <2.0 (3.2) |
| yceC | Similar to tellurium resistance protein | 2.5 (<2.0) |
| yceD | Similar to tellurium resistance protein | <2.0 |
| yceE | Similar to tellurium resistance protein | <2.0 |
| yceF | Similar to tellurium resistance protein; probable transporter | <2.0 |
| yceG | Conserved protein | <2.0 |
| yceH | Similar to toxic anion resistance protein | <2.0 |
| ydbS | Conserved membrane protein | <2.0 (3.2) |
| ydbT | Conserved membrane protein | 2.1 (3.0) |
| yeaA | Conserved protein | <2.0 |
| ydjP | Similar to chloroperoxidase | <2.0 |
| ydjO | Function unknown and unique | <2.0 |
| yfhK | Similar to B. subtilisywsB | 6.1 |
| yfhL | Function unknown and unique | 2.5 (<2.0) |
| yfhM | Similar to epoxide hydrolase | 3.0 |
| yjbC | Conserved protein | 3.8 (<2.0) |
| yjbD (spx) | Glutaredoxin family; disruption bypasses the ClpXP requirement for ComK expression | 4.8 |
| yjoB | Similar to FtsH | 4.3 (25) |
| yknW | Conserved membrane protein | 4.4 (2.2) |
| yknX | Conserved protein | <2.0 |
| yknY | Probable ABC transport system ATP-binding protein | 2.6 |
| yknZ | Probable ABC transport system permease protein | <2.0 (2.8) |
| yoaF | Function unknown and unique | 2.0 (2.0) |
| yoaG | Similar to prophage protein | − (2.4) |
| yobJ | Function unknown and unique | <2.0 (4.0) |
| yozO | Conserved protein | <2.0 (<2.0) |
| yqeZ | Conserved membrane protein | − (9.5) |
| yqfA | Conserved protein | 15.1 (6.7) |
| yqfB | Function unknown and unique | 8.1 (5.6) |
| yqjL | Function unknown and unique | 6.6 |
| yrhH | Similar to methyltransferase | <2.0 (<2.0) |
| ysdB | Conserved protein | <2.0 (10) |
| ythP | Similar to ABC transport system ATP-binding protein | <2.0 (31) |
| ythQ | Function unknown and unique | <2.0 (16) |
| yuaF | Function unknown and unique | <2.0 (89) |
| yuaG | Conserved protein | <2.0 |
| yuaI | Probable acetyltransferase | 2.5 (11) |
| yvlA | Function unknown and unique | 3.9 (2.7) |
| yvlB | Conserved protein | 2.8 |
| yvlC | Conserved protein | <2.0 (2.1) |
| yvlD | Conserved membrane protein | <2.0 (2.3) |
| ywaC | Similar to GTP-pyrophosphokinase | <2.0 (<2.0) |
| ywbL | Putative transporter | <2.0 |
| ywbM | Conserved protein | <2.0 |
| ywbN | Conserved protein | <2.0 (<2.0) |
| ywnJ | Function unknown and unique | <2.0 |
| ywoA | Bacitracin resistance protein | 3.7 (<2.0) |
| ywrE | Conserved membrane protein | <2.0 (11) |
| yxjJ | Conserved protein | <2.0 |
| yxjI | Function unknown and unique | <2.0 (12) |
| yxzE | Function unknown and unique | 2.5 (<2.0) |
Microarray data were obtained from early-stationary-phase cultures of strains WN715 and WN716 grown in liquid R medium (21).
SigW-dependent genes compiled from the DBTBS database (http://dbtbs.hgc.jp/; 44).
Annotated functions taken from the Bacillus subtilis genome database BSORF (http://bacillus.genome.ad.jp/).
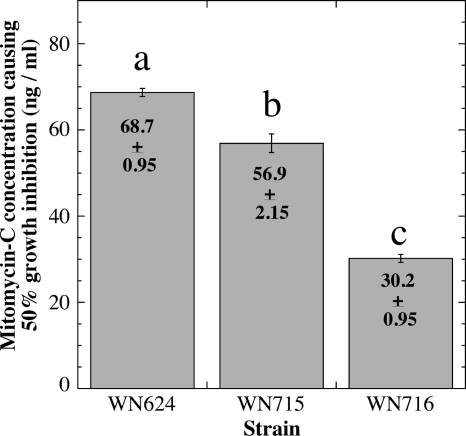
Fig. 4.
Sensitivities of strains WN624, WN715, and WN716 to mitomycin C. The concentration of mitomycin C causing a 50% reduction in the growth rate was determined for each strain. The data are shown as averages ± standard deviations (n = 3). The lowercase letters above the bars denote significant differences (ANOVA; P < 0.05).
Mutations in uvrB and addA.
Examination of SNPs in the WN716 genome identified missense mutations in the uvrB and addA genes (Table 2). The uvrB gene encodes the UvrB subunit of the UvrABC excinuclease involved in nucleotide excision repair of DNA damage, and addA encodes a component of the B. subtilis AddAB helicase-nuclease complex involved in general homologous recombination and in recombinational DNA repair (38, 39). Because mutants lacking UvrB or AddA are known to exhibit increased sensitivity to the DNA-damaging agent mitomycin C, we predicted that WN716 might also exhibit this phenotype. This prediction was tested by growing strains WN624, WN715, and WN716 in liquid LB medium containing various concentrations of mitomycin C and comparing their sensitivity profiles, presented here as the concentration of mitomycin C that inhibited the growth rate of cells by 50% (Fig. 4). Strain WN716 exhibited a modest but statistically significant 2.3-fold increase in mitomycin C sensitivity compared to ancestral strain WN624 (Fig. 4). However, strain WN715 also exhibited a 1.2-fold increase in mitomycin C sensitivity compared to the ancestor, so it is difficult to attribute mitomycin C sensitivity to mutations in uvrB and/or addA in this case. In any event, the increase in mitomycin C sensitivity observed was much less severe than would be expected of mutations completely inactivating either UvrB or AddA (15, 30).
Genome-sequencing analysis suggested that WN716 accumulated a substantial number of mutations within the first ∼1,800 generations of evolution. Earlier data indicated that the bulk 624A population exhibited an ∼50-fold increase in the rate of spontaneous mutation to rifampin resistance (Rifr) during evolution, most of which had occurred within the first 2,000 generations, strongly suggesting that the evolving populations were accumulating cells with mutator phenotypes (19). Measurement of the frequency of mutation to Rifr in pure cultures of strains WN624, WN715, and WN716 showed that the strains mutated to Rifr at frequencies of 3.1 × 10−9, 4.6 × 10−7, and 4.7 × 10−6, respectively. Thus, the spontaneous-mutation frequencies in evolved strains WN715 and WN716 had increased ∼150-fold and ∼1,500-fold, respectively, over that of the ancestral strain, WN624, in the course of less than 2,000 generations of evolution. Such high mutation frequencies strongly suggested that the evolved strains had sustained mutations in genes conferring a mutator phenotype, as has previously been observed in laboratory bacterial-evolution experiments (50). Interestingly, the Escherichia coli equivalents of the uvrB and addA gene products have both been implicated in stationary-phase hypermutagenesis (9). The AddA protein contains 6 helicase motifs important for activity (52), but the E522K mutation found in strain WN716 was not located in any of these regions (data not shown).
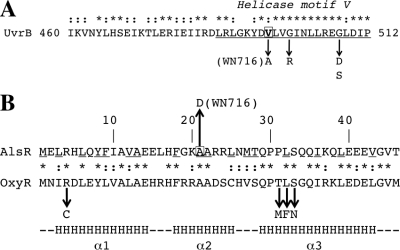
UvrB is an ATP-dependent DNA helicase (25), and examination of the deduced amino acid sequence of UvrB revealed that the V498A mutation in WN716 was located within highly conserved helicase motif V (Fig. 5A). In E. coli UvrB, nearby mutations in motif V (G502R and G509D/S) were shown to affect UvrB helicase activity and formation of the UvrABC preincision complex (25). In UvrB proteins from several bacterial species, amino acid 498 in motif V is often, but not always, valine (Fig. 5A) (51). Despite the fact that valine and alanine both carry small hydrophobic side groups, alanine-scanning mutagenesis experiments have revealed numerous V-to-A amino acid changes affecting protein activity (22). Taken together, the data suggest that the mutations in uvrB and/or addA might be partially responsible for the mutator phenotype observed in strain WN716. Genetic reconstruction experiments will test this notion directly.
Fig. 5.
(A) Deduced amino acid sequence of the B. subtilis UvrB protein. Helicase motif V (12) is underlined and labeled. Mutations altering UvrB helicase and incision activity (G502R, G509D, and G509S) (25) are indicated by downward arrows. Valine 498 is boxed, and the V498A mutation in strain WN716 is indicated. (B) Deduced N-terminal amino acid sequences of AlsR and OxyR. The underlined amino acids are highly conserved among LysR family proteins (53). Alanine 21 in AlsR is boxed, and the A21D mutation in AlsR is indicated by the upward arrow. The downward arrows denote mutations in the OxyR HTH region leading to decreased DNA binding (16). The bottom line indicates the position and extent of alpha-helices (α1, α2, and α3) in the LysR HTH region (53). In both panels, the asterisks and colons denote identical and conserved amino acids. See the text for details.
Mutation in alsR.
Strain WN716 was previously shown to be unable to produce the fermentation product acetoin in R medium, and microarray experiments showed that the alsSD (acetoin) operon was severely downregulated in WN716 (21). The alsS and alsD genes encode the enzymes α-acetolactate synthase and acetolactate decarboxylase, respectively (36). Divergently transcribed from alsSD is the alsR gene that encodes AlsR, a LysR family positive regulator of alsSD transcription; disruption of alsR was previously shown to result in severe downregulation of alsSD transcription and acetoin production (36). In this work, the WN716 genome sequence was found to contain a mutation in the alsR gene, which was predicted to result in the amino acid change A21D in the AlsR protein sequence (Tables 2 and 3). In AlsR, residue A21 is located within the helix-turn-helix (HTH) DNA-binding motif and is a highly conserved amino acid among all LysR family proteins (53) (Fig. 5B). Mutations in the HTH region of another LysR family protein, OxyR, have been shown to reduce or eliminate binding of the mutant OxyR proteins to their cognate DNA-binding sites (16). Collectively these observations suggest that the A21D mutation in WN716 AlsR may be directly responsible for downregulation of alsSD transcription and loss of acetoin production. Experiments to test this notion are currently in progress.
Mutations in biosynthetic pathways.
Sequence analysis also uncovered a number of missense mutations in several biosynthetic (argH, bioF, carA, citB, and hutU) or glutamate-transport (gltP) genes, inactivation of which would be predicted to result in nutritional auxotrophies, in addition to the trpC marker present in all strains used here (Table 2). Note that mutation in the aconitase gene citB also leads to sporulation deficiency, as well as glutamate auxotrophy (5) (Table 2). Growth of strains WN624, WN715, and WN716 on SMM plates containing various combinations of arginine, biotin, glutamate, histidine, and tryptophan showed that both strains WN624 and WN715 had retained the original trpC2 marker and had not gained additional auxotrophy for other nutrients (Table 5). In contrast, growth of WN716 on SMM supplemented with all five nutrients (tryptophan, biotin, histidine, arginine, and glutamate) failed to rescue its auxotrophic phenotype (Table 5), indicating that strain WN716 carried at least one other auxotrophy in addition to those tested. Therefore, in this case, genomic sequence analysis fell short of identifying all mutations responsible for the multiply auxotrophic phenotype of strain WN716.
Table 5.
Growth of B. subtilis strains on supplemented Spizizen minimal mediuma
| Strain | Growth on Spizizen minimal medium supplemented with: |
|||||
|---|---|---|---|---|---|---|
| BGHT | AGHT | ABHT | ABGT | ABGH | ABGHT | |
| WN624 | + | + | + | + | − | + |
| WN715 | + | + | + | + | − | + |
| WN716 | − | − | − | − | − | − |
A, arginine; B, biotin; G, glutamate; H, histidine; T, tryptophan; +, growth; −, no growth.
SNPs located in intergenic regions.
It was reasoned that SNPs occurring in intergenic regions could affect gene expression through alterations in regulatory sequences, such as promoters, binding sites for transcription factors, or transcription terminators. Thus, an attempt was made to locate the 11 intergenic SNPs found in strain WN716 within known or putative regulatory sites using B. subtilis databases. Of the 11 SNPs originally identified as located in intergenic regions, no discernible regulatory feature could be identified in 5 cases; furthermore, one SNP was actually located in the last codon of the pcp gene, resulting in an H-to-L amino acid change (Table 6). The remaining 5 SNPs were located in putative regulatory sites. One SNP was located in the previously characterized Zur box C2, located upstream from the yciC gene encoding a putative zinc transport protein (10); however, the SNP did not change yciC expression in WN716 on the microarray (Table 6). A second SNP was located in a putative AraR-binding site located between the divergently transcribed nudF and yqkF genes, encoding an ADP-ribose pyrophosphatase and a putative oxidoreductase, respectively. This SNP did not result in a dramatic change in nudR or yqkF mRNA levels on the microarray (Table 6), and indeed, it is difficult to envision how or why nudR or yqkF would be subject to AraR regulation, because the AraR repressor specifically regulates genes involved in arabinose and hemicellulose metabolism (35). A third SNP was located within a putative Fur box located between recO, encoding a recombination/DNA repair protein, and era, encoding a GTP-binding protein essential for cell growth; again, no evidence for altered mRNA levels in WN716 was detected by microarray, and the recO and era genes were not identified as part of the Fur regulon (1). A fourth SNP was located in a putative MntR-binding site between the hutG (formiminoglutamase) and hutM (histidine permease) genes; these genes have not been identified as part of the MntR regulon (26), nor were their mRNA levels altered in WN716 on microarrays. The fifth SNP was located between the convergent yyaJ and maa genes encoding a putative transporter and a probable maltose o-acetyltransferase (Table 6). The last SNP was located in the stem region of the putative transcriptional terminator for maa and was predicted to lower the base-pairing stability of the stem from −17 to −13 kcal/mol. In this case, a slight (∼2-fold) decrease in the mRNA levels was observed in strain WN716 on the microarrays (Table 6). Taken together, the 11 SNPs detected in intergenic regions were not found to correlate with significant gene expression changes or with known phenotypic changes in strain WN716.
Table 6.
SNPs found in intergenic regions in WN716a
| SNP | Position | Intergenic region | Comment | Expression in WN716 (microarray)b | Serverc used or reference |
|---|---|---|---|---|---|
| T to G | 119844 | rplA-rplJ | No feature found | −2.8 (rplA) | S, D, M |
| −2.6 (rplJ) | |||||
| A to T | 211093 | ybcI-ybcL | No feature found | −1.1 (ybcI) | S, D |
| −0.7 (ybcL) | |||||
| A to T | 286973 | pcp-ybcU | Mutation in last codon of pcp; CAC(H) to CTC(L) | −0.1 (pcp) | S |
| − (ybcU) | |||||
| G to A | 365358 | yciB-yciC | Mutation in Zur box C2 upstream from yciC | − (yciB) | S, D |
| −0.4 (yciC) | 10 | ||||
| G to A | 688754 | yeaC-yeaD | No feature found | −0.1 (yeaC) | S, D |
| −4.7 (yeaD) | |||||
| T to C | 953843 | ygzA-ygaJ | No feature found | −2.2 (ygzA) | S, D |
| −0.9 (ygaZ) | |||||
| T to C | 2458407 | nudF-yqkF | Divergent genes; SNP located in a putative AraR-binding site | −1.8 (nudF) | S, D |
| −0.6 (yqkF) | |||||
| A to G | 2609075 | recO-era | SNP located within a putative Fur box | −1.7 (recO) | S, D |
| −0.6 (era) | |||||
| G to A | 2838039 | csbX-yrbE | No feature found | +0.48 (csbX) | S, D |
| −0.6 (yrbE) | |||||
| T to C | 4046515 | hutG-hutM | SNP located within a putative MntR-binding site | −0.4 (hutG) | S, D |
| −0.6 (hutM) | |||||
| T to C | 4194778 | yyaJ-maa | Convergent genes. SNP located in stem of putative maa transcription terminator; reduces ΔG from −17 to −13 kcal/mol | −2.7 (yyaJ) | S, D, M |
| −2.3 (mma) |
SNPs located within intergenic regions were first located in Subtilist; the intergenic regions were then imported into DBTBS and searched for putative promoters or binding sites, for accessory transcription factors, and for transcriptional terminators. If the SNP was found in a terminator stem-loop, MFOLD was used to calculate the potential change in stability.
Microarray data are from reference 21. −, gene was not spotted on the microarray.
Abbreviations for internet servers: S, Subtilist (http://genolist.pasteur.fr/SubtiList/); D, DBTBS (http://dbtbs.hgc.jp/); M, MFOLD (http://mfold.rna.albany.edu/).
Linking genomic changes with fitness.
Continued evolution under constant environmental conditions can lead to massive loss of genetic material, a phenomenon often referred to as genomic erosion (27). In support of this notion, we previously observed that cells from populations 624A and 624B that had been propagated for 6,000 generations in R medium had sustained a >9.7-kb deletion in the ppsABCDE operon encoding biosynthesis of the nonessential antibiotic plipastatin (18). In this study, optical mapping revealed that there had been no discernible large-scale genomic changes in either WN715 or WN716 in the ∼1,800 generations since their descent from WN624, suggesting that the source of WN716's increased competitive fitness likely was small mutations, including SNPs and/or indels, rather than large deletions, translocations, or inversions in the genome. The optical-mapping data supported our strategy of using 454 pyrosequencing to discover small mutations for the source of WN716's enhanced fitness.
Strain WN716 cells grow in R medium as long nonmotile filaments, and previous microarray experiments demonstrated downregulation of at least 45 genes, including all the flagellar operons and several lyt genes encoding autolysins (21). Because flagellar operons and lyt genes belong to the sigD regulon (42), their downregulation might have been due to mutation in the sigD gene itself. However, no mutation in the sigD coding sequence was identified in WN716, either by 454 pyrosequencing (this study) or by directed Sanger sequencing (21). Interestingly, 454 pyrosequencing did identify numerous mutations in sigD-dependent genes, such as the lytD, fliI, fliL, fliR, and motA genes (Table 2), and as discussed above, a frameshift mutation in fliR could possibly exert a polar effect on the expression of the downstream sigD gene itself. Furthermore, mutations were identified in genes belonging to several biosynthetic pathways in strain WN716 (Table 5). One explanation for these two observations could be that, because there is little or no selective pressure for maintaining functional motility or biosynthetic pathways in WN716 evolving under constant, nutrient-rich conditions, these genes are rapidly accumulating mutations, i.e., are in the process of becoming pseudogenes (17, 24).
In contrast, sequence analysis did identify a +1 frameshift mutation that would potentially inactivate the sigW gene encoding σW, which is known to control a regulon of antibiotic and bacteriocin resistance genes (2–4). Phenotypic characterization of strain WN716 (Fig. 3 and 4) and comparison of microarray data obtained for WN716 and an authentic sigW mutant (Table 4) strongly suggests that strain WN716 indeed appears to be defective in σW function.
Loss of either (or both) sigD- and sigW-directed functions would be selectively advantageous for strain WN716, because maintenance of motility, chemotaxis, and resistance to multiple antibiotics is energetically costly and confers no selective advantage during shake flask propagation in a nutrient-rich environment in the absence of nutrient concentration gradients or niche competitors. In support of this notion, large-scale flux analyses demonstrated that the metabolic efficiency of B. subtilis cells was actually improved in sigD or sigW knockout mutants (8). Thus, knockout of the sigD and/or sigW regulon would be predicted to result in a higher growth rate (i.e., enhanced fitness) of WN716 in R medium.
Strain WN716 was previously observed to have substantially lost both sporulation ability and competence for genetic transformation; furthermore, microarray analysis resulted in the identification of several genes encoding early sporulation and competence functions, transcription of which appeared to be severely downregulated (21). Although the exact cause of the sporulation deficiency of strain WN716 remains unclear at this time, mutations were identified in several candidate genes affecting sporulation. In addition to genes directly involved in spore formation (cotX and spsI, encoding a spore coat protein and a polysaccharide, respectively), we found mutations in genes involved in both competence and sporulation initiation. For example, a point mutation in phrE may affect sporulation and/or competence by limiting the ability of PhrE to assert negative control over the RapE phosphatase (34). A polarity-changing point mutation in minD could affect sporulation by disrupting septum placement during sporulation (43); however, microscopic examination of strain WN716 cells failed to reveal minicell production (data not shown). Finally, a mutation in the citB gene, encoding the Krebs cycle enzyme aconitase, was identified by 454 pyrosequencing (Table 2). Mutations inactivating aconitase are known to block sporulation at its earliest stage (5, 45). It seems reasonable to speculate that each of these mutations, either individually or in combination, could contribute to strain WN716's lack of genetic competence and its inability to sporulate. Future efforts will concentrate on obtaining direct experimental evidence for this supposition.
Despite the failure of optical mapping to reveal gross genomic changes, 454 pyrosequencing identified several small genomic changes that had occurred during the evolution of B. subtilis cells under relaxed selective pressure for sporulation. In particular, mutations were identified in the genome of WN716 that allowed testing of specific predicted phenotypes; in one case (mutation in sigW), the predicted phenotype was confirmed, while in other cases (motility, competence for transformation, sporulation, and acetoin biosynthesis), mutations were identified in genes which potentially led to the observed phenotype but which remain to be experimentally tested. In the case of auxotrophy, the loss of prototrophy was previously observed to be a consequence of evolution in nutrient-rich R medium in all evolving B. subtilis populations tested (19). Examination of population 624A had revealed that at generation 1,000 (before strain WN716 swept the culture), essentially 100% of the cells were prototrophic (aside from tryptophan auxotrophy conferred by the trpC2 marker present in the ancestral strain), but by generation 2,000 (after the sweep), prototrophy had decreased dramatically to only 0.1% of the cells in the population (19). In the present study, whole-genome sequencing of strain WN716 revealed five mutations in four biosynthetic pathway genes, but an additional mutation(s) responsible for its multiply auxotrophic phenotype clearly remains to be discovered.
In conclusion, whole-genome sequencing revealed that strain WN716 had sustained a number of mutations that, individually or collectively, could contribute to its selective advantage in R medium. An important future step is to identify which mutated genes are responsible for WN716's selective advantage by producing targeted mutations inactivating specific genes and testing the resulting mutants in pairwise competition experiments (21, 33). These experiments are currently in progress.
ACKNOWLEDGMENTS
We thank Bill Farmerie at the UF ICBR NextGen sequencing group for his help with the 454 pyrosequencing and SNP/indel detection, John Helmann for helpful discussions, Mike Roberts for generous donation of B. subtilis subsp. spizizenii strains, and the four anonymous reviewers for insightful comments.
Much of the work described in this communication was performed as part of an undergraduate course, MCB4934 “Bacterial Genome Sequencing,” under the direction of J.C.D., E.W.T., and W.L.N.
This work was supported in part by grants from the NASA Astrobiology, Exobiology, and Evolutionary Biology program (NNX08AO15G) to W.L.N. and from the Course, Curriculum and Laboratory program at the National Science Foundation (0920151) to J.C.D. and E.W.T.
Footnotes
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Baichoo N., Wang T., Ye R., Helmann J. D. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613–1629 [DOI] [PubMed] [Google Scholar]
- 2. Butcher B. G., Helmann J. D. 2006. Identification of Bacillus subtilis σW-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol. Microbiol. 60:765–782 [DOI] [PubMed] [Google Scholar]
- 3. Cao M., Bernat B. A., Wang Z., Armstrong R. N., Helmann J. D. 2001. FosB, a cysteine-dependent fosfomycin resistance protein under the control of σW, an extracytoplasmic-function σ factor in Bacillus subtilis. J. Bacteriol. 183:2380–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao M., et al. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443–457 [DOI] [PubMed] [Google Scholar]
- 5. Craig J. E., Ford M. J., Blaydon D. C., Sonenshein A. L. 1997. A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J. Bacteriol. 179:7351–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elena S. F., Lenski R. E. 2003. Evolution experiments with microorganisms: the dynamics and genetic basis of adaptation. Nat. Rev. Genet. 4:457–469 [DOI] [PubMed] [Google Scholar]
- 7. Errington J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Mol. Biol. Rev. 57:1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fischer E., Sauer U. 2005. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat. Genet. 37:636–640 [DOI] [PubMed] [Google Scholar]
- 9. Foster P. L. 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42:373–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gabriel S. E., Miyagi F., Gaballa A., Helmann J. D. 2008. Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur. J. Bacteriol. 190:3482–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giongo A., Tyler H. L., Zipperer U. N., Triplett E. W. 2010. Two genome sequences of the same bacterial strain, Gluconacetobacter diazotrophicus PAl 5, suggest a new standard in genome sequence submission. Stand. Genomic Sci. 2:309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gorbalenya A. E., Koonin E., Donchenko A. P., Blinov V. M. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair, and expression of DNA and RNA. Nucleic Acids Res. 17:4713–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graumann P.(ed.). 2007. Bacillus: cellular and molecular biology. Caister Academic Press, Wymondham, United Kingdom [Google Scholar]
- 14. Hansen T. 2003. Is modularity necessary for evolvability? Remarks on the relationship between pleiotropy and evolvability. Biosystems 69:83–94 [DOI] [PubMed] [Google Scholar]
- 15. Kooistra J., Vosman B., Venema G. 1988. Cloning and characterization of a Bacillus subtilis transcription unit involved in ATP-dependent DNase synthesis. J. Bacteriol. 170:4791–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kullik I., Stevens J., Toledano M. B., Storz G. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol. 177:1285–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence J. G., Hendrix R. W., Casjens S. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9:535–540 [DOI] [PubMed] [Google Scholar]
- 18. Maughan H., Birky C. W., Nicholson W. L. 2009. Transcriptome divergence and the loss of plasticity in Bacillus subtilis after 6,000 generations of evolution under relaxed selection for sporulation. J. Bacteriol. 191:428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maughan H., et al. 2006. The population genetics of phenotypic deterioration in experimental populations of Bacillus subtilis. Evolution 60:686–695 [PubMed] [Google Scholar]
- 20. Maughan H., Masel J., Birky C. W., Nicholson W. L. 2007. The roles of mutation accumulation and selection in loss of sporulation in experimental populations of Bacillus subtilis. Genetics 177:937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maughan H., Nicholson W. L. 2011. Increased fitness and the alteration of metabolic pathways during Bacillus subtilis laboratory evolution. Appl. Environ. Microbiol. 77:4105–4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mercante J., Suzuki K., Cheng X., Babitzke P., Romeo T. 2006. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J. Biol. Chem. 281:31832–31842 [DOI] [PubMed] [Google Scholar]
- 23. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Mira A., Ochman H., Moran N. A. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589–596 [DOI] [PubMed] [Google Scholar]
- 25. Moolenaar G. F., Visse R., Ortiz-Buysse M., Goosen N., van de Putte P. 1994. Helicase motifs V and VI of the Escherichia coli UvrB protein of the UvrABC endonuclease are essential for the formation of the preincision complex. J. Mol. Biol. 240:294–307 [DOI] [PubMed] [Google Scholar]
- 26. Moore C. M., Helmann J. D. 2005. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 8:188–195 [DOI] [PubMed] [Google Scholar]
- 27. Moran N. A., McLaughlin H. J., Sorek R. 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323:379–382 [DOI] [PubMed] [Google Scholar]
- 28. Moszer I. 1998. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 430:28–36 [DOI] [PubMed] [Google Scholar]
- 29. Moszer I., Glaser P., Danchin A. 1995. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology 141:261–268 [DOI] [PubMed] [Google Scholar]
- 30. Munakata N. 1977. Mapping of the genes controlling excision repair of pyrimidine photoproducts in Bacillus subtilis. Mol. Gen. Genet. 156:49–54 [Google Scholar]
- 31. Nakamura L. K., Roberts M. S., Cohan F. M. 1999. Relationship of Bacillus subtilis clades associated with strains 168 and W23: a proposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int. J. Syst. Bacteriol. 49:1211–1215 [DOI] [PubMed] [Google Scholar]
- 32. Ordal G. W., Márquez-Magaña L. M., Chamberlin M. J. 1992. Motility and chemotaxis in Bacillus subtilis, p. 765–784In Sonenshein A., Hoch J. A., Losick R.(ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, DC [Google Scholar]
- 33. Palsson B. 2011. Adaptive laboratory evolution. Microbe 6:69–74 [Google Scholar]
- 34. Perego M., Hoch J. A. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases, p. 473–481In Sonenshein A. L., Hoch J. A., Losick R.(ed.). Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 35. Raposo M. P., Inácio J. M., Mota L. J., de Sá-Nogueira I. 2004. Transcriptional regulation of genes encoding arabinan-degrading enzymes in Bacillus subtilis. J. Bacteriol. 186:1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Renna M. C., Najimudin N., Winik L. R., Zahler S. A. 1993. Regulation of the Bacillus subtilis alsS, aslD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roesch L. W. F., et al. 2009. Influence of fecal sample storage on bacterial community diversity. Open Microbiol. J. 3:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanchez H., Carrasco B., Ayora S., Alonso J. C. 2007. Homologous recombination in low dC+dG Gram-positive bacteria. Top. Curr. Genet. 17:27–52 [Google Scholar]
- 39. Sanchez H., Carrasco B., Ayora S., Alonso J. C. 2007. Dynamics of DNA double-strand break repair in Bacillus subtilis, p. 43–66In Graumann P.(ed.), Bacillus: cellular and molecular biology. Caister Academic Press, Wymondham, United Kingdom [Google Scholar]
- 40. Schaeffer P., Millet J., Aubert J.-P. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schöbel S., Zellmeier S., Schumann W., Wiegert T. 2004. The Bacillus subtilis σW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 52:1091–1105 [DOI] [PubMed] [Google Scholar]
- 42. Serizawa M., et al. 2004. Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and Northern blotting analyses. Gene 329:125–136 [DOI] [PubMed] [Google Scholar]
- 43. Sharp M. D., Pogliano K. 2002. MinCD-dependent regulation of the polarity of SpoIIIE assembly and DNA transfer. EMBO J. 21:6267–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sierro N., Makita Y., de Hoon M. J. L., Nakai K. 2008. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36:D93–D96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sonenshein A. L. 2002. The Krebs citric acid cycle, p. 151–162In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 46. Sonenshein A. L., Hoch J. A., Losick R.(ed.). 2002. Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 47. Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. U. S. A. 44:1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stragier P., Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297–341 [DOI] [PubMed] [Google Scholar]
- 49. Strauch M. A., Hoch J. A. 1993. Transition-state regulators: Sentinels of Bacillus subtilis post-exponential gene expression. Mol. Microbiol. 7:337–342 [DOI] [PubMed] [Google Scholar]
- 50. Tanaka M. M., Bergstrom C. T., Levin B. R. 2003. The evolution of mutator genes in bacterial populations: the roles of environmental change and timing. Genetics 164:843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Theis K., Chen P. J., Skorvaga M., Van Houten B., Kisker C. 1999. Crystal structure of UvrB, a DNA helicase adapted for excision repair. EMBO J. 18:6899–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yeeles J. T. P., Gwynn E. J., Webb M. R., Dillingham M. S. 2011. The AddAB helicase-nuclease catalyses rapid and processive DNA unwinding using a single Superfamily 1A motor domain. Nucleic Acids Res. 39:2271–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zaim J., Kierzek A. M. 2003. The structure of full-length LysR-type transcriptional regulators: modeling of the full-length OxyR transcription factor dimer. Nucleic Acids Res. 31:1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]