Abstract
Surface water can be contaminated by bacteria from various sources, including manure from agricultural facilities. Attachment of these bacteria to soil and organic particles contributes to their transport through the environment, though the mechanism of attachment is unknown. As bacterial attachment to human tissues is known to be correlated with antibiotic resistance, we have investigated here the relationship between bacterial attachment to environmental particles and antibiotic resistance in agricultural isolates. We evaluated 203 Escherichia coli isolates collected from swine facilities for attachment to quartz, resistance to 13 antibiotics, and the presence of genes encoding 13 attachment factors. The genes encoding type I, EcpA, P pili, and Ag43 were detected, though none was significantly related to attachment. Quartz attachment was positively and significantly (P < 0.0038) related to combined resistance to amoxicillin/streptomycin/tetracycline/sulfamethazine/tylosin/chlortetracycline and negatively and significantly (P < 0.0038) related to combined resistance to nalidixic acid/kanamycin/neomycin. These results provide clear evidence for a link between antibiotic resistance and attachment to quartz in agricultural isolates. We propose that this may be due to encoding by the responsible genes on a mobile genetic element. Further exploration of the relationship between antibiotic resistance and attachment to environmental particles will improve the understanding and modeling of environmental transport processes, with the goal of preventing human exposure to antibiotic-resistant or virulent microorganisms.
INTRODUCTION
During rainfall or irrigation, manure-borne bacteria can be transported to surface waters via soil and organic particles (38). The transport of these bacteria in surface water impacts their ultimate location and survival in the environment (6, 10) and thus the likelihood of exposure to humans (17). Previous studies have found that the fraction of Escherichia coli attached to eroded particles suspended in saturation excess flow ranges from >1% to 49% (34, 43, 44), while the attached fraction in streams has been reported to be between 20% and 55% (8, 27). Concentrations of bacteria in stream sediments can be 10 to 10,000 times higher than the concentration in the overlying waters (3), highlighting the importance of understanding interactions between bacteria and sediment particles, so that the risk to public health can be properly evaluated (14, 30).
Despite the importance of bacteria-sediment interactions, limited understanding exists of the bacterial properties that govern attachment and the resulting particulate-mediated environmental transport. Researchers who have attempted to understand bacterial attachment to quartz, a model environmental particle, have obtained mixed results, possibly due to the limited sample size used in these studies. Bolster et al. measured the physical properties of 12 E. coli isolates and found a significant relation between attachment and cell width and sphericity (5). Contrastingly, Foppen et al.'s study of 54 E. coli isolates found no relation between attachment and cell sphericity, motility, zeta potential, aggregation or lipopolysaccharide composition (16). Bacteria carry genes that encode various types of pili, fimbriae, and other surface proteins that mediate attachment to other bacteria, host tissues, or other surfaces (18). An attempt to attribute quartz attachment to these surface features, instead of a cell's chemical or physical properties, found a significant relation with the expression of the Ag43 attachment factor or flagella, but the study considered only 6 isolates (31). However, a separate study of 20 E. coli isolates that investigated 22 attachment and motility factors, including Ag43, type I pili, and afimbrial adhesion (Afa), concluded that there was no statistically significant link to any of the queried properties (16). Thus, the underlying genetic basis for bacterial attachment to quartz, and other environmental particles, remains unidentified.
The U.S. agricultural industry uses more than 10 million pounds of antibiotics each year (20, 41) for growth promotion, disease prevention, and disease eradication purposes. The United States National Swine Survey found that 57% of feeds contain antibiotic levels three or four times above recommended levels (13), and others have reported that up to 90% of some administered antibiotics pass through the animal's digestive systems and are excreted (37). A comparison of surface and ground water samples collected up- or down-gradient of swine farms showed that E. coli from the down-gradient samples had increased resistance to certain antibiotics (41). Therefore, it is not surprising that swine manure is a reservoir for transferable antibiotic resistance genes (4). E. coli isolates from wild small mammals collected on swine farms have increased antimicrobial resistance relative to small mammals from other environments, providing another potential mechanism for dispersal of these bacteria (1).
Intriguingly, certain attachment factors associated with attachment to human tissues have been associated with antibiotic resistance in clinical isolates. For example, Liverell's study showed that antibiotic-resistant bacteria have increased adherence to human intestinal cells relative to antibiotic-sensitive strains (28). Ampicillin resistance has been associated specifically with P pili and Dr-type attachment factors (2, 24, 25), and tobramycin resistance has been associated with afimbrial adhesion, S fimbriae, and P pili (2). This relationship is not surprising when one considers that mobile genetic elements carrying both attachment and resistance-associated genes have been found in E. coli (29, 39). However, to the best of our knowledge, this association between resistance and attachment has been noted only for clinical isolates.
Manure from agricultural facilities is a major source of bacteria leading to impaired water quality. Therefore, understanding the attachment properties of these manure-borne bacteria will provide insight into the mechanisms of environmental transport and the resulting risk to public health. The goal of this study is to assess the relationship between attachment and antibiotic resistance in agricultural E. coli isolates. To achieve this goal, we evaluated the association between quartz attachment and resistance to 13 antibiotics in 203 isolates collected from swine facilities. Additionally, we queried these strains for the presence of genes encoding 13 different attachment factors and then related the presence of these genes to attachment to quartz and antibiotic resistance.
MATERIALS AND METHODS
Isolate collection and enumeration.
Swine waste samples were collected from five swine facilities in Iowa between November 2008 and April 2009. The farms were selected to encompass a range of management practices: two facilities did not use antibiotics at all, while three facilities administered antibiotics at subtherapeutic levels. Freshly excreted manure samples were collected from three locations, while samples from the remaining two locations were collected from a deep pit and lagoon.
Samples were collected in two sterile 1-liter containers, stored on ice for transport to the laboratory, and processed within 12 h of collection. Manure samples were homogenized by mixing 1 g manure with 9 ml of phosphate-buffered water (Hach Co., Loveland, CO) for 10 min. E. coli was isolated and enumerated by membrane filtration, EPA Method 1603 (48), on modified mTEC agar (47). Approximately 100 typical and atypical E. coli colonies were selected from each location and preserved in 25% glycerol frozen stocks at −80°C for further analysis.
Antibiotic resistance assays.
Antibiotic susceptibility was determined by the agar dilution procedure technique (9). Standard powders of the following antimicrobial agents were obtained from Sigma-Aldrich (St. Louis, MO): amoxicillin (AMX), ampicillin (AMP), chloramphenicol (CMP), chlortetracycline (CTC), erythromycin (ERY), gentamicin (GEN), kanamycin (KAN), nalidixic acid (NAL), neomycin (NEO), streptomycin (STP), sulfamethazine (SMZ), tetracycline (TET), and tylosin (TYL). Antibiotics for susceptibility testing were selected based on their utilization in human medicine, veterinary medicine, or both (7, 46). One or more representatives from the following classes of antimicrobials were selected: penicillins (AMP and AMX), aminoglycosides (GEN, NEO, STP, and KAN), macrolides (ERY and TYL), quinolones (NAL), sulfonamides (SMZ), tetracyclines (CTC and TET), and other (CMP). CMP is not a macrolide, but it functions similarly by affecting the 50S ribosomal subunit (45). While the selected antibiotics are important in treating bacterial infections, not all target Gram-negative bacteria (i.e., TYL, CMP, and ERY).
Antibiotic-infused Mueller-Hinton agar was mixed according to the guidelines proposed by the Clinical and Laboratory Standards Institute (9) for all antimicrobial agents except tylosin, which was dissolved in methanol and diluted in distilled water as previously described (26). The E. coli isolates were tested for resistance using the MICs presented in Table 1. All antibiotics tested fell within MIC quality control ranges that were established by the CLSI for E. coli ATCC 29522. The E. coli lab strains MG1655, BW25113, ATCC 8677, ATCC 9637, and ATCC 25922 were used as negative controls.
Table 1.
MICs for antimicrobial agents tested against E. coli isolatesa
| Antimicrobial agent | MIC (μg/ml) |
|
|---|---|---|
| MIC range | MIC90 | |
| Amoxicillin (AMX) | 16–48 | 48 |
| Ampicillin (AMP) | 16–48 | 48 |
| Chloramphenicol (CMP) | 16–48 | 48 |
| Chlortetracycline (CTC) | 16–48 | 48 |
| Erythromycin (ERY) | 15–30 | 30 |
| Gentamicin (GEN) | 8–24 | 24 |
| Kanamycin (KAN) | 32–96 | 96 |
| Naladixic acid (NAL) | 16–48 | 48 |
| Neomycin (NEO) | 8–24 | 24 |
| Tetracycline (TET) | 8–24 | 24 |
| Tylosin (TYL) | 16–48 | 48 |
| Streptomycin (STP) | 12–22.5 | 22.5 |
| Sulfamethazine (SMZ) | 256–768 | 768 |
The MIC90 was used to determine resistance.
Isolates were grown overnight in 10 ml of Mueller-Hinton broth (MHB) in 15-ml conical tubes at 37°C in a reciprocating shaker water bath (Thermo Scientific, Waltham, MA). Strains were diluted to a 0.5 McFarland standard prior to inoculation of the antibiotic dilution plates. One microliter of each strain was aseptically transferred to plates containing one of each of the 13 antimicrobial agents at the MIC level. All strains were tested in triplicate and plated on Mueller-Hinton agar and Mueller-Hinton agar II, as controls. Plates were allowed to dry prior to closing and inverting for incubation at 37°C for 16 to 22 h. Following incubation, each strain was recorded as present or absent.
E. coli attachment to quartz.
Quartz or silica sand particles passing through American Society for Testing and Materials (ASTM) sieve number 120 but remaining on sieve number 230 were selected for the attachment assay (38). The very fine sand (63 to 125 μm) was washed in distilled water and dried for 1 h in an oven at 105°C. Using the mass of quartz in 1 ml, 1.405 g cm−3, with an assumed density of 2.6 g cm−3, an average radius of 63 μm, and a volume of 1.035 × 10−6 cm3, the total number of particles of quartz was calculated. Assuming that the particles are spherical in shape, the surface area of one particle was determined to be 4.95 × 10−2 mm2 (2.5 × 1010 μm2 ml−1); a mass of 0.4 g (which corresponds to a total surface area of 12,500 mm2) of quartz provided sufficient surface area for all E. coli isolates to attach.
Each strain was transferred into sterile 15-ml conical tubes containing Mueller-Hinton broth (DifcoTM, Detroit, MI) for growth at 37°C in a reciprocal shaking water bath (Thermo Scientific, Waltham, MA) for 12 hours. Samples were removed and centrifuged for 5 min at 2,500 rpm in a refrigerated centrifuge (Eppendorf, Hauppauge, NY; model 5702 R) at 4°C. The supernatant was discarded, and 10 ml of phosphate-buffered water was added to each pellet. The pellet was resuspended by vortexing. The resuspended cells were diluted to a 0.5 McFarland standard (approximately 1.0 × 108 CFU ml−1) according to the Clinical and Laboratory Standards Institute (January 2006) using phosphate-buffered water. The actual initial E. coli concentration was determined by EPA Method 1603, consisting of CFU enumeration. Forty milliliters of the diluted bacteria cultures were aseptically added to the 50-ml conical tubes containing quartz, vortexed, and placed horizontally on an orbital shaker for 20 min at 80 rpm. After shaking, the conical tubes were placed vertically in racks, and the quartz particles were allowed to settle via gravity for 5 min, sufficient time for all particles to settle as determined by Stoke's equation. After settling, 1 ml of supernatant was removed and serially diluted to 1 × 106 in 9 ml of phosphate-buffered water. The total E. coli concentration in the supernatant was enumerated using EPA Method 1603 and recorded as the unattached fraction. The percent attached E. coli was computed as the total number of CFU minus the number of unattached CFU divided by the total number of CFU. For quality control assessment, 15 strains were analyzed in duplicate. The average percentage error in these duplicate attachment measurements was 7.68 ± 0.07.
The effect of mannose was investigated by performing the attachment assay with or without 0.2% (wt/vol) d-mannose in the phosphate-buffered water (36).
Assessment for attachment factor and virulence genes.
Two hundred and three strains that exhibited positive attachment and for which a complete set of resistance data was generated were selected for PCR analysis. Genomic DNA was isolated from cells grown in liquid Luria broth medium at 37°C overnight and pelleted by centrifugation. Genomic DNA extraction was performed using the DNeasy tissue kit according to the manufacturer's instructions (Qiagen, Valencia, CA). The concentration was determined using the Qubit nucleic acid assay system (Invitrogen, Carlsbad, CA). All genomic DNA stocks were stored at −20°C and diluted to 0.1 μg ml−1 for PCR analysis. For colony PCR, cells were obtained directly from frozen glycerol stocks, transferred to 100 μl of sterile nanopure water, and boiled at 95°C for 10 min.
All primers have been used previously in genotyping studies and are listed in Table 2, except for the Ag43-specific primers, which were designed by our lab based on the MG1655 sequence. Primer sequences for virulence factors were obtained from the following sources: hemolysin A (2), shiga toxin I (21), shiga toxin II (21), cytotoxic necrotizing factor I (2), aerobactin (2), surface antigens 1, 2, 3, 4, 5, 6, 14, and 17 (19), and enterotoxin A (21) but are not listed, as none of these virulence factors was detected in our isolate collection.
Table 2.
Primers for assessing the presence of genes encoding known attachment factors
| Purpose or attachment factor (queried gene) | Positive controlb | Primer | Reference or source | Product size (bp) |
|---|---|---|---|---|
| Verification of E. coli (wecA)a | MG1655 | GGTGTTCGGCAAGCTTTATCTCAG | 2 | 763 |
| GGTTAAATTGGGGCTGCCACCACG | ||||
| P pili (pap) | pPAP5 plasmid | GACGGCTGTACTGCAGGGTGTGGCG | 2 | 328 |
| ATATCCTTTCTGCAGGGATGCAATA | ||||
| Type I pili (fim) | MG1655 | CGACGCATCTTCCTCATTCTTCT | 35 | 721 |
| ATTGGTTCCGTTATTCAGGGTTGTT | ||||
| S fimbriae (sfa) | CFT073 | CTCCGGAGAACTGGGTGCATCTTAC | 2 | 410 |
| CGGAGGAGTAATTACAAACCTGGCA | ||||
| Colonization factor antigen I (CFA/I gene) | NA | GCTCTGACCACAATGTTGA | 20 | 364 |
| TTACACCGGATGCAGAATA | ||||
| Colonization factor antigen III (CFA/III gene) | NA | GCCTTCTGGAAGTCATCAT | 20 | 437 |
| TGCCACATACTCCCAGTTA | ||||
| F1C fimbriae (foc) | CFT073 | GGTGGAACCGCAGAAAATAC | 32 | 388 |
| GAACTGTTGGGGAAAGAGTG | ||||
| Dr hemagglutinin (Dr) | SM298 plasmid | GCCAACTGACGGACGCAGCAC | 35 | 229 |
| CCCCAGCTCCCGACATCGTTTTT | ||||
| Afimbrial adhesion I (afa) | NA | GCTGGGCAGCAAACTGATAACTCTC | 2 | 750 |
| CATCAAGCTGTTTGTTCGTCCGCCG | ||||
| E. coli common pilus (ecpA) | NA | TGAAAAAAAAGGTTCTGGCAATAGC | 40 | 483 |
| CGCTGATGATGGAGAAAGTGAA | ||||
| F41 pili (F41) | NA | GCATCAGCGGCAGTATCT | 21 | 380 |
| GTCCCTAGCTCAGTATTATCACCT | ||||
| K99 pili (K99) | NA | TATTATCTTAGGTGGTATGG | 21 | 314 |
| GGTATCCTTTAGCAGCAGTATTTC | ||||
| F17 pili (F17) | NA | CGGAGCTAATACTGCATCAACC | 21 | 615 |
| TGTTGATATTCCGTTAACCGTAC | ||||
| Ag 43 (flu) | MG1655 | ATGCCTCGAGATGAAACGACATCTGAATACCTG | This work | 1,650 |
| ATGCAAGCTTGTCAATGGCACCGTTCAGCACAGTG |
wecA was used to confirm strain identity as E. coli.
NA, not available.
All strains were confirmed as E. coli using wecA-specific primers (2). Optimal annealing temperatures were identified using a gradient PCR for each gene by the use of genomic DNA from a positive control. Twenty-microliter reactions were performed in 96-well plates with the following components: 0.5 μl bacterial DNA or boiled cells, 10 μl 2× Taq PCR master mix (Qiangen, Valencia, CA), 0.2 μl each primer, and 9.1 μl RNase-free water. Amplification was conducted in a Bio-Rad iCycler thermocycler (Bio-Rad, Hercules, CA) under the following conditions: initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 30 s, and extension at 72°C for 3 min, and a final extension at 72°C for 7 min. For the gradient PCR, annealing temperatures were set from 45°C to 63°C. Positive controls were utilized for P pili, type I pili, S fimbriae, F1C fimbriae, and Dr hemagglutinin genes. These templates were obtained as follows: MG1655 and CFT073 were obtained from American Type Culture Collection (ATCC), pPAP5 (2) was obtained from Scott Hultgren (Washington University, St. Louis, MO), and SM298 was obtained from Steve Moseley (University of Washington). We did not have a positive control for EcpA, but the identity of the PCR product was confirmed by DNA sequencing.
The PCR product was analyzed on 1% Tris-acetate-EDTA (TAE) agarose with SYBR Safe DNA gel stain (Invitrogen, Carlsbad, CA) using an 8-ul PCR product and a 1-kb Plus DNA ladder as a standard (Invitrogen, Carlsbad, CA). Gels were documented using a Fotodyne Minivisionary photo system.
Statistical analysis.
Statistical analyses were performed using the Statistical Analysis JMP 8 software (SAS Institute; 2008). All data were confirmed as normally distributed using the statistic index. A t test was conducted to analyze the difference between E. coli resistance and sensitivity as a function of median percent attachment. Fisher's exact test (11) was used to analyze the difference between E. coli resistance and sensitivity as a function of the presence of type 1, EcpA, Ag43, or P pili attachment factors. For the association between quartz attachment and attachment factors as well as the association between antibiotic resistance and attachment factors, 13 comparisons were conducted separately. According to the multiplicity of determinations (Bonferroni correction), the adjusted P value was set at 0.0038 (0.05 divided by 13 comparisons).
RESULTS
Prevalence of antibiotic resistance.
In order to characterize our isolate collection, we assessed our strains for their sensitivity to 13 antimicrobial agents. These agents were grouped according to their importance to humans, as determined by the World Health Organization: critically important (GEN, AMP, ERY, AMX, NAL, STP), highly important (TET, SMZ, KAN, CMP), and others (TYL, CTC, NEO) (7).
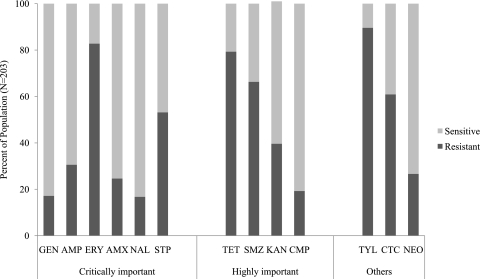
Antibiotic resistance was observed in the following increasing order (Fig. 1): NAL (16.75%), GEN (17.17%), CMP (19.21%), AMX (24.63%), NEO (26.6%), AMP (30.54%), KAN (39.61%), STP (53.14%), CTC (60.89%), SMZ (66.29%), TET (79.31%), ERY (82.76%), and TYL (89.6%). According to the classes of antibiotics, isolates showed a higher level of resistance to macrolides (ERY and TYL), which is to be expected since macrolides do not target Gram-negative cells, and the lowest resistance to aminoglycosides (GEN, NEO, STP, and KAN). Of the 203 isolates, there was one isolate resistant to all antibiotics and only two isolates were sensitive to all agents (data not shown).
Fig. 1.
Distribution of sensitivity and resistance to each antibiotic agent. Antibiotics are presented according to their importance ranking by the WHO. S, sensitive; R, resistant.
Antibiotic resistance is associated with quartz attachment.
Other researchers have previously noted a relation between attachment to human tissues and antibiotic resistance (28). We aim to determine if there is also a relationship between attachment to environmental particles, such as quartz, and antibiotic resistance. Therefore, we grouped the population according to antibiotic sensitivity and compared the median quartz attachment of the sensitive group to that of the resistant group (Fig. 2).
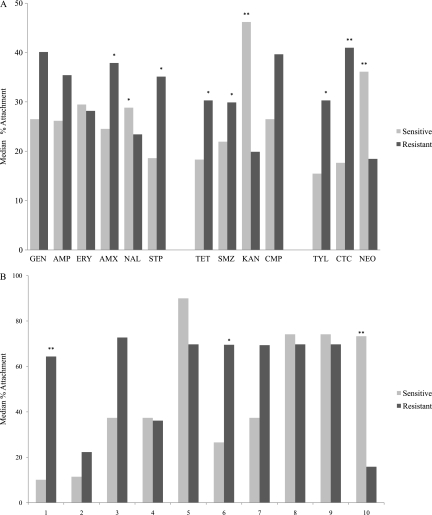
Fig. 2.
Association of quartz attachment and antibiotic resistance. P values were determined by t test (*, P < 0.1; **, P < 0.0038). (A) Antibiotics are sorted according to importance ranking, as described in the legend to Fig. 1. (B) Antibiotic combinations are sorted as groups showing a possibly significant increase in quartz attachment relative to the antibiotic-sensitive group (lane 1) or to the antibiotic-resistant group (lane 10), according to the rankings shown in panel A, and as six-antibiotic combinations selected randomly (lanes 2 to 9). Lanes 1, AMX/STP/TET/SMZ/TYL/CTC; 2, GEN/AMP/TET/SMZ/KAN/TYL; 3, ERY/AMX/CTC/NEO/NAL/STP; 4, AMP/SMZ/ERY/CTC/NAL/CMP; 5, AMX/STP/GEN/AMP/ERY/CMP; 6, AMX/STP/TET/AMP/ERY/CMP; 7, AMX/STP/TET/SMZ/ERY/CMP; 8, AMX/STP/TET/GEN/ERY/CMP; 9, AMX/STP/TET/AMP/ERY/GEN; 10, NAL/KAN/NEO.
Six of the antibiotic-resistant groups showed a possibly significant increase in quartz attachment relative to the corresponding antibiotic-sensitive group (Fig. 2A): AMX (37.9% versus 24.5%, P = 0.098), STP (35.1% versus 18.6%, P = 0.005), TET (30.3% versus 18.3%, P = 0.03), SMZ (29.9% versus 21.9%, P = 0.054), TYL (30.3% versus 15.5%, P = 0.017), and CTC (41.0% versus 17.7%, P = 3.59E−06). When applying the most stringent significance criteria, a P value cutoff of 0.05 adjusted for a multiplicity of determinations to 0.0038, only CTC showed a significant increase in quartz attachment between antibiotic-resistant and antibiotic-sensitive groups. In contrast to the trend observed with CTC, the attachment for the strains resistant to KAN (P = 1.0E−06), NEO (P = 1.2E−04), or NAL (P = 0.053) was lower than for sensitive strains (Fig. 2A). KAN and NEO met the adjusted significance criteria.
We used the single-antibiotic results to select combinations of antibiotics for further analysis. Since AMX, STP, TET, SMZ, TYL, and CTC each met the possibly significant cutoff of 0.1, we analyzed strains that were resistant to these six antibiotics. As a control, eight other randomly selected combinations of 6 antibiotics were also considered. Figure 2B shows the resistant pattern of antibiotic combinations in the isolates, showing a possibly significant increase in quartz attachment relative to the antibiotic-sensitive group or to the antibiotic-resistant group, as shown in Fig. 2A, and six-antibiotic combinations selected randomly. The combination of the above six antibiotics (AMX/STP/TET/SMZ/TYL/CTC) showed a significant increase of quartz attachment in antibiotic-resistant groups (P = 0.000567) (Fig. 2B). We similarly analyzed the 3 antibiotics that had a negative association with attachment (NAL, KAN, and NEO). The combination of NAL/KAN/NEO decreased quartz attachment in antibiotic-resistant groups significantly (P = 0.0012) (Fig. 2B).
Attachment cannot be attributed to known attachment factors.
We screened 203 isolates from five facilities for the genes encoding 13 attachment factors: P pili, type I pili, S fimbriae, colonization factor antigens I and III, F1C fimbriae, Dr hemagglutinin, afimbrial adhesion I, E. coli common pilus (EcpA), F41 pili, K99 pili, F17 pili, and Ag43. Genes associated with only four of these attachment factors were detected in our isolate pool: EcpA, type I pili, Ag43, and P pili (Table 3). A total of 150 strains (73.9%) carried genes encoding EcpA, 122 strains (60.1%) carried genes encoding type I, 28 strains carried genes encoding Ag43 (13.8%), and only 4 (1.97%) carried genes encoding P pili. An additional 247 strains were queried for the P pili-encoding gene by the use of colony PCR, but no additional strains were identified. Additionally, we screened our isolate collection for key virulence-associated genes, but none of these virulence factors was detected (data not shown).
Table 3.
Relationship between the presence of genes encoding known attachment factors and the observed quartz attachment
| Group according to genotype | No. | Median attachment (%) | P value relative to strains without any known attachment factorsa | P value relative to all strainsa |
|---|---|---|---|---|
| All strains | 203 | 28.5 | 0.304 | |
| No known attachment factors | 22 | 31.2 | 0.304 | |
| Type I (any) | 122 | 26.4 | 0.174 | 0.198 |
| EcpA (any) | 150 | 25.0 | 0.181 | 0.210 |
| P pili (any) | 4 | 30.8 | 0.316 | 0.357 |
| Ag43 (any) | 28 | 29.3 | 0.438 | 0.364 |
| Type I (only) | 23 | 39.6 | 0.232 | 0.049 |
| EcpA (only) | 49 | 31.1 | 0.472 | 0.276 |
| Ag43 (only) | 1 | 85.1 | ND | ND |
| Type I + EcpA (only) | 77 | 22.7 | 0.054 | 0.044 |
| P pili + type I (only) | 1 | 6.02 | ND | ND |
| P pili + type I + EcpA (only) | 3 | 36.0 | 0.495 | 0.427 |
| Type I + Ag43 (only) | 6 | 29.8 | 0.441 | 0.316 |
| EcpA + Ag43 (only) | 9 | 54.8 | 0.330 | 0.180 |
| Type I + EcpA + Ag43 (only) | 12 | 19.9 | 0.137 | 0.158 |
P values were determined by t test. ND, not determined.
For these 203 strains, quartz attachment ranged from 0.84% to 99.87%, with a median value of 28.6%. Note that the attachment assay has a measurement error of 7.7%, determined by repeated analysis of 15 strains. In an attempt to attribute quartz attachment to the presence of one or more of these attachment-associated genes, strains were grouped according to their genotype (Table 3). For example, 22 strains carry genes that encode no known attachment factors, 122 strains carry genes that encode type I pili, and 4 strains carry genes that encode P pili. Strains were also grouped according to the presence of other attachment factors. For example, of the 122 strains with genes encoding type I pili, 23 encode only type I pili, 77 also encode EcpA but not P pili or Ag43, and none encodes all four attachment factors. The median percent attachment of each of these groups was compared to that of the general population and to that of the group with no known attachment factors. Note that because the type I/P pili-only group and Ag43-only group each contained just one strain, the statistical significance was not assessed. While some comparisons met the potentially significant cutoff of 0.05, none met the adjusted cutoff.
As a secondary test for the importance of the type I pili, we repeated the attachment assay for select strains that do or do not carry genes that encode type I pili in the presence of d-mannose, where exogenous d-mannose can inhibit the binding of type I pili (36). For strains that do not carry genes that encode type I pili, the presence of d-mannose during the attachment assay decreased the median attachment 1.8 (±0.1)-fold (n = 4) but did not eliminate attachment. For strains not carrying genes encoding type I pili, quartz attachment was not mannose sensitive: the median attachment value increased 1 (±1)-fold (n = 4) in the presence of mannose. Thus, we conclude that quartz attachment cannot be attributed to any of these four attachment factors, though type I pili may play a contributing role.
Antibiotic resistance is associated with genes encoding attachment factors.
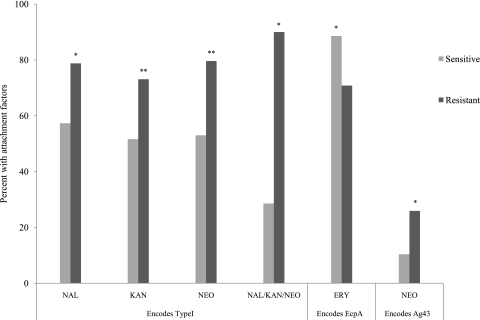
The results shown in Fig. 2 indicate a clear relationship between attachment and antibiotic resistance. While we cannot attribute attachment to any of the queried attachment factors, we looked for a relationship between antibiotic resistance and the genes encoding attachment factors. Specifically, we compared the percentage of antibiotic-resistant strains with genes encoding the focal attachment factor to the percentage of antibiotic-sensitive strains with genes encoding the focal attachment factor. For example, the type I attachment factor is encoded by the genes of 78.8% of NAL-resistant strains, but by those of only 57.3% of NAL-sensitive strains. Meaning, NAL-resistant isolate genes more frequently encode type I pili than those of NAL-sensitive isolates. Figure 3 shows those antibiotics for which a significant difference between the resistant and sensitive populations was detected in regard to type I, EcpA, and Ag43. Strains were treated similarly whether they contained a given attachment factor only or a combination of attachment factors.
Fig. 3.
Relationship between antibiotic resistance and the presence of genes encoding known attachment factors; data shown only for possibly significant relations. Significance was determined by Fisher's exact test (*, P < 0.1; **, P < 0.0038).
The type I attachment factor shows the most significant associations. Specifically, the isolates resistant to KAN (P = 0.002), NEO (P = 0.001), NAL (P = 0.015), or the combination of these three antibiotics (P = 0.018) carried the type I gene more often than the corresponding sensitive isolates (Fig. 3). When the P value is adjusted to 0.0038 to account for multiplicity, the association with NAL and the combination group do not meet significance criteria, but the associations with KAN and NEO do. There was no relationship between resistance to other antibiotics and the presence of genes encoding the type I attachment factor (data not shown). Contrastingly, strains that were sensitive to ERY were more likely to carry genes that encode EcpA (P = 0.034) (Fig. 3). Strains with genes encoding Ag43 showed higher resistance to NEO (P = 0.008). Finally, strains with P pili showed a broad increase in resistance to all the antibiotics (except NAL) and the combination of AMX/STP/TET/SMZ/TYL/CTC. However, given the small size of the population (4 strains) with genes encoding P pili, it was difficult to satisfy the significance criteria (data not shown). Thus, our analysis shows that there is a relation between resistance to certain antibiotics and the presence of genes encoding certain attachment factors.
DISCUSSION
Here we have queried the relationship between three properties in a set of agricultural E. coli isolates: attachment to quartz particles, antibiotic resistance, and the presence of genes known to encode attachment factors. This work was motivated by the fact that antibiotic resistance and attachment factors can be encoded together on mobile genetic elements (23), leading to the proposition of a significant relationship between the attachment phenotype and antibiotic resistance. This relationship has previously been observed with clinical isolates (2, 24, 25); the work presented here is the first investigation of agricultural isolates. We found that resistance to 6 of the 13 tested antibiotics is significantly and positively associated with quartz attachment (Fig. 2A); resistance to their combinations was as well (P = 0.00057) (Fig. 2B). These six antibiotics are AMX, STP, TET, SMZ, TYL, and CTC. Three other antibiotics (NAL, KAN, and NEO), and their combination, are also significantly associated with quartz attachment, though the fact that attachment is negatively associated with resistance was not expected (Fig. 2A and B).
Our underlying hypothesis is that genes encoding antibiotic resistance and attachment are linked on mobile genetic elements, and the combination of six antibiotics (AMX/STP/TET/SMZ/TYL/CTC) showed a significant increase of quartz attachment in antibiotic-resistant groups (Fig. 2B), which supports this proposed linkage. To assess the validity of this hypothesis, we related the presence of known attachment-encoding genes to the observed antibiotic resistance. Of the attachment factors detected in our isolate collection, type I, Ag43, and P pili showed significant relations with resistance to at least one antibiotic (Fig. 3). The relation of P pili with AMP resistance in our collection is consistent with previous results with clinical isolates (2) (data not shown). Note that the lack of a statistically significant relation with P pili may be due to the fact that very few strains in our collection carry genes that encode P pili. It should be clarified that nearly all of the attachment factors queried in this study are known for their ability to mediate attachment to mammalian tissues. While some of the carbohydrates known to mediate binding of these attachment factors have been detected in soil samples (12, 15), it seems unlikely that these tissue-specific attachment factors mediate attachment to soil particles and even more unlikely that they mediate attachment to quartz. Instead, we propose that the attachment factors responsible for mediating attachment to quartz or other environmental particles, though possibly associated with type I pili, remain unidentified.
While previous studies have related quartz attachment with the expression of the Ag43 attachment factor, other studies were unable to replicate this relationship (16), and Ag43 was not related with attachment in our study. There are two possible explanations for this discrepancy. First is the difference in sample size between these two studies: Lutterodt et al.'s analysis utilized less than 10 strains (31), while our comparison includes more than 200. A second explanation is that neither Ag43 nor type I pili is directly responsible for quartz attachment, but instead this is an indirect effect, possibly due to regulatory cross talk between attachment factors. The model attachment factor systems, particularly P pili and type I, as well as the flagellation system, are known to engage in regulatory cross talk (22, 42, 49). We propose that the as yet unidentified attachment factor(s) responsible for quartz attachment could be subject to regulation by other attachment systems, possibly explaining our observed negative relationship of quartz attachment with certain antibiotics.
We did observe that quartz attachment was somewhat mannose sensitive in strains with genes encoding type I pili but not in strains lacking the type I genes. This suggests that the type I pili may play a contributing role in quartz attachment, though the type I genes do not meet our criteria for significant association with attachment.
The pattern of antibiotic resistance seen in our isolate collection provides an interesting comparison for other studies. For example, Moore et al.'s analysis of 12 bacterial isolates collected from rivers and lakes in Northern Ireland showed that 17%, 75%, and 75% of isolates were resistant to GEN, TET, and ERY, respectively (33). In our isolates, the respective resistances were a strikingly similar 17%, 79%, and 83%. Contrastingly, our isolates showed a higher prevalence of resistance to certain antibiotics than that reported in a study of human stool samples by the National Antimicrobial Resistance Monitoring System (NARMS) (7). While the 2005 NARMS report identified 0% of the isolate collection as resistant to KAN, 4.2% resistant to AMX, and 9.3% resistant to NAL, our isolate collection showed resistant populations of 40%, 25%, 17%, respectively. This difference is even more striking considering that our criteria for determining resistance were more stringent than those used in the NARMS study.
In conclusion, we have shown that there is a relationship between antibiotic resistance and attachment to quartz, a model environmental particle. We were unable to attribute attachment to any known attachment factors. Therefore, we propose that quartz attachment is mediated by another, as yet unidentified surface protein or surface feature. We did find that the presence of certain attachment factors is significantly related with resistance to certain antibiotics, providing supporting evidence that these features may be encoded on shared mobile genetic elements. These results highlight the importance of studying these properties in agriculturally derived bacteria, as transport of these bacteria through ground and surface waters can result in the exposure of human populations. While we did not detect known virulence factors in our isolate collection, this potential for human exposure would be even more concerning if the strains did contain virulence factors. The identification of the specific surface features responsible for sediment attachment is critical to understanding and modeling environmental transport processes and preventing exposure of human populations to virulent, antibiotic-resistant bacteria.
ACKNOWLEDGMENTS
Funding for this work was provided by Iowa State University.
We thank Tyler Myers, Abigail Jensen, Brittany Rover, Meredith Breton, and Kara Moeller for assistance with DNA preparation and PCR. We also thank Lisa Nolan and Oscar G. Gomez for helpful discussion.
Footnotes
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Allen S. E., Boerlin P., Janecko N., Lumsden J. S., Jardine1 C. 2011. Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in southern Ontario, Canada. Appl. Environ. Microbiol. 77:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arisoy M., Rad A. Y., Akin A., Akar N. 2008. Relationship between susceptibility to antimicrobials and virulence factors in paediatric Escherichia coli isolates. Int. J. Antimicrob. Agents 31:S4–S8 [DOI] [PubMed] [Google Scholar]
- 3. Bai S., Lung W. S. 2005. Modeling sediment impact on the transport of fecal bacteria. Water Res. 39:5232–5240 [DOI] [PubMed] [Google Scholar]
- 4. Binh C. T. T., Heuer H., Kaupenjohann M., Smalla K. 2008. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol. Ecol. 66:25–37 [DOI] [PubMed] [Google Scholar]
- 5. Bolster C. H., Haznedaroglu B. Z., Walker S. L. 2009. Diversity in cell properties and transport behavior among 12 different environmental Escherichia coli isolates. J. Environ. Qual. 38:465–472 [DOI] [PubMed] [Google Scholar]
- 6. Burton G. A., Gunnison D., Lanza G. R. 1987. Survival of pathogenic bacteria in various fresh-water sediments. Appl. Environ. Microbiol. 53:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. CDC. 2008. National Antimicrobial Resistance Monitoring System for enteric bacteria (NARMS): human isolates final report, 2005. U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 8. Characklis G. W., et al. 2005. Microbial partitioning to settleable particles in stormwater. Water Res. 39:1773–1782 [DOI] [PubMed] [Google Scholar]
- 9. CLSI. 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10. Davies C. M., Long J. A. H., Donald M., Ashbolt N. J. 1995. Survival of fecal microorganisms in marine and fresh-water sediments. Appl. Environ. Microbiol. 61:1888–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dawson-Saunders B., Trapp R. G. 1990. Basic clinical biostatistics, p. 148–152. Appleton-Lange, Stamford, CT [Google Scholar]
- 12. Derrien D., Marol C., Balabane M., Balesdent J. 2006. The turnover of carbohydrate carbon in a cultivated soil estimated by 13C natural abundances. Eur. J. Soil Sci. 57:547–557 [Google Scholar]
- 13. Dewey C. E., Cox B. D., Straw B. E., Bush E. J., Hurd H. S. 1997. Associations between off-label feed additives and farm size, veterinary consultant use, and animal age. Prev. Vet. Med. 31:133–146 [DOI] [PubMed] [Google Scholar]
- 14. Droppo I. G., et al. 2009. Dynamic existence of waterborne pathogens within river sediment compartments. Implications for water quality regulatory affairs. Environ. Sci. Technol. 43:1737–1743 [DOI] [PubMed] [Google Scholar]
- 15. Fischer H., Meyer A., Fischer K., Kuzyakov Y. 2007. Carbohydrate and amino acid composition of dissolved organic matter leached from soil. Soil Biol. Biochem. 39:2926–2935 [Google Scholar]
- 16. Foppen J. W., Lutterodt G., Roling W. F. M., Uhlenbrook S. 2010. Towards understanding inter-strain attachment variations of Escherichia coli during transport in saturated quartz sand. Water Res. 44:1202–1212 [DOI] [PubMed] [Google Scholar]
- 17. Fries J. S., Characklis G. W., Noble R. T. 2008. Sediment-water exchange of Vibrio sp. and fecal indicator bacteria: implications for persistence and transport in the Neuse River Estuary, North Carolina, U. S. A.. Water Res. 42:941–950 [DOI] [PubMed] [Google Scholar]
- 18. Garcia M. I., LeBouguenec C. 1996. Role of adhesion in pathogenicity of human uropathogenic and diarrhoeogenic Escherichia coli. Bull. Inst. Pasteur 94:201–236 [Google Scholar]
- 19. Ghosal A., Bhowmick R., Nandy R. K., Ramamurthy T., Chatterjee N. S. 2007. PCR-based identification of common colonization factor antigens of enterotoxigenic Escherichia coli. J. Clin. Microbiol. 45:3068–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghosh S., LaPara T. M. 2007. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 1:191–203 [DOI] [PubMed] [Google Scholar]
- 21. Guler L., Guenduez K., Ok U. 2008. Virulence factors and antimicrobial susceptibility of Escherichia coli isolated from calves in Turkey. Zoonoses Public Health 55:249–257 [DOI] [PubMed] [Google Scholar]
- 22. Holden N. J., et al. 2006. Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiology 152:1143–1153 [DOI] [PubMed] [Google Scholar]
- 23. Johnson T. J., Siek K. E., Johnson S. J., Nolan L. K. 2005. DNA sequence and comparative genomics of pAPEC-O2-R, an avian pathogenic Escherichia coli transmissible R plasmid. Antimicrob. Agents Chemother. 49:4681–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kachroo B. B. 2001. Association between antibiotic resistance and the expression of Dr adhesin among uropathogenic Escherichia coli. Chemotherapy 47:97–103 [DOI] [PubMed] [Google Scholar]
- 25. Karami N., Hannoun C., Adlerberth I., Wold A. E. 2008. Colonization dynamics of ampicillin-resistant Escherichia coli in the infantile colonic microbiota. J. Antimicrob. Chemother. 62:703–708 [DOI] [PubMed] [Google Scholar]
- 26. Kaukas A., Hinton M., Linton A. H. 1988. The effect of growth-promoting antibiotics on the fecal enterococci of healthy young chickens. J. Appl. Microbiol. 64:57–64 [DOI] [PubMed] [Google Scholar]
- 27. Krometis L. A. H., et al. 2007. Intra-storm variability in microbial partitioning and microbial loading rates. Water Res. 41:506–516 [DOI] [PubMed] [Google Scholar]
- 28. Livrelli V., et al. 1996. Adhesive properties and antibiotic resistance of Klebsiella, Enterobacter, and Serratia clinical isolates involved in nosocomial infections. J. Clin. Microbiol. 34:1963–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopes L. M., et al. 2005. Heterogeneity among strains of diffusely adherent Escherichia coli isolated in Brazil. J. Clin. Microbiol. 43:1968–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luna G. M., et al. 2010. Extraintestinal Escherichia coli carrying virulence genes in coastal marine sediments. Appl. Environ. Microbiol. 76:5659–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lutterodt G., Basnet M., Foppen J. W. A., Uhlenbrook S. 2009. The effect of surface characteristics on the transport of multiple Escherichia coli isolates in large scale columns of quartz sand. Water Res. 43:595–604 [DOI] [PubMed] [Google Scholar]
- 32. Mitsumori K., Terai A., Yamamoto S., Yoshida O. 1998. Identification of S, F1C and three PapG fimbrial adhesins in uropathogenic Escherichia coli by polymerase chain reaction. FEMS Immunol. Med. Microbiol. 21:261–268 [DOI] [PubMed] [Google Scholar]
- 33. Moore J. E., et al. 2010. Determination of total antibiotic resistance in waterborne bacteria in rivers and streams in Northern Ireland: can antibiotic-resistant bacteria be an indicator of ecological change? Aquat. Ecol. 44:349–358 [Google Scholar]
- 34. Muirhead R. W., Collins R. P., Bremer P. J. 2005. Erosion and subsequent transport state of Escherichia coli from cowpats. Appl. Environ. Microbiol. 71:2875–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nowrouzian F., Adlerberth I., Wold P. E. 2001. P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol. Infect. 126:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Old D. C., Duguid J. P. 1970. Selective outgrowth of fimbriate bacteria in static liquid medium. J. Bacteriol. 103:447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Onan L. J., LaPara T. M. 2003. Tylosin-resistant bacteria cultivated from agricultural soil. FEMS Microbiol. Lett. 220:15–20 [DOI] [PubMed] [Google Scholar]
- 38. Pachepsky Y. A., et al. 2008. Strain-dependent variations in attachment of E. coli to soil particles of different sizes. Int. Agrophysics 22:61–66 [Google Scholar]
- 39. Pukall R., Tschape H., Smalla K. 1996. Monitoring the spread of broad host and narrow host range plasmids in soil microcosms. FEMS Microbiol. Ecol. 20:53–66 [Google Scholar]
- 40. Rendon M. A., et al. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U. S. A. 104:10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sapkota A. R., Curriero F. C., Gibson K. E., Schwab K. J. 2007. Antibiotic-resistant enterococci and fecal indicators in surface water and groundwater impacted by a concentrated swine feeding operation. Environ. Health Perspect. 115:1040–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simms A. N., Mobley H. L. T. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect. Immun. 76:4833–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soupir M. L., Mostaghimi S. 2011. Escherichia coli and enterococci attachment to particles in runoff from highly and sparsely vegetated grassland. Water Air Soil Pollut. 216:167–178 [Google Scholar]
- 44. Soupir M. L., Mostaghimi S., Dillaha T. 2010. Attachment of Escherichia coli and enterococci to particles in runoff. J. Environ. Qual. 39:1019–1027 [DOI] [PubMed] [Google Scholar]
- 45. Sutcliffe J. A. 2005. Improving on nature: antibiotics that target the ribosome. Curr. Opin. Microbiol. 8:534–542 [DOI] [PubMed] [Google Scholar]
- 46. USDA-APHIS. 2007. Antimicrobial resistance issues in animal agriculture. USDA, Washington, DC. [Google Scholar]
- 47. U.S. EPA. 2000. Improved enumeration methods for the recreational water quality indicators: enterococci and Escherichia coli. EPA/821/R-97/004. U.S. EPA, Office of Science and Technology, Washington, DC [Google Scholar]
- 48. U.S. EPA. 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). EPA 821-R-02-023. U.S. EPA, Office of Science and Technology, Washington DC [Google Scholar]
- 49. Xia Y., Gally D., Forsman-Semb K., Uhlin B. E. 2000. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 19:1450–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]