Abstract
A major hurdle in the application of therapeutic peptides is their rapid degradation by peptidases. Thioether bridges effectively protect therapeutic peptides against breakdown, thereby strongly increasing bioavailability, enabling oral and pulmonary delivery and potentially significantly optimizing the receptor interaction of selected variants. To efficiently select optimal variants, a library of DNA-coupled thioether-bridged peptides is highly desirable. Here, we present a unique cell surface display system of thioether-bridged peptides and successfully demonstrate highly selective screening. Peptides are posttranslationally modified by thioether bridge-installing enzymes in Lactococcus lactis, followed by export and sortase-mediated covalent coupling to the lactococcal cell wall. This allows the combinatorial optimization and selection of medically and economically highly important therapeutic peptides with strongly enhanced therapeutic potential.
INTRODUCTION
Surface display of peptides is a powerful technology for selecting peptides with desired properties from combinatorial libraries. A peptide display system comprises a scaffold—e.g., phage (39), bacterium (10), ribosome (25), or the yeast Saccharomyces cerevisiae (4)—for the displayed peptide and a link between the displayed peptide and the encoding DNA. Selection involves several rounds of target binding, washing, elution of specific binders, and then amplification of the selected peptides. Each round results in increasing enrichment of binders over the nonbinders. To date phage display is the best known display system.
Bacterial cell surface display has been developed for Gram-negative and Gram-positive bacteria (22). In the latter case, cell surface display relies on the translational fusion of a peptide to an LPXTG cell wall-anchoring motif such as that of the Lactococcus lactis PrtP protease (15, 38). This anchoring mechanism requires processing by a sortase for covalent anchoring of the peptide to the peptidoglycan of the bacterial cell wall (36). In this way the peptide and the encoding DNA are linked allowing selection, followed by rapid identification.
Various peptide display libraries have been designed, including linear peptides, disulfide-linked cyclic peptides (40), and chemically modified peptides (1), for instance, peptides coupled to an organic core (11) and peptides with nonnatural amino acids (8). This has resulted in the identification of various useful peptides, including therapeutically effective peptides (20). In nature, the majority of therapeutic (poly)peptides bears some form of posttranslational modification (PTM) which modulates the peptide's function (41). Combining display technology and true posttranslational modification of peptides, a system which to the best of our knowledge has not yet been reported, would be a powerful extension of current display possibilities.
A particularly important PTM which may have impact on therapeutic peptide development is the introduction of lanthionines and methyllanthionines, thioether-bridged amino acids, e.g., Ala-S-Ala and Abu-S-Ala, where Abu is amino butyric acid. (Methyl)lanthionines have recently been introduced chemically in translated peptides (37). (Methyl)lanthionine rings occur in special peptide antibiotics, so called lantibiotics (3, 6), and in lantipeptides (23). These PTMs are formed in the propeptide in two enzymatic steps which are induced by an N-terminal leader peptide. First, a dehydratase catalyzes the dehydration of serines and threonines to dehydroalanines and dehydrobutyrines, respectively. In a second step a cyclase couples the dehydroamino acids stereo- and regiospecifically to cysteines, yielding mesolanthionine and β-methyllanthionine. In these special residues a d-amino acid and an l-amino acid are linked via a thioether bridge. The combined presence of a d-amino acid and a thioether bridge strongly increases the resistance against peptidase action. The well-known lantibiotic nisin is produced by some Lactococcus lactis strains. Nisin biosynthesis involves the action of the dehydratase NisB and the cyclase NisC, resulting in two dehydroalanines, one dehydrobutyrine, one lanthionine, and four methyllanthionines. The fully modified prenisin is secreted into the medium via the dedicated transporter NisT.
Our laboratory and the van der Donk group have demonstrated that choosing the position of serines/threonines/cysteines allows the enzymatically controlled introduction of thioether bridges in a variety of leader peptide fusion peptides unrelated to lantibiotics (5, 27). In this way the position(s), number, and sizes of rings in a single peptide can be designed. This has significant impact on therapeutic peptide development since thioether links are more stable than peptide bonds and disulfide bridges. For example, the thioether-bridged form of the potentially highly important therapeutic peptide cyclic angiotensin-(1-7) [cAng-(1-7)] was fully resistant against degradation by angiotensin-converting enzyme (ACE), exhibited significantly more resistance against peptidases than its linear counterpart, and had 34-fold enhanced bioavailability (14). Both oral and pulmonary delivery could be used for cAng-(1-7) (7), and it had more effective receptor interaction (14). Modulated receptor interaction may involve enhanced receptor specificity and enhanced effectivity or both. For example, thioether-bridged somatostatin is more specific (28); thioether enkephalin is more specific and dramatically more effective (31).
Hence, exploiting the nisin biosynthesis enzymes opens the way for engineering and selection of novel bioactive peptides. To this end a selection/screening system is urgently needed. In this paper we are the first to demonstrate cell surface display of thioether- and dehydroamino acid-containing posttranslationally modified peptides. The L. lactis host organism provides the nisin biosynthesis and export machinery for introduction of the thioether linkages and a scaffold for the displayed peptide. As model peptides, nisin and angiotensin-(1-7) are translationally fused to an LPXTG cell wall-anchoring motif of the L. lactis PrtP protease. After sortase-mediated covalent attachment of the peptide to the bacterial cell wall, the peptide and its encoding DNA are linked, allowing selection/screening for thioether-bridged peptides with the desired properties. As an example, we generated a random thioether-bridged peptide display library and demonstrated the successful, exclusive selection of one thioether peptide using the model target protein streptavidin (Strep). Combining posttranslational modification of peptides with bacterial cell surface display provides a powerful tool for identifying stabilized peptides with optimal receptor interactions as well as entirely novel bioactive peptides with desired properties.
MATERIALS AND METHODS
Bacterial strains and plasmid vectors.
The host strain L. lactis NZ9000 was used for the expression of the modification enzymes and peptides. Strains and plasmids used are listed in Table 1. L. lactis was grown at 30°C in M17 broth (Oxoid) containing 0.5% (wt/vol) glucose (GM17) and, when necessary, supplemented with chloramphenicol (5 μg/ml) and/or erythromycin (5 μg/ml) for plasmid selection. For protein production, stationary-phase cultures were inoculated (20-fold diluted) on minimal medium and immediately induced with nisin or on M17 broth as described above and induced with nisin at an optical density at 600 nm (OD600) of about 0.4.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| L. lactis strains | ||
| NZ9700 | nisABTCIPRKEFG | 18 |
| NZ9000 | MG1363 derivative; nisRK+ | 19 |
| Plasmids | ||
| pNZnisA-E3 | pNZ8048 derivative, nisA, Emr | 16 |
| pIL3BTC | Pnis nisBTC, Cmr | 31 |
| pIL3BC | Pnis nisBC, Cmr | 17 |
| pILT | Pnis nisT, Cmr | Laboratory collection |
| pTB1 | pNZ8048 derivative, Pnis with the nisin prepeptide fused to IEGR-Strep tag-3C-PrtP cwa1789–1912a | This study |
| pTB2 | pNZ8048 derivative, Pnis plus the leader peptide and angiotensin-(1-7) fused to PrtP cwa1789–1912; Emr | This study |
| pTB3 | pNZ8048 derivative, Pnis plus the leader peptide and angiotensin-(1-7) fused to IEGR-Strep Tag-3C-PrtP cwa1789–1912; Emr | This study |
| pTB5 | pNZ8048 derivative, Pnis plus the nisin prepeptide fused to PrtP cwa1789–1912; Emr | This study |
| pTB5-tr1 | pTB5 derivative with the truncated cell wall anchor PrtP cwa1789–1905; Emr | This study |
| pTB5-tr2 | pTB5 derivative with the truncated cell wall anchor PrtP cwa1789–1876; Emr | This study |
| pTB5-tr3 | pTB5 derivative with the truncated cell wall anchor PrtP cwa1789–1831; Emr | This study |
| pTB5-tr4 | pTB5 derivative with the truncated cell wall anchor PrtP cwa1789–1811; Emr | This study |
| pRND-X3 | pTB5 derivative encoding a peptide library with an N-terminus consisting of leader-ISXXXCNMKTATCHCSIHVSK, in which the X are randomized amino acids | This study |
| pRND-X3-1 | pRND-X3-derivative encoding a peptide with an N-terminal sequence of leader-ISNMVCNMKTATCHCSIHVSK | This study |
| pTB9 | pRND-X3-1 derivative with a termination codon behind the encoding part for leader-ISNMVCNMKTATCHCSIHVSK | This study |
IEGR, the factor Xa cleavage site; PrtP cwa1789-1912, amino acids 1789 to 1912 of the cell wall anchor (cwa) of the L. lactis PrtP protease. Notation for truncated cell wall anchors follows this model.
General molecular biology.
Unless specified otherwise, all protein analysis and standard recombinant DNA techniques were performed as described previously (35) or as specified by the manufacturers. Enzymes and buffers were purchased from New England BioLabs, Finnzymes, or Roche. Electrotransformation of L. lactis was carried out as described previously (12) using a Bio-Rad Gene Pulser (Bio-Rad). Nucleotide sequence analyses were performed by BaseClear (Leiden, the Netherlands). Total protein extracts from cell cultures for SDS-PAGE were prepared as described by Piard et al. (30).
Library construction.
We constructed a random thioether-bridged peptide library of the form SerXXXCys (where X stands for any amino acid and where Ser-Cys form the thioether bridge after posttranslational dehydration of Ser and coupling of the formed dehydroalanine to Cys). A fusion PCR approach was used to engineer the randomized thioether ring with plasmid pTB5 as a template. A 5′PCR fragment was amplified with primers NisA.fw and RND.rev (see Table S1 in the supplemental material). The 3′ fragment was amplified with oligonucleotides anchor.fw and anchor.rev (see Table S1). Both amplification products were purified and fused by 30 cycles of primerless PCR. The assembled PCR product was purified and subjected to 15 cycles of PCR with the outside primers NisA.fw and Anchor.rev to allow cloning. The amplified full-length product was digested and ligated into plasmid pTB5 previously digested with BglII and HindIII, yielding pRND-X3. The ligation mixture was electroporated to L. lactis NZ9000(pIL3BTC) to generate a library of random thioether-bridged peptides on the lactococcal cell surface. The library size was estimated to be 11,000 independent clones based on extrapolation of the number of transformants in small-scale platings.
Library screening.
An aliquot of the RND-X3 library (≈108 cells) was inoculated in 50 ml of GM17 containing both erythromycin and chloramphenicol (GM17EmCm). The culture was grown at 30°C until an OD600 of about 0.4. At this point RND-X3 peptide production was induced by adding 1/1,000 volume of filtered (0.45-μm pore size) medium from an overnight-grown culture of the nisin-producing L. lactis NZ9700. Incubation was continued overnight. Library cells were collected by centrifugation, washed once with phosphate-buffered saline ([PBS] 50 mM NaH2PO4, 300 mM NaCl, pH 7.3), and resuspended in PBS until an OD600 of ≈1.4 to 1.5. To reduce the number of nonspecifically binding peptides from the initial library, a 2-ml library cell suspension was incubated for 30 min at room temperature with 200 μl of biotin-saturated streptavidin-coated magnetic beads (Dynabeads and MyOne Streptavidin T1; Invitrogen). The unbound cells in the supernatant were collected and resuspended in 10 ml of PBS plus 0.2% Tween 20 (PBST)–1% bovine serum albumin (BSA). For selection of streptavidin binders, 200 μl of streptavidin-coated magnetic beads was added and incubated under rotation at room temperature for 1 h. Streptavidin-binding cells were collected by magnetic-bead assisted cell sorting (MACS), washed 8-fold with 1 ml of PBST-BSA, resuspended in 50 ml of GM17EmCm, and grown overnight at 30°C. The next day the selected culture was inoculated 20-fold diluted into fresh GM17EmCm to repeat the selection procedure. After two rounds of MACS individual clones were picked for analysis.
Whole-cell enzyme-linked immunosorbent assay (ELISA) for detection of surface-displayed peptides.
Control L. lactis NZ9000 cells and L. lactis NZ9000 cells containing plasmid pIL3BTC and harboring the display vectors were grown with and without nisin for induction of peptide anchor production. After production, cells were collected by centrifugation, washed three times with PBS (58 mM Na2HPO4·2H2O, 17 mM Na2H2PO4·H2O, 68 mM NaCl, pH 7.4) and then resuspended to an OD600 of ≈20. Aliquots of 50 to 200 μl of these cell suspensions were incubated with a 1,000-fold diluted rabbit anti-nisin leader antibody solution in a final volume of 1 ml PBS plus 0.5% BSA at room temperature for 1 h under rotation. After three washes with PBS, the displayed peptide was visualized by incubation with alkaline phosphatase-conjugated goat anti-rabbit IgG (1:10000) and p-nitrophenyl phosphate (0.5 mg/ml) as substrate. The absorbance was determined at 410 nm after a suitable time period, which is a measure for the number of displayed peptides.
For detection of streptavidin binding, cells displaying selected peptides were incubated for 1 h at room temperature under rotation with 4,000-fold diluted horseradish peroxidase (HRP)-conjugated streptavidin (Pierce) in 1 ml of PBST-BSA (PBS plus 0.2% Tween 20 and 1% BSA). Cells were washed three times with PBST-BSA. Streptavidin binding peptide was visualized by the addition of 1 ml of 1× ABTS-H2O2 [0.1 mg/ml 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) in 0.05 M citric acid, pH 4.5, and 1% H2O2] horseradish peroxidase substrate solution. After a suitable period, absorbance readings were taken at 410 nm.
Antimicrobial assay.
A GM17 agar plate with an extensively washed SDS-polyacrylamide (PAA) gel or spots of TB5-producing L. lactis NZ9000 cells was covered with a 200-fold diluted sample of L. lactis MG1363 or NZ9000 strain in 0.5% top agar with 0.1 mg/ml trypsin. Trypsin is required for cleaving off the nisin leader, yielding active nisin. The agar plates were incubated overnight at 30°C.
Proteolytic removal of displayed peptide from the cell and peptide purification.
Lactococcal surface display vectors pTB1 and pTB3 were constructed to facilitate purification and analysis. These plasmids encoded a human rhinovirus 3C protease recognition sequence (PreScission protease; GE Healthcare) to release the N-terminal part encoding leader peptide, nisin, and Strep tag II from the lactococcal cell surface. The Strep tag II sequence was included for purification purposes.
L. lactis NZ9000 cells displaying TB1 or TB3 were subjected to 16% trichloroacetic acid (TCA) for 30 min on ice. Cells were washed twice with 1 ml of acetone, dried in a Speed Vac, and resuspended in 0.25 ml of 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1 mg of lysozyme, and 0.1 mg of mutanolysin. Cell wall digestion was performed at 37°C for 1 h. Cells were collected by centrifugation and washed twice with PBS. The cells were resuspended in 1 ml of PreScission protease cleavage buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]) with 40 U of PreScission protease (GE Healthcare) and incubated at 4°C for 24 to 48 h under rotation.
After protease digestion the supernatant was collected, and the streptavidin-tagged N-terminal parts of the TB1 and TB3 peptides were purified using a Strep-Tactin spin column purification kit (IBA, GmbH) according to the instructions of the manufacturer. Peptides in the eluate were precipitated with 10% TCA and dissolved in water.
Mass spectrometry.
Mass spectra were recorded with a Voyager DE PRO matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer in the linear mode.
Western analysis.
In immunoblots, leader peptide fusion peptides were detected with rabbit polyclonal anti-leader peptide antibodies (16). Leader-Ang-(1-7) fusion peptides were detected with mouse anti-cAng-(1-7) polyclonal antibodies. Alkaline phosphatase-conjugated goat anti-rabbit IgG with nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) as a substrate solution (Sigma) was used for detection.
Antibodies.
Polyclonal antibodies against cAng-(1-7) and natural Ang-(1-7) [nAng-(1-7)] were raised in mice. Keyhole limpet hemocyanin (KLH) was coupled to the N terminus of cAng-(1-7) and nAng-(1-7). Mice were three times intraperitoneally immunized with 25 μg of angiotensin peptide conjugated to KLH. For boosting, complete Freund's adjuvant was included in the first immunization while Freund's incomplete adjuvant was used for the second and third immunizations.
RESULTS
Lactococcal surface display vector for thioether peptides.
The encoding DNA sequence of the 124-amino-acid LPXTG cell wall-anchoring sequence of the L. lactis PrtP protease was translationally fused by PCR to the 3′ end of the nisA gene in pNZnisA-E3, yielding the display vector pTB5. The display vector comprises the sequences for the inducible NisA promoter, nisin leader peptide, nisin, the LPXTG cell wall-anchoring motif of the L. lactis PrtP protease, and the erythromycin resistance marker (Fig. 1). The display vector was electroporated to L. lactis NZ9000 containing pIL3BTC, which provides the nisin biosynthesis enzymes NisB and NisC and the transporter NisT (33) (Table 1).
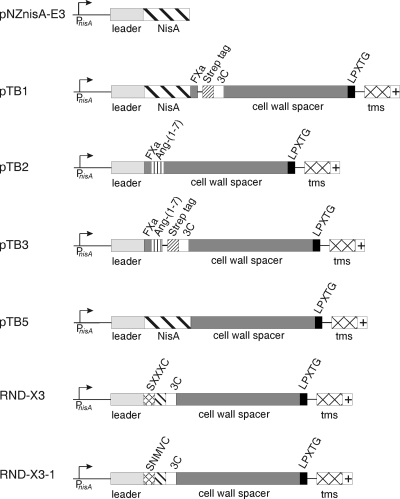
Fig. 1.
Genetic organization of the display vectors used in this study. PnisA, nisin-inducible promoter; leader, coding sequence for the nisin leader peptide; nisA, coding sequence pronisin; FXa, codons for factor Xa recognition site; SXXXC, sequence containing codons for randomized thioether ring, where X stands for any amino acid; Strep tag, codons for streptavidin recognition sequence; 3C, codons for human rhinovirus protease recognition sequence; cell wall spacer and cell wall anchor, codons for the cell wall spacer and cell wall anchor of L. lactis PrtP protease (amino acids 1789 to 1912) (13, 15); LPXTG motif, codons for the sortase recognition sequence; crossed box (tms), codons for the hydrophobic membrane-spanning sequence; +, codons for the charged tail (sequence, KRKQREE).
Surface display of nisin.
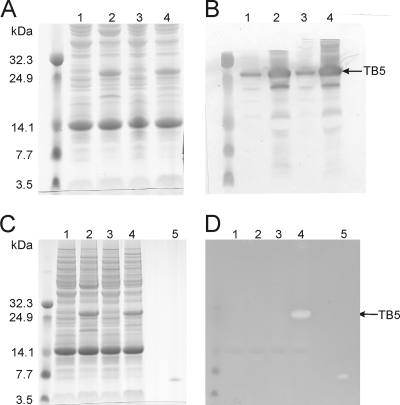
First, we verified whether intact TB5 was expressed. L. lactis NZ9000 cells with plasmid pTB5 and with or without pIL3BTC were grown in the absence or presence of inducing nisin to allow production of TB5. Proteins in total cell extracts were separated by SDS-PAGE and visualized by Coomassie staining (Fig. 2A). Comparing cell extracts from induced cultures with those from uninduced cultures showed the significantly higher presence in the induced culture of a protein band migrating at 28 kDa. Western blot analysis with anti-nisin leader antibodies identified this protein as the nisin anchor fusion protein TB5 as seen by the strong immunoreactive signal (Fig. 2B). The observed molecular mass of TB5 was higher than the theoretical molecular mass of 18 kDa, indicating covalent attachment of peptidoglycan fragments to TB5. All together, lactococcal cells containing the constructed display vector, pTB5, clearly directed the production of a significant amount of TB5 when cells were induced with nisin.
Fig. 2.
Prenisin in TB5 is modified by both the dehydratase and cyclase. Coomassie-stained 12% SDS-PAA gel (A and C) and Western blot analysis with anti-nisin leader peptide antibodies (B) or overlay with nisin-sensitive L. lactis NZ9000 strain with 0.1 mg/ml trypsin (D) on total protein extracts of L. lactis NZ9000 with the following plasmids: lanes 1, pTB5 uninduced; lanes 2, pTB5 induced; lanes 3, pTB5 and pIL3BTC, uninduced; lanes 4, pTB5 and pIL3BTC, induced; lanes 5, supernatant from NZ9000 with pIL3BTC and pNZnisA-E3. The halo in lane 4 of panel D at the site of the induction-dependent protein band proves that the prenisin part of the fusion protein is modified by NisB and NisC.
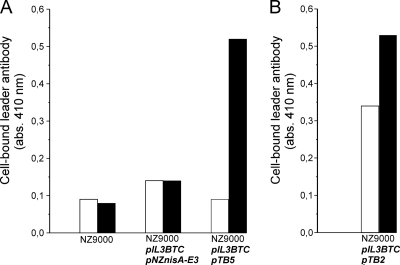
Next, lactococcal cell surface display of TB5 was evaluated with a whole-cell ELISA using anti-nisin leader antibodies. The results summarized in Fig. 3A showed a positive color response for the induced L. lactis NZ9000 with plasmid pTB5 and with pIL3BTC, whereas only a background color response was seen for the control cultures without pTB5. Furthermore, prenisin lacking the cell wall anchor motif [Fig. 3A, NZ9000(pIL3BTC/pNZnisA-E3)] did not remain attached to the lactococcal cells. Hence, the anchor moiety of TB5 provided the signals for covalent attachment of TB5 to the peptidoglycan layer, thereby displaying prenisin on the lactococcal cell surface.
Fig. 3.
Cell surface location of the N terminus of the fusion peptide construct. Whole-cell ELISA on L. lactis NZ9000 cells for detection of surface-displayed prenisin anchor fusion protein (A) and angiotensin-(1-7) anchor fusion protein (B). Rabbit anti-nisin leader antibodies were allowed to bind to lactococcal cells displaying TB5 or TB2. Alkaline phosphatase-conjugated goat anti-rabbit IgG was added, and a color was generated by the addition of p-nitrophenylphosphate. White bars, uninduced cells; black bars, induced cells. A typical experiment is depicted. The experiment was repeated in more than three independent replicates with differences of ≤15%.
The surface-displayed nisin is posttranslationally modified.
Is the prenisin part of TB5 modified by the dehydration and cyclization enzymes NisB and NisC? Modification of prenisin within TB5 was evaluated with an antimicrobial activity assay. In this assay nisin or derivatives of it, such as TB5, are covered with a nisin-sensitive L. lactis strain in the presence of trypsin. Trypsin can liberate nisin by removing the leader peptide, which ends with an arginine, and by cleaving after nisin's C-terminal lysine 34. Growth inhibition of the nisin-sensitive L. lactis strain, visible as a clearing zone or halo, indicated modification of nisin since nisin exhibits antimicrobial activity when at least the first three N-terminal rings are formed and the nisin leader peptide is removed (34) (Fig. 2D, lane 5). Only the TB5 produced in the presence of the modification enzymes NisB and NisC inhibited growth of the indicator strain, seen as a conspicuous clearing zone (halo) in the trypsin-containing overlay (Fig. 2D, lane 4), whereas no halo was found with the TB5 produced in the absence of these enzymes (Fig. 2D, lane 2). Taken together, these data demonstrated that NisB and NisC modified the prenisin part within TB5 such that nisin or an active nisin variant could be released from TB5 by trypsin.
When whole cells that express TB5 were overlaid, results were obtained that are fully consistent with the above results (see Fig. S1 in the supplemental material). Antimicrobial activity was found for TB5-producing lactococcal cells which contained modification and transport enzymes. Since the nisin leader is accessible only to trypsin (see Fig. S1) from the outside, active nisin is displayed on the lactococcal cell surface.
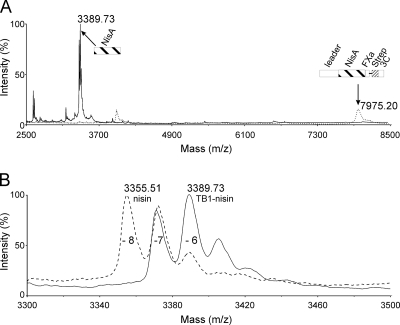
To facilitate purification and analysis, a Strep tag II sequence and a human rhinovirus 3C protease recognition sequence were inserted into plasmid pTB5, yielding display vector pTB1 (Fig. 1). MALDI-TOF spectrometry of the purified N-terminal part of TB1 was executed to analyze modification of the nisin part within TB1. The results summarized in Fig. 4 clearly and consistently demonstrate posttranslational modification. The MALDI-TOF spectrum in Fig. 4A (dotted line) showed the highest mass peak around 7,975 Da, which corresponds to the full-length N-terminal TB1 part, approximately 6-fold dehydrated, as schematically indicated in the Fig. 4A. This peak disappeared when the sample was treated with trypsin, while the nisin mass peak emerged around 3,389 Da (Fig. 4A, solid line). Analysis of the nisin mass peak from TB1 showed the 7- and 6-fold dehydration mass peaks, whereas the 8-fold dehydration peak is lacking (Fig. 4B, solid line). The last mass peak is typically found in the spectrum of secreted nisin (Fig. 4B, interrupted line). In conclusion, the data convincingly demonstrate that the cell surface-exposed prenisin is 7- to 6-fold dehydrated.
Fig. 4.
Cell surface-displayed prenisin is 6- to 7-fold dehydrated. MALDI-TOF spectrometry of nisin anchor fusion peptide TB1. (A) Purified TB1 (dotted line)and purified TB1 treated with trypsin (solid line) are shown. (B) Nisin part of TB1 treated with trypsin (solid line) and nisin from the supernatant of L. lactis NZ9000 containing pIL3BTC and pNZnisA-E3 treated with trypsin (dotted line) are shown. The numbers of dehydrations are indicated by −8, −7, −6, etc. The relevant peptide fragment is schematically shown.
Taken together, NisB and NisC modified the nisin anchor fusion protein TB5 such that after liberation with trypsin, active nisin was observed with a nisin-sensitive strain in an overlay on an SDS-PAA gel and on whole cells. Furthermore, MALDI-TOF analysis demonstrated that nisin within TB1 was 7- to 6-fold dehydrated.
Surface display and modification of angiotensin-(1-7).
The nisin coding sequence in plasmid pTB5 was replaced by a DNA sequence encoding a factor Xa recognition site and angiotensin-(1-7) (DRVSIHC), yielding surface display vector pTB2 (Fig. 1). Induced cells, with pIL3BTC, expressing TB2 generated a positive color response when incubated with anti-leader peptide antibodies in a whole-cell ELISA (Fig. 3B). This demonstrated that the leader peptide was accessible from the outside, and thus angiotensin-(1-7) was displayed on the lactococcal cell surface.
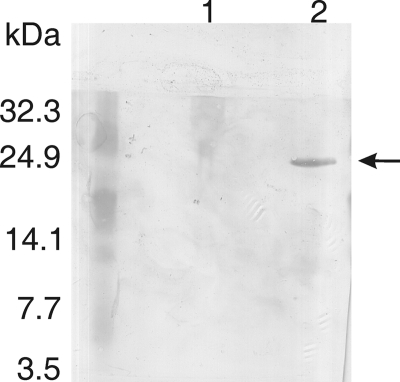
To determine whether angiotensin-(1-7) within TB2 was enzymatically modified, Western blot analysis was performed using antibodies specific for the cyclized analog of angiotensin-(1-7) (data not shown). A Coomassie-stained SDS-PAA gel with cell extracts of L. lactis NZ9000(pTB2) with or without pIL3BTC showed that the angiotensin-(1-7) anchor peptide TB2 was equally well produced in both strains (data not shown). Western analysis on these cell extracts showed that in the presence of modification enzymes, TB2 generated an immunoreactive signal with anti-cyclic-angiotensin-(1-7) antibodies (Fig. 5, lane 2), whereas no signal was found with nonmodified TB2 (Fig. 5, lane 1), indicating that the displayed angiotensin-(1-7) was cyclized.
Fig. 5.
Cell surface-displayed angiotensin-(1-7) is thioether bridged. Western blot analysis with specific anti-cyclic-angiotensin-(1-7) antibodies on total protein extracts separated on a 12% SDS-PAA gel of L. lactis NZ9000 with plasmids pTB2 (lane 1) and pTB2 and pIL3BTC (lane 2).
We subsequently verified modification of the displayed angiotensin-(1-7) by MALDI-TOF analysis. To that end we designed plasmid pTB3 encoding leader peptide, factor Xa recognition sequence, angiotensin-(1-7), Strep tag II sequence, and a human rhinovirus 3C protease recognition sequence to facilitate purification and analysis (Fig. 1). A clear mass peak of 5,398.8 Da was found which corresponded to the singly dehydrated N-terminal part of TB3 and matched the calculated mass of 5,400 Da (see Fig. S2 in the supplemental material). Furthermore, no addition of 1-cyano-4-dimethylaminopyridinium tetrafluoroborate (CDAP) to TB3 was observed, which means the absence of a free cysteine residue. This proves the involvement of the cysteine in thioether bridge formation (see Fig. S3). These results clearly corroborated modification of the angiotensin-(1-7)-containing N-terminal part of TB3. Taken together, these results obtained by an antibody specific for cAng-(1-7) and by mass spectrometry clearly demonstrate that thioether-bridged angiotensin-(1-7) is displayed on the cell surface of L. lactis.
Thioether-bridged peptide library screening.
The described display system gives us the unique possibility of designing and selecting combinatorial thioether-bridged peptide libraries. To demonstrate this concept, we constructed a lactococcal thioether peptide display library, termed RND-X3 (Fig. 1; Table 1, pRND-X3). The sequence of the displayed peptide was such that it favors enzymatic thioether ring formation (33). Three randomized amino acids were flanked by a serine and a cysteine, which allows enzyme-catalyzed dehydration of the serine and cyclase-catalyzed coupling of the resulting dehydroalanine to the cysteine. The use of NNS codons for library construction allows all possible amino acids at each position between the serine and the cysteine, yielding a library with a theoretical protein diversity of 8,000 variants. The estimated library size was about 11,000 independent clones. Relatively small libraries, when well designed, can yield interesting and valuable lead sequences (24).
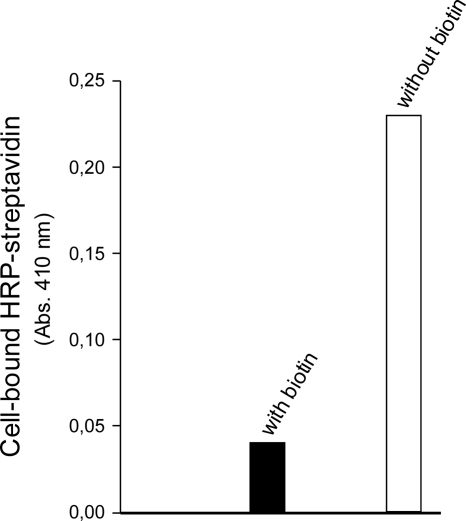
We screened the library for streptavidin binders. After two rounds of magnetic selection (MACS) using streptavidin-coated magnetic beads, selected bacteria were plated for analysis. Analysis of the selected clones exclusively yielded one single sequence, S-MNV-C. The absence of sequence variation within the analyzed selected peptides is likely due to the combination of the conformational constraints imposed by the thioether bridge and the requirements for streptavidin binding. Limited sequence variation was also observed within leads from cyclic peptide phage libraries with small ring sizes (9, 26). Lactococcal cells displaying this MNV-containing polypeptide generated a positive color response in a whole-cell ELISA, proving binding to streptavidin. Furthermore, in the presence of biotin, binding was abolished, demonstrating the specificity for the biotin binding site (Fig. 6).
Fig. 6.
Display of the selected streptavidin binding peptide RND-X3-1. RND-X3-1 is described in Table 1. Whole-cell ELISA on L. lactis NZ9000(pIL3BTC) displaying RND-X3-1 with or without coincubation with biotin. The displayed peptide was detected with HRP-conjugated streptavidin with ABTS-H2O2 as a substrate solution. Black bar, RND-X3-1-displaying cells with coincubation of 50 μM biotin; open bar, RND-X3-1-displaying cells. The experiment was repeated in more than three independent replicates with differences of ≤15%.
Analyses with MALDI-TOF revealed that CDAP did not add to the MNV-containing peptide, which precludes the presence of an unmodified cysteine, thus proving that a thioether bridge was present (see Fig. S4 in the supplemental material). Strikingly, the identical MNV sequence, constrained by a disulfide bond, was selected on streptavidin from an Escherichia coli-based 15-amino-acid random peptide display library (2). Hence, a thioether-bridged peptide with affinity for streptavidin can be most convincingly selected from the completely novel lactococcal thioether peptide display library reported here. In conclusion, we clearly demonstrate that a highly innovative lactococcal library of cell surface-displayed thioether-bridged peptides can be successfully screened for a specific ligand.
DISCUSSION
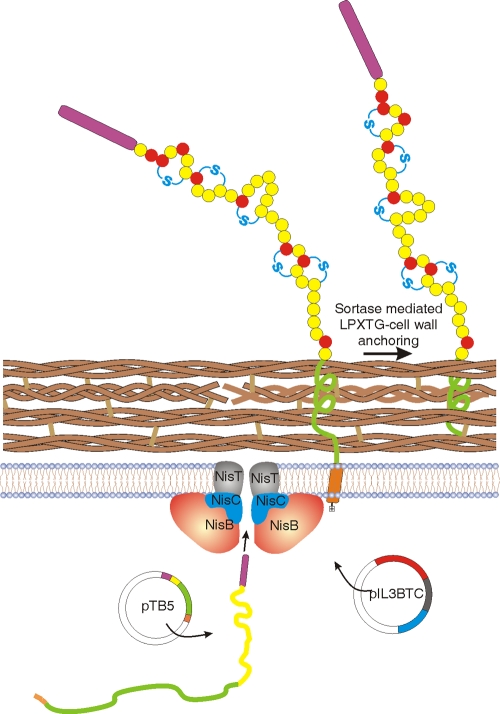
Here, we present a bacterial surface display system (Fig. 7) which exposes thioether-bridged peptides. By the maintained link between the thioether-bridged peptide, which is covalently coupled to the cell wall of the producing bacteria, and the encoding DNA, receptor-binding peptides selected from a library can be readily identified. We demonstrated this concept by constructing a thioether-bridged peptide library and screening for streptavidin binding peptides. Two rounds of magnetic selection were sufficient to select a thioether-bridged peptide with affinity for streptavidin.
Fig. 7.
Cartoon of bacterial cell surface display of thioether-bridged prenisin. Plasmid pIL3BTC codes for the modification enzymes NisB (dehydratase; red) and NisC (cyclase; blue) and the transporter NisT (black). The display vector pTB5 encodes the prepeptide (purple, leader;; yellow, peptide) translationally fused to the LPXTG-containing cell wall anchor (green). The prepeptide anchor fusion is synthesized in the cytoplasm. The N-terminal leader peptide (purple) is recognized by the modification enzymes, allowing modification of the propeptide; NisB-dehydrated amino acids are shown in red; NisC-cyclized thioether bridges are shown in blue. The modified prepeptide anchor fusion is translocated over the cell membrane, retained within the cell membrane via its C-terminal hydrophobic membrane anchor (orange) and charged tail (colorless), after which sortase-mediated covalent attachment of the cell wall anchor via its LPXTG motif to the peptidoglycan takes place.
By imposing a conformational constraint, thioether bridges in peptides strongly confer proteolytic resistance and may modulate receptor interaction. By introducing thioether bridges in peptides, the receptor interaction may become much more specific and strongly enhanced, but it may also be completely lost, making the utility of the display and screening system presented here obvious. In addition to the presented example, other formats can be envisioned. For example, this system allows the efficient screening of diverse libraries, as witnessed by the surface display of thioether-bridged peptides as different as the pentacyclic nisin and the entirely unrelated monocyclic angiotensin-(1-7) which are extracellularly accessible. Furthermore, it allows the combinatorial optimization of the position(s) of thioether bridges in a specific selected peptide with respect to receptor interaction. In a second step a specific position(s) may be (semi)randomized to further fine-tune the receptor interaction. We are currently exploring these and other possibilities.
Several therapeutic peptides are composed of a distinct receptor-binding domain and a signal-transducing domain, including commercially interesting ligands of class B G protein-coupled receptor (GPCRs) (29). Rapid deactivation of these ligands by endogenous protease hinders therapeutic administration. Hence, an improved receptor-binding domain can be combined with the signal-transducing domain, resulting in an improved agonist. Alternatively, the signal-transducing domain can be omitted, and improved receptor binding can be exploited for developing a strong antagonist. Radiolabeled thioether-stabilized specific receptor-binding peptides are excellent diagnostic and potentially radiotherapeutic tools (21).
The longest peptide reported to be translocated across the membrane by NisT was 70 amino acids long, composed of prenisin extended with nisin-(1-14) (32). Here, we measured the surprising membrane translocation of the 203-amino-acid-long nisin anchor peptide TB3 by NisT. Supplementary studies indicated a role of the C-terminal transmembrane segment and positively charged sequence in the stability of this polypeptide (the complete membrane anchor is needed for stable display of NisB- and NisC-modified peptide [see supplemental data and Fig. S5 and S6 in the supplemental material]).
The surface-displayed nisin in the present study was dehydrated 6- and 7-fold instead of the normal 8-fold. Likely, Ser33 lacks dehydration. This residue already escapes dehydration in 10% of the cases in wild-type nisin A. Furthermore, serines are less readily dehydrated than threonines (32, 33). The antimicrobial activity of the displayed nisin proves thioether bridge formation and at least the presence of nisin rings A, B, and C. Apparently, the relatively long sequence extension at the C terminus of prenisin has little effect on the activity of the NisB dehydratase and NisC cyclase. The 7-fold dehydration proves that at least two dehydroamino acids are present in the displayed nisin. Display and screening of peptides that contain dehydroalanine and dehydrobutyrine may also lead to the selection of important dehydroamino acid-containing peptides. These amino acids are special by their planar shape and may be essential for bioactivity of peptides (32). Furthermore, dehydroamino acids are reactive Michael acceptors, allowing specific coupling of other molecules (13). The unique display and screening system presented here will also be a powerful tool for the discovery of entirely new, stable, and specific ligands which have no similarity to existing bioactive peptides.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Angelini A., Heinis C. 2011. Post-translational modification of genetically encoded polypeptide libraries. Curr. Opin. Chem. Biol. 15:355–361 [DOI] [PubMed] [Google Scholar]
- 2. Bessette P. H., Rice J. J., Daugherty P. S. 2004. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng. Des. Sel. 17:731–739 [DOI] [PubMed] [Google Scholar]
- 3. Bierbaum G., Sahl H.-G. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2–18 [DOI] [PubMed] [Google Scholar]
- 4. Boder E. T., Wittrup K. D. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553–557 [DOI] [PubMed] [Google Scholar]
- 5. Chatterjee C., Patton G. C., Cooper L., Paul M., A. van der Donk W. 2006. Engineering dehydro amino acids and thioethers into peptides using lacticin 481 synthetase. Chem. Biol. 13:1109–1117 [DOI] [PubMed] [Google Scholar]
- 6. Chatterjee C., Paul M., Xie L., A. van der Donk W. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633–684 [DOI] [PubMed] [Google Scholar]
- 7. de Vries L., et al. 2010. Oral and pulmonary delivery of thioether-bridged angiotensin-(1-7). Peptides 31:893–898 [DOI] [PubMed] [Google Scholar]
- 8. Dwyer M. A., Lu W., Dwyer J. J., Kossiakoff A. A. 2000. Biosynthetic phage display: a novel protein engineering tool combining chemical and genetic diversity. Chem. Biol. 7:263–274 [DOI] [PubMed] [Google Scholar]
- 9. Giebel L. B., et al. 1995. Screening of cyclic peptide phage libraries identified ligands that bind streptavidin with high affinities. Biochemistry 34:15430–15435 [DOI] [PubMed] [Google Scholar]
- 10. Hansson M., Samuelson P., Gunneriusson E., Ståhl S. 2001. Surface display on gram positive bacteria. Comb. Chem. High Throughput Screen. 4:171–184 [DOI] [PubMed] [Google Scholar]
- 11. Heinis C., Rutherford T., Freund S., Winter G. 2009. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 5:502–507 [DOI] [PubMed] [Google Scholar]
- 12. Holo H., Nes I. F. 1995. Transformation of lactococcus by electroporation. Methods Mol. Biol. 47:195–199 [DOI] [PubMed] [Google Scholar]
- 13. Humphrey J. M., Chamberlin R. 1997. Chemical synthesis for natural product peptides: coupling methods for the incorporation of noncoded amino acids into peptides. Chem. Rev. 97:2243–2266 [DOI] [PubMed] [Google Scholar]
- 14. Kluskens L. D., et al. 2009. Angiotensin-(1-7) with thioether bridge: an angiotensin-converting enzyme-resistant, potent angiotensin-(1-7) analog. J. Pharmacol. Exp. Ther. 328:849–854 [DOI] [PubMed] [Google Scholar]
- 15. Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. 1988. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl. Environ. Microbiol. 54:231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuipers A., et al. 2004. NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with nonlantibiotic peptides. J. Biol. Chem. 279:22176–22182 [DOI] [PubMed] [Google Scholar]
- 17. Kuipers A., et al. 2006. Sec-mediated transport of posttranslationally dehydrated peptides in Lactococcus lactis. Appl. Environ. Microbiol. 72:7626–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuipers O. P., Beerthuyzen M. M., Siezen R. J., de Vos W. M. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactisRequirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281–291 [DOI] [PubMed] [Google Scholar]
- 19. Kuipers O. P., de Ruyter P. G., Kleerebezem M., de Vos W. M. 1997. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 15:135–140 [DOI] [PubMed] [Google Scholar]
- 20. Ladner R. C., Sato A. K., Gorzelany J., de Souza M. 2004. Phage display-derived peptides as therapeutic alternatives to antibodies. Drug Discov. Today 9:525–529 [DOI] [PubMed] [Google Scholar]
- 21. Lee S., Xie J., Chen X. 2010. Peptide-based probes for targeted molecular imaging. Biochemistry 49:1364–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee Y., Choi J. H., Xu Z. 2003. Microbial cell-surface display. Trends Biotechnol. 21:45–52 [DOI] [PubMed] [Google Scholar]
- 23. Li B., et al. 2010. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 107:10430–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mao H., et al. 2010. Spatially addressed combinatorial protein libraries for recombinant antibody discovery and optimization. Nat. Biotechnol. 28:1195–1202 [DOI] [PubMed] [Google Scholar]
- 25. Mattheakis L. C., Bhatt R. R., Dower W. J. 1994. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc. Natl. Acad. Sci. U. S. A. 91:9022–9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McLafferty M. A., Kent R. B., Ladner R. C., Markland W. 1993. M13 bacteriophage displaying disulfide-constrained microproteins. Gene 128:29–36 [DOI] [PubMed] [Google Scholar]
- 27. Moll G. N., Kuipers A., Rink R. 2010. Microbial engineering of dehydro-amino acids and lanthionines in non-lantibiotic peptides. Antonie Van Leeuwenhoek 97:319–333 [DOI] [PubMed] [Google Scholar]
- 28. Ösapay G., et al. 1997. Lanthionine somatostatin analogs: synthesis, characterization, biological activity and enzymatic studies. J. Med. Chem. 40:2241–2251 [DOI] [PubMed] [Google Scholar]
- 29. Parthier C., Reedtz-Runge S., Rudolph R., Stubbs M. T. 2009. Passing the baton in class B GPCRs: peptide hormone activation via helix induction? Trends Biochem. Sci. 34:303–310 [DOI] [PubMed] [Google Scholar]
- 30. Piard J. C., et al. 1997. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J. Bacteriol. 179:3068–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rew Y., et al. 2002. Synthesis and biological activities of cyclic lanthionine enkephalin analogues: delta-opioid receptor selective ligands. J. Med. Chem. 45:3746–3754 [DOI] [PubMed] [Google Scholar]
- 32. Rink R., et al. 2007. Production of dehydroamino acid-containing peptides by Lactococcus lactis. Appl. Environ. Microbiol. 73:1792–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rink R., et al. 2005. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry 44:8873–8882 [DOI] [PubMed] [Google Scholar]
- 34. Rink R., et al. 2007. Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C-terminal truncation. Appl. Environ. Microbiol. 73:5809–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Schneewind O., Mihaylova-Petkov D., Model P. 1993. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 12:4803–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seebeck F. P., Ricardo A., Szostak J. W. 7 June 2011, posting date. Artificial lantipeptides from in vitro translations. Chem. Commun. (Camb.) doi:10.1039/c0cc05663d. [DOI] [PubMed]
- 38. Siezen R. J. 1999. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Van Leeuwenhoek 76:139–155 [PubMed] [Google Scholar]
- 39. Smith G. P. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317 [DOI] [PubMed] [Google Scholar]
- 40. Smith G. P., Petrenko V. A. 1997. Phage display. Chem. Rev. 97:391–410 [DOI] [PubMed] [Google Scholar]
- 41. Walsh G., Jefferis R. 2006. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24:1241–1252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.