Abstract
Methane release from seafloor sediments is moderated, in part, by the anaerobic oxidation of methane (AOM) performed by consortia of archaea and bacteria. These consortia occur as isolated cells and aggregates within the sulfate-methane transition (SMT) of diffusion and seep-dominant environments. Here we report on a new SMT setting where the AOM consortium occurs as macroscopic pink to orange biofilms within subseafloor fractures. Biofilm samples recovered from the Indian and northeast Pacific Oceans had a cellular abundance of 107 to 108 cells cm−3. This cell density is 2 to 3 orders of magnitude greater than that in the surrounding sediments. Sequencing of bacterial 16S rRNA genes indicated that the bacterial component is dominated by Deltaproteobacteria, candidate division WS3, and Chloroflexi, representing 46%, 15%, and 10% of clones, respectively. In addition, major archaeal taxa found in the biofilm were related to the ANME-1 clade, Thermoplasmatales, and Desulfurococcales, representing 73%, 11%, and 10% of archaeal clones, respectively. The sequences of all major taxa were similar to sequences previously reported from cold seep environments. PhyloChip microarray analysis detected all bacterial phyla identified by the clone library plus an additional 44 phyla. However, sequencing detected more archaea than the PhyloChip within the phyla of Methanosarcinales and Desulfurococcales. The stable carbon isotope composition of the biofilm from the SMT (−35 to −43‰) suggests that the production of the biofilm is associated with AOM. These biofilms are a novel, but apparently widespread, aggregation of cells represented by the ANME-1 clade that occur in methane-rich marine sediments.
INTRODUCTION
Marine sediments contain an estimated 500 to 10,000 Gt (1 Gt = 1015 g) of methane carbon, primarily in the form of gas hydrate (26). In the past, this reservoir may have been a significant greenhouse gas source to the atmosphere (8). Presently, however, oceanic methane contributions (including that from marine gas hydrate) account for <2% of the global atmospheric methane flux (39), with most sedimentary methane being microbially oxidized to carbon dioxide in anoxic sediments by the anaerobic oxidation of methane (AOM). Globally, AOM consumes ∼90% of the methane produced in marine sediments (39).
Environmental genomic and stable isotope studies have linked methane-oxidizing archaea (clades ANME-1, -2, and -3) to AOM (5, 32). These anaerobic methanotrophs often form aggregates with sulfate-reducing Deltaproteobacteria, where they are believed to symbiotically reduce sulfate and oxidize methane by the net reaction SO42− + CH4 → HS− + HCO3− + H2O.
AOM consortia depend on sulfate and methane and thus typically occur at the sulfate-methane transition (SMT), a biogeochemical horizon where seawater sulfate and methane from the underlying anoxic sediments converge. SMT environments are classified as diffusion or seep-dominant environments (1). Diffusion-dominant SMTs are well-defined, occur as deep as several meters below the surface, and have moderate methane concentrations (∼1 mM) and reduced sulfate concentrations (2). In these environments, AOM aggregates consist of fewer than 106 cells cm−3 and consume methane at a rate of 0.001 to 1 nmol cm−3 day−1 (29, 46). In contrast to diffusion-dominant environments, advective seep-dominant environments, such as the Hydrate Ridge offshore of Oregon and the Black Sea, have near-surface SMTs. The advective flow of methane in these environments often overwhelms the AOM biofilter. For example, anaerobic processes consume only 37% of methane from the Haakon-Mosby mud volcano (30), leaving a large proportion of the methane to escape into the overlying water. Seep-dominant environments contain ample sulfate and methane that support biofilms or mats with cell densities of the AOM aggregates that exceed 1010 cells cm−3 (20). AOM occurs in both diffusion and seep-dominant SMT environments; however, the microbial mechanisms and processes may be different for each environment (1).
Within each SMT environment, gradients of sulfate and methane are the controlling factors for the types of anaerobic methanotrophs present. For example, members of the ANME-1 clade are typically found in areas with high methane and limited sulfate concentrations (12). Examples include a methane-rich brine pool in the Gulf of Mexico (23) and microbial mats in the Black Sea (20, 40). In contrast, members of the ANME-2 and ANME-3 clades are found in areas with more sulfate and lower methane concentrations, such as in the top 10 cm of sediments at cold seeps (19).
We have found macroscopic biofilms from subseafloor fractures intersecting SMTs at depths of between 0.5 and 19 m below the seafloor (mbsf) in gas hydrate-bearing continental margins of the northeastern Pacific Ocean and the Indian Ocean. The objective of this study was to determine the cell concentrations, phylogenetic relationships, and the isotopic composition of the microbes from this novel fracture-dominant SMT environment. These objectives were achieved by sampling visible biofilm from fractures in split sediment cores for molecular analysis, stable isotope analysis, or epifluorescent microscopy.
MATERIALS AND METHODS
Site descriptions.
Macroscopic pink to orange subseafloor biofilms (typically 0.5 cm3 per occurrence) were collected from sediment fractures that were within or near the SMT in three marine methane-bearing settings: Hydrate Ridge (offshore of Oregon), northern Cascadia Margin (NCM; offshore of Vancouver Island, Canada), and the Indian Ocean (offshore of India) (Fig. 1; Table 1).
Fig. 1.
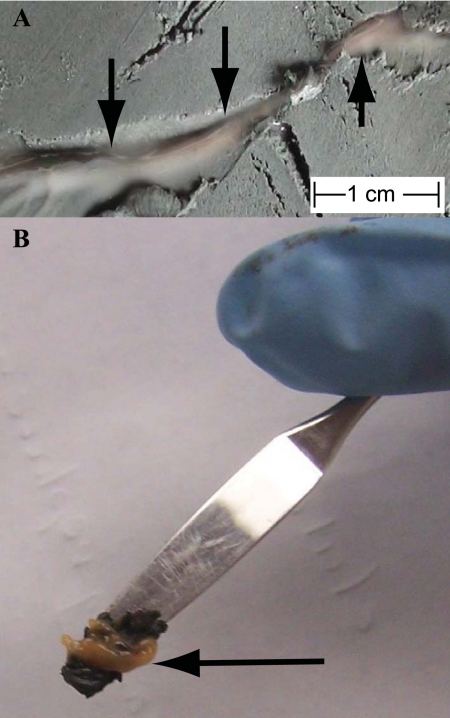
Photograph of a biofilm collected from a fracture in the Indian Ocean at 19 m below the seafloor (Table 1). Arrows point to the orange biomass. (A) Biofilm within the fracture before sampling; (B) biofilm that has been scraped out of the fracture using a spatula. The spatula blade is approximately 2 cm long.
Table 1.
Summary of biofilms examined in this study, including the analyses that were performed on each sample
| Location | Site | Analysisa | Latitude | Longitude | Biofilm depth (mbsf) | SMT (mbsf) |
|---|---|---|---|---|---|---|
| Northern Cascadia Margin | 1325B | Isotopic | 48°39.150′N | 126°59.650′W | 1.35 | 1.4 |
| 1327D | Isotopic | 48°42.484′N | 126°51.367′W | 7.2 | 7.6 | |
| Stn06 | CL, tR | 48°40.194′N | 126°50.945′W | 3.84 | 5.02 | |
| Stn18 | tR | 48°39.690′N | 126°55.244′W | 2.87 | 2.85 | |
| Stn19 | PC, CL, tR | 48°39.640′N | 126°55.105′W | 2.15 | 2.29 | |
| Hydrate Ridge | 1251A | Microscopic | 44°34.219′N | 125°4.452′W | 0.47 | 0.5 |
| Indian Ocean | 20A | CL, tR | 15°48.567′N | 81°50.576′E | 18 | 16.5 |
| 18A | PC, CL, tR | 19°09.145′N | 85°46.375′E | 19.8 | 19.8 |
Isotopic, isotopic composition of carbon; PC, PhyloChip; CL, clone library; tR, terminal restriction fragment length polymorphism (t-RFLP); microscopic, epifluorescent image obtained during Ocean Drilling Program Expedition leg 201 by Mark Delwiche and used for cell counts.
Samples from NCM were collected during Integrated Ocean Drilling Program (IODP) expedition 311 (EXP 311) and Pacific Geosciences Center (PGC) cruise PGC2008007 (referred to here as PGC0807). During PGC0807, the active cold seep Bullseye Vent was sampled. Bullseye Vent is part of a larger seep field covering an area of about 2 by 4 km on the midslope of the NCM. Four large seeps varying from 10 to several 100 m in diameter have been identified within this seep field (42).
Hydrate Ridge is a 25- by 15-km ridge in the Cascadia accretionary complex. Drilling in this area has verified the presence of methane hydrate (51) and active vents that expel methane into the overlying water column, creating bubble plumes hundreds of meters high (21, 47). A biofilm was collected from the Hydrate Ridge area during the coring of Ocean Drilling Program (ODP) leg 204 site 1251A (45). Site 1251A is characterized by a shallow (0.5-mbsf) SMT (45).
In the Indian Ocean, biofilm samples were collected from cores obtained from the Krishna-Godavari (KG) Basin (site 20A) and the Mahanadi Basin (site 18A) during National Gas Hydrate Program expedition 01 (NGHP01). The KG Basin is in a passive margin setting that shows a thick alluvium sediment accumulation deposited since the Miocene in a growth fault environment (13). Sediment input in this region has been dominated by the Krishna and Godavari River systems producing sedimentation rates of ∼20 to 25 cm ka−1 (37). The high sedimentation rate and rich source of organic carbon (1.5 to 2.0 wt% total organic carbon) make the site a good candidate for methane generation in shallow subsurface sediments (22).
Sample collection.
Biofilm and proximal sediments (∼10 g of sediment adjacent to but lacking visible biofilm and here referred to as “reference” samples) were collected during NGHP01 and PGC0807 and preserved for molecular analysis (Table 1). Samples were retrieved using an advanced piston core during NGHP01 or by piston core during the PGC0807 expedition. The IODP guidelines for obtaining high-quality microbiology cores were adhered to on both expeditions. The cores were split onboard, and if a biofilm was identified in the split core a sterile spatula was used to transfer the biofilm into a sterile 2-ml microcentrifuge tube. During NGHP01, the tubes were immediately frozen at −80°C. During the PGC0807 expedition, 1 ml of RNA Later was added to the microcentrifuge tubes, which were kept at 4°C for 24 h and stored at −80°C thereafter. Biofilm samples for stable carbon isotope analysis collected during EXP 311 were preserved at −20°C. Biofilms obtained for epifluorescent microscopy were collected during ODP leg 204 at site 1251A. Cells were counted from a photomicrograph of the stained biofilm taken aboard the JOIDES Resolution immediately after the biofilm was collected. The direct count procedure is detailed in the initial reports of ODP leg 204 (45), but briefly, cells were suspended in a solution containing 0.01% acridine orange and 0.25% formaldehyde, stained for 5 min, and then filtered through a 0.2-μm-pore-size filter. The area of the image (1.2 × 10−2 mm2) was calculated on the basis of the scale bar shown in the photomicrograph (Figure F23 in reference 45). These counts estimate the cellular abundance in the biofilm, assuming that 10 mg of the biofilm cells was evenly distributed on the filter (M. Delwiche, personal communication).
Geochemistry.
Pore water analysis was performed on whole-round core samples collected during EXP 311 (NCM) and NGHP01 (Indian Ocean). The surface of each whole-round core sample was carefully scraped with a clean spatula to remove potential contamination from seawater and sediment smearing in the borehole. The cleaned sediment was placed into a titanium squeezer, modified after the stainless-steel squeezer of Manheim and Sayles (24). Gauge pressures up to 15 MPa were applied using a laboratory hydraulic press to extract pore water. Interstitial water was passed through a prewashed Whatman no. 1 filter fitted above a titanium screen, filtered through a 0.2-μm-pore-size Gelman polysulfone disposable filter, and subsequently extruded into a precleaned (10% HCl) 50-ml plastic syringe attached to the bottom of the squeezer assembly (41). During PGC0807, pore water for sulfate analysis was extracted by pressure squeezing (∼3 bar) and filter sterilized with 0.2-μm-pore-size acrodisc polyethersulfone (PES) syringe filters (Pall Corporation) (35).
The sulfate (SO42−) concentration was measured with a Dionex DX-120 (EXP 311) or a Metrohm 861 (NGHP01 and PGC0807) advanced ion chromatograph (35, 41). Removal of H2S was done by nitrogen bubbling of the sample (EXP 311) or by adding Cd(NO3)2 aliquots (NGHP01 and PGC0807) immediately after interstitial water collection.
Samples tested for pore water methane concentration were collected as 3-cm3 sediment plugs from split cores and stored at −20°C in 20-ml serum vials sealed with 1-cm-thick butyl rubber septa. Methane concentrations were determined by the headspace equilibration technique and gas chromatography-flame ionization detection using a Hewlett-Packard 6890 Plus (GC3) apparatus during EXP 311 and NGHP01 and a Shimadzu GC-14A apparatus during PGC0807.
Samples for stable carbon isotope analysis of the biofilm were placed in silver cups, acidified with 10% HCl to dissolve all carbonaceous material, dried at 50°C for 24 h, and then combusted with a Fisons EA-1100 elemental analyzer (EA) interfaced to a Thermo Finnigan Delta Plus isotope ratio mass spectrometer. The carbon isotope ratios are reported in the standard δ notation relative to the Vienna PeeDee Belemnite (V-PDB) standard.
Nucleic acid extraction and purification.
Total DNA was extracted from reference samples (10 g) using a PowerMax Soil DNA extraction kit (Mo Bio Laboratories, Carlsbad, CA). The Mo Bio protocol was modified initially by suspending the sediment in artificial seawater (6 g MgSO4, 30 g NaCl, 2 g KCl liter−1) with occasional shaking at room temperature for 1 h prior to using a PowerMax Soil kit. The sediment/seawater mixture was centrifuged at 8,000 × g for 10 min, and then the sediment was transferred into the bead-beating tubes of the PowerMax Soil kit, along with two 0.25-in. steel balls (MP Biomedicals, Solon, OH), and subsequently the extraction procedure followed PowerMax Soil kit instructions. DNA was extracted from biofilm samples (∼1 g) using a PowerSoil DNA extraction kit (Mo Bio Laboratories) according to the manufacturer's recommendations. The DNA eluted from all Mo Bio kits was concentrated to 50 μl using a Montage PCR spin column (Millipore, Billerica, MA). The amount of DNA in each sample was measured using a Qubit apparatus (Invitrogen, Inc., Carlsbad, CA) according to the manufacturer's recommendations.
t-RFLP analysis.
To determine microbial diversity between samples, terminal restriction fragment length polymorphism (t-RFLP) analysis of bacterial and archaeal rRNA genes was performed on samples from both India and NCM. Each DNA suspension was amplified using the general bacterial primers 27F-FAM and 926R (11) or general archaeal primers 21F-FAM and 958R (6) in a MasterCycler thermocycler (Eppendorf, Westbury, NY). Each 20-μl PCR mixture contained 1.25 units of AmpliTaq Gold LD (ABI, Foster City, CA), 1× PCR buffer, 4 mM MgCl2, 800 μM each deoxynucleoside triphosphate, 0.5 μM each primer, and 8 μg of bovine serum albumin. PCR conditions consisted of an initial denaturation step of 5 min at 95°C, followed by 35 cycles of 40 s at 95°C, 40 s at 50°C, and 60 s at 72°C and a final elongation step of 5 min at 72°C (43). Thirty-five cycles were required to amplify the low levels of DNA in the reference samples. This is consistent with other studies with samples with low biomass (43). Products were combined from three PCR runs per DNA sample and purified with Montage PCR spin columns (Millipore, Billerica, MA).
One hundred nanograms of amplified and combined DNA products was digested using the restriction enzyme HaeIII (28). Digestions were run according to the manufacturer's specifications (Fermentas, Glen Burnie, MD) by incubating the restriction digest for 3 h at 37°C, followed by heat inactivation at 80°C for 20 min. The size of the restricted samples was determined by capillary gel electrophoresis using an ABI Prism 3100 genetic analyzer at the Oregon State University Center for Genome Research and Biotechnology (CGRB).
Clone library construction and phylogenetic analysis.
Clones were created from both biofilm samples from India (sites 18A and 20A) and the biofilm sample from site PGC0807-C19. Bacterial clone libraries were constructed using nonlabeled bacterium-specific forward 27F and universal reverse 1492R primers that target the 16S rRNA gene (14). The archaeal 16S rRNA gene was amplified using archaeon-specific forward 21F and universal reverse 1492R primers (48). PCR amplification of the 16S rRNA gene used the same protocol used for t-RFLP analysis. PCR products of the correct size were cloned using pCR 2.1 TOPO vector (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Clones from each library were sequenced at the CGRB or at the Genome Center at Washington University (St. Louis, MO).
Sequence analysis was performed using Geneious (9) and Mothur (44) freeware. Sequences with >95% similarity were assigned to the same phylotype (43). Representative phylotypes were then sequenced in the reverse direction. The sequences were aligned in Mothur and then imported into FastTree to create a phylogenetic tree (36). The tree was visualized on the Interactive Tree of Life (ITOL) website (http://itol.embl.de/). In addition, phylotypes were digested in silico with HaeIII, and the end fragment sizes were compared to those of terminal restriction fragments (TRFs) obtained from t-RFLP analysis.
PhyloChip microarray analysis.
PhyloChip, version G3, microarray analysis sample preparation and data analysis were performed on biofilm and reference samples from site 18A (India) and site PGC0807-C19 (NCM) as previously described (15), except that 100 ng of internal spike DNA, 200 ng of bacterial 16S rRNA gene amplicons, and 50 ng of archaeal 16S rRNA gene amplicons were hybridized to the array.
Nucleotide sequence accession numbers.
The nucleotide sequences of the rRNA gene clones were deposited in the GenBank database under the following accession numbers: HQ700668 to HQ700686 and HQ711382 to HQ711400.
RESULTS
Geochemistry.
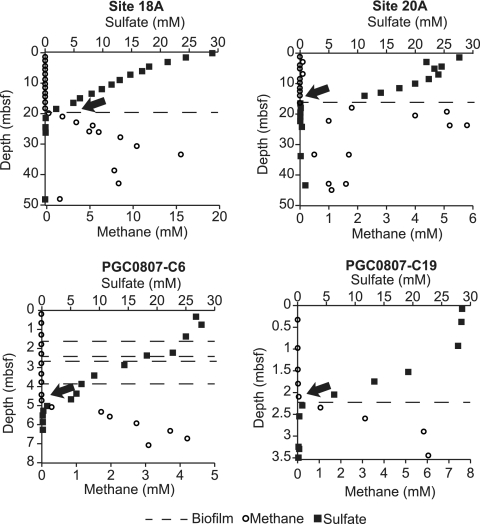
The sulfate and methane profiles for each of the study sites are illustrated in Fig. 2. The SMT depth is defined as the depth where sulfate and methane concentrations converge at minimum values. The depth of the SMT in the Indian Ocean sediments was 18 to 20 mbsf, and at the NCM it ranged from 1.4 to 5 mbsf. The depth of the biofilm occurrence typically corresponded to the SMT depth (Fig. 2). However, site PGC0807-C6 also had three biofilm occurrences above the SMT. The δ13C of the biofilm samples ranged from −35 to −43‰.
Fig. 2.
Sulfate (squares) and methane (circles) profiles depicting the SMT depth (noted by arrows). Dashed lines show the depths where biofilm was recovered from fractures in the respective cores. The depth is measured in meters below the seafloor (mbsf), and the scale varies for the different sites shown.
Nucleic acid extractions and cell counts.
DNA was extracted from the biofilm and reference samples collected from the NCM and offshore India. Concentrations of DNA extracted from the reference samples ranged from 0.2 to 2.5 ng g−1 of sediment, whereas concentrations of total DNA extracted from the biofilms ranged from 27 to 119 ng g−1 (wet weight). On the basis of the DNA yield from the biofilm and reference samples and with the assumption of an average DNA content of 2 fg cell−1 (4, 48), between 1.3 × 107 to 5.9 × 107 cells g−1 and 1 × 105 to 1.3 × 105 cells g−1 were present in biofilm and reference samples, respectively. The cellular abundance counted from the epifluorescent image of a biofilm taken from the Hydrate Ridge sample was 2.9 × 108 cells g−1.
Molecular phylogenetic analysis.
Microbial community fingerprints recovered from the DNA assemblages of the biofilm and reference samples were examined by PCR-mediated t-RFLP analysis. Identification of taxa is more difficult with t-RFLP than with other methods; however, a benefit of this approach is that it allows comparison of the prominent DNA sequences obtained from all of the samples. A total of 133 archaeal TRFs and 148 bacterial TRFs were detected. Fifty-two archaeal and 56 bacterial TRFs were shared between the biofilm and reference samples. The 16S rRNA gene sequences obtained from clone libraries were digested in silico and then compared to the TRF sequences. This enabled the identification of peaks in the t-RFLP electropherogram that corresponded to the ANME-1 clade, the Thermoplasmatales, and the Crenarchaeales TRFs (see Fig. S1 in the supplemental material). We were unable to identify any of the other peaks in the electropherograms using this method and must rely on the clone library and PhyloChip results for additional data.
A total of 228 bacterial clones and 161 archaeal clones were sequenced from the biofilm samples. A similarity analysis with a 95% similarity cutoff indicated 33 distinctive bacterial phylotypes and 19 distinctive archaeal phylotypes. Full 16S rRNA gene sequences were obtained for each of the representative phylotypes and used in phylogenetic analysis.
The major bacterial groups recovered from the biofilm were the Deltaproteobacteria and Chloroflexi, representing 71% and 10% of the clones, respectively. In addition, candidate division JS1, candidate division WS3, Gammaproteobacteria, Deferribacteres, and OP11 were represented in smaller quantities (21% of the clones) (see Fig. S2 in the supplemental material). The biofilms from NCM were mainly composed of Deltaproteobacteria, while the biofilms from the Indian Ocean were composed of all previously mentioned phyla. Within the Deltaproteobacteria, sequences were related to putative sulfate-reducing bacteria (SRB). The sequence slm_bac_110 was the most frequently obtained SRB phylotype (38% of total clones), and this sequence is closely related to the HydGH-Bac13 sequence (96.6% similarity) detected in a cold seep from Hydrate Ridge (GenBank accession number AM229187). Sequences that clustered within the Chloroflexi phylum did not have a dominant phylotype, with one clone detected for each phylotype, all clustering with the Dehalococcoides class. One phylotype (sequence slm_bac_40) was detected in the Deferribacteres phylum and had a 97.6% similarity to a phylotype that was detected in the Kazan mud volcano (33).
The archaeal diversity within the biofilm contained both Euryarchaeota and Crenarchaeota for both NCM and India. Euryarchaeota sequences were related to ANME-1, Thermoplasmatales, and Methanosarcinales, representing 73%, 11%, and 2% of total archaeal clones, respectively. The Crenarchaeota sequences were related to Desulfurococcales and Crenarchaeales, representing 10% and 3% of the total archaeal clones, respectively (see Fig. S3 in the supplemental material). Four phylotypes of ANME-1 were detected; however, two sequences (slm_arc_201 and slm_arc_400) were the most abundant, making up 94% of the ANME phylotypes. The sequence slm_arc_201 was predominantly found in the NCM (29 clones from NCM, 1 clone from India), while the sequence slm_arc_400 was exclusively found in Indian samples (42 clones). The sequence slm_arc_201 was closely related to the sequence Arch125 (99%) detected in Aarhus Bay (3). The sequence slm_arc_400 was related to the sequence GBa1r013 (95.6%) detected in the Guaymas Basin (50). Sequences slm_arc_400 and slm_arc_201 are 94% similar to each other. No other ANME group was detected.
PhyloChip.
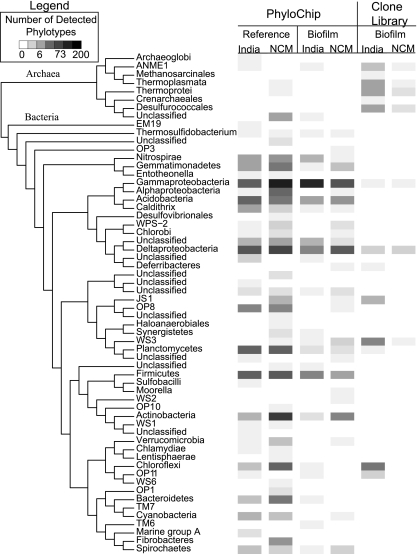
A total of 132, 491, 178, and 193 phylotypes were detected on the PhyloChip from the NCM biofilm, NCM reference, Indian Ocean biofilm, and Indian Ocean reference samples, respectively (Fig. 3). All seven bacterial phyla that were detected in the clone library were also detected in the PhyloChip. In addition, 44 phyla were detected on the PhyloChip but not in the clone library. Of the additional 44 phyla detected, 9 were unclassified phyla. Only five archaeal phyla were detected, one of which was an unclassified phylum. The PhyloChip detected a member of the Archaeoglobi that was not detected in the clone library. Although the PhyloChip detected ANME-1 cells in the biofilm and reference samples from India, it failed to detect Methanosarcinales and Desulfurococcales. The general observation here that the PhyloChip detected more taxa than a library composed of a few hundred clones is consistent with the findings of prior research where the two methods were compared in an analysis of microbial populations in urban aerosols, subsurface soil, and subsurface water samples that contained 417, 485, and 253 clones, respectively (7).
Fig. 3.
Phylogenetic tree of archaeal and bacterial phyla detected using PhyloChip and clone library analyses of reference and biofilm samples acquired from different locations examined in this study. The number of phylotypes that were detected in each classification using the respective analytical method are shown using a heat map.
DISCUSSION
Unique macroscopic subseafloor biofilms found in fractures in NCM and Indian Ocean sediments intersecting the SMT were characterized by culture-independent techniques. Most biofilms reported in our study were located at the SMT, a geochemical indicator of AOM (5, 38). The presence of the ANME-1/SRB consortia within the biofilm and the SMT suggest that this biofilm is affiliated with AOM. At site PGC0807-C6, biofilm was found above the SMT (Fig. 2). However, the DNA concentration of that biofilm (0.5 ng g−1) was similar to that of the surrounding sediment that lacked biofilm (0.2 to 2.5 ng g−1 of sediment). While we do not know the biological characteristics of these non-SMT biofilms, as molecular analyses were not performed, it is plausible that these are remnant biofilms from a time when the methane flux was higher and the SMT was located nearer the seafloor. This explanation is consistent with the highly dynamic nature of methane seepage at this site.
Detection of more 13C-depleted values for the biofilm (−35 to −43‰) than the surrounding sediment (−25.7‰) (18) provides further evidence that the biofilm was synthesized within an active SMT. Assimilation of 13C-depleted methane carbon or autotrophic fixation of CO2 by the AOM consortium results in 13C-depleted microbial biomass (53). However, previous isotopic values for lipids from the AOM consortia are typically more 13C depleted (−60‰) (1) and more consistent with the isotopic value of the carbon in the methane itself. Metabolism of the organic carbon coupled to sulfate reduction could produce the observed isotopic values (1). It is also possible that during sampling of the biofilms significant amounts of surrounding sediment were mixed with the biofilm, thereby generating a mixed sediment-biofilm isotopic ratio.
Three molecular techniques were used to determine the composition of the microbial communities: t-RFLP, clone libraries, and PhyloChip. Each technique depends upon the efficiency of DNA extraction from cells and then amplification of the resulting DNA. t-RFLP is a rapid and relatively inexpensive technique useful in identifying trends of dominant taxa but is not sensitive to species-level differences and multiple taxa can be represented by individual TRFs (10). Both clone libraries and the PhyloChip are more labor-intensive but provide a much higher resolution of closely related taxa. The PhyloChip, version G3, is a parallel hybridization microarray technology with probes for 59,000 microbial taxa (15). For each taxon there are additional probes that have mismatches yielding a total of 1,100,000 probes. Thus, known taxa and closely related taxa can be detected simultaneously, and the array is more sensitive to rare microbes than typical sequencing approaches (7). Our results were consistent with this observation. As a probe-based approach, the PhyloChip has limitations in detecting novel organisms. Archaeal 16S rRNA gene databases are notoriously underrepresented compared to bacterial 16S rRNA gene databases. Therefore, clone libraries can sometimes detect novel archaeal taxa that would be missed by any array technology.
t-RFLP was performed on all of the samples, allowing a direct comparison. An in silico approach of comparing TRFs to the clone library was able to identify only three of the phyla that were detected in the clone libraries and on the PhyloChip (see Fig. S1 in the supplemental material). Takishita et al. (49) used the same in silico approach and were able to identify TRFs that correspond to the ANME-2 group from a cold seep environment. Our investigation was able to detect TRFs that correspond to ANME-1 in all biofilm samples, and this finding was consistent with the findings of both of the other molecular ecology methods used.
The presence of ANME-1 is significant because microbial biogeography studies have shown a variety of environmental factors that influence the distribution of the AOM consortia. From studies conducted to date, the main controlling factors appear to be the availability of methane and sulfate (27). For instance, bioreactor studies have shown that ANME-1 isolates are enriched in high-methane, low-sulfate environments, while ANME-2 isolates are present in high-sulfate, low-methane environments (12). Field studies of sediments overlying a methane-rich brine pool in the Gulf of Mexico and microbial mats of the Black Sea offer additional evidence for the localization of ANME groups according to sulfate and methane concentrations (23). In the Black Sea mats, the outer (lower-methane) black portion is primarily composed of ANME-2, while the inner (higher-methane) pink portion is composed of ANME-1 (40). The biofilms reported herein are also pink to orange and contain ANME-1, suggesting some commonalities between the fracture-dominant environments and the conditions that foster growth of the Black Sea mats. The dominant sequences that were recovered from both NCM and Indian Ocean biofilms were similar to sequences found in advective cold seeps, where there is typically an ample supply of methane (33).
Our results show that 107 to 108 cells cm−3 can occur within these fracture-dominant environments. In diffusion-dominant environments, the AOM consortium densities are typically low, with 106 cells cm−3 (29). In contrast, seep-dominant environments such as Eel River Basin (16, 31), Hydrate Ridge (5, 20), the Black Sea (25, 40), and the Gulf of Mexico (23) contain >1010 AOM consortium cells cm−3. For example, in the Black Sea the AOM consortium produces macroscopic biomass seen as columnar structures in the methane-laden anoxic bottom waters that extend as much as 4 m into the water (20, 25). Thus, despite energetic constraints, the AOM consortia have been shown to produce macroscopic quantities of biomass in seep-dominant environments. The question, then, is how can AOM produce macroscopic quantities of biofilm in fracture-dominant environments where the physical and energetic constraints are presumably more austere than in seep-dominant environments?
Field observations of microbes in the subsurface indicate that the sediment grain size can be a controlling factor for cell densities. Inagaki et al. (17) observed that ash layers from the Sea of Okhotsk contained significantly higher biomass concentrations than intervening clay layers. Likewise, Phelps et al. (34) also observed lower cell density and activity in clay-rich sediment than sandy sediment. The ash and sand layers that harbor greater cell densities have larger grain sizes than the clay layers. The mechanism that allows higher cell densities is unknown, although Inagaki et al. (17) hypothesized that habitable space and/or fluid flow and delivery of electron acceptors and donors could explain the higher levels of cells observed in locations with greater pore space. The fractured systems that we have studied may function in a manner similar to geological media with large pore space available for microbial colonization. Still other explanations for variable cell densities in subsurface media include water activity or the mineralogy of the sediment (48, 52).
The AOM consortia are restricted to environments with both sulfate and methane. Seep-dominant SMT environments have higher sulfate and methane concentrations that occur coincident with a higher abundance of cells involved with AOM. We have not measured methane or sulfate flux in the fractures where we collected the biofilms; however, fractures in the sediment may be preferred flow paths for fluids (54). If advective flux of methane and sulfate occurs in these environments, most would probably travel through the fractures, making them the most likely location for higher biomass.
On the basis of the preceding discussion and observations of biofilms in fractures, we postulate that fractures are pathways for advective methane flux. Support for gas migration through the NCM fractures comes from geophysical measurements obtained during EXP 311 at Bullseye Vent, the location of some of the biofilms that we collected. Electrical resistivity tools imaged several steeply dipping fractures with elevated resistivity values caused by high gas hydrate saturation in the fractures (41). That these fractures are preferred flow paths for fluid advection is also supported by observations of methane bubbles at the seafloor and acoustic images of bubble plumes rising from the seafloor near our NCM biofilm sampling sites (55). The model presented by Riedel et al. (41) describes Bullseye Vent as a complex subsurface network of fractures partially filled with gas hydrate and feeding methane upwards toward the seafloor. Such fractures may increase habitable space that is otherwise lacking in sediments and provide a supply of methane that creates a favorable condition for higher biomass accumulation. The biofilms from the Indian Ocean were also collected from what appeared to be fractures; however, there is no geophysical evidence for fracturing.
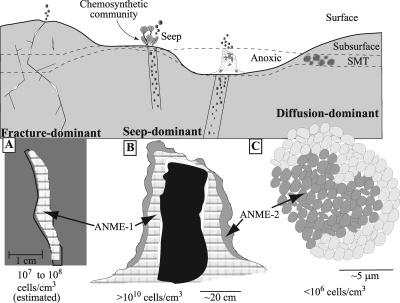
AOM consortia occur in environmental settings that range from diffusion to seep-dominant SMTs (1). Fracture-dominant SMT settings contain macroscopic biofilms comprised of ANME-1 (Fig. 4A). The distribution of ANME groups is, in part, controlled by methane and sulfate concentrations (12). In the Black Sea mats, the outer portion (lower methane concentrations) is primarily composed of ANME-2, while the inner portion (higher methane concentrations) is composed of ANME-1 (40) (Fig. 4B). The Black Sea is a seep-dominant SMT, and biomass can exceed 1010 cells cm−3. In contrast, diffusion-dominant SMTs typically contain microcolonies of ANME-2 and their sulfate-reducing symbiont (Fig. 4C).
Fig. 4.
Conceptual model of biomass and ANME groups found in fracture-, seep-, and diffusion-dominant sulfate-methane environments. (A) Representation of a biofilm in a fracture; (B) representation of the Black Sea microbial mats; (C) representation of the microcolonies found in diffusion-dominant environments.
In summary, it appears that fracture-dominant SMT constitutes a previously unreported setting where the AOM consortia reside and produce macroscopic biofilms. Their presence may be associated with characteristics typical of fractures in the seafloor and as such may represent an alternative condition where AOM may occur (i.e., not diffusive or seep dominant). If indeed these fractures are conduits for methane flow, we would also expect to see higher AOM rates in a subsurface fractured SMT than a subsurface diffusive-dominant SMT. Additional studies will be needed to verify this hypothesis.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Energy, Office of Fossil Energy, National Energy Technology Laboratory. Samples were collected as a part of the National Gas Hydrate Program cruise 01 (NGHP01) in 2006 and the Pacific Geosciences Center cruise 007 (PGC007; this article is ESS contribution number 20110050) in 2008. Jayson Hieter, who assisted with the study, was supported by an undergraduate research fellowship through the Subsurface Biosphere Initiative at Oregon State University. Part of this work was performed under the Climate Sciences Scientific Focus Area at the Lawrence Berkeley National Laboratory, supported in part by the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
We thank the entire NGHP01 and PGC007 scientific parties and vessel crews for assisting us with the collection of samples. We appreciate Jayson Hieter for assistance with DNA extraction and subsequent molecular analysis. We also thank Mark Delwiche for the epifluorescence image taken of the biofilm from Hydrate Ridge.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Alperin M. J., Hoehler T. M. 2009. Anaerobic methane oxidation by archaea/sulfate-reducing bacteria aggregates. 1. Thermodynamic and physical constraints. Am. J. Sci. 309:869–957 [Google Scholar]
- 2. Alperin M. J., Hoehler T. M. 2010. The ongoing mystery of sea-floor methane. Science 329:288–289 [DOI] [PubMed] [Google Scholar]
- 3. Aquilina A., et al. 2010. Biomarker indicators for anaerobic oxidizers of methane in brackish-marine sediments with diffusive methane fluxes. Org. Geochem. 41:414–426 [Google Scholar]
- 4. Bakken L. R., Olsen R. A. 1989. DNA content of soil bacteria of different cell size. Soil Biol. Biochem. 21:789–793 [Google Scholar]
- 5. Boetius A., et al. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626 [DOI] [PubMed] [Google Scholar]
- 6. DeLong E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeSantis T. Z., et al. 2007. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 53:371–383 [DOI] [PubMed] [Google Scholar]
- 8. Dickens G. R. 2003. Rethinking the global carbon cycle with a large, dynamic and microbially mediated gas hydrate capacitor. Earth Planet. Sci. Lett. 213:169–183 [Google Scholar]
- 9. Drummond A., et al. 2009. Geneious, version 4.7 Biomatters Ltd., Auckland, New Zealand [Google Scholar]
- 10. Dunbar J., Ticknor L. O., Kuske C. R. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edlund A., Hardeman F., Jansson J. K., Sjoling S. 2008. Active bacterial community structure along vertical redox gradients in Baltic Sea sediment. Environ. Microbiol. 10:2051–2063 [DOI] [PubMed] [Google Scholar]
- 12. Girguis P. R., Cozen A. E., DeLong E. F. 2005. Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl. Environ. Microbiol. 71:3725–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta S. K. 2006. Basin architecture and petroleum system of Krishna Godavari Basin, east cost of India. Leading Edge 25:830–837 [Google Scholar]
- 14. Harrison B. K., Zhang H., Berelson W., Orphan V. J. 2009. Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California). Appl. Environ. Microbiol. 75:1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hazen T. C., et al. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208 [DOI] [PubMed] [Google Scholar]
- 16. Hinrichs K. U., Hayes J. M., Sylva S. P., Brewer P. G., DeLong E. F. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805 [DOI] [PubMed] [Google Scholar]
- 17. Inagaki F., et al. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69:7224–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaneko M., Shingai H., Pohlman J. W., Naraoka H. 2010. Chemical and isotopic signature of bulk organic matter and hydrocarbon biomarkers within mid-slope accretionary sediments of the northern Cascadia margin gas hydrate system. Mar. Geol. 275:166–177 [Google Scholar]
- 19. Knittel K., Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63:311–334 [DOI] [PubMed] [Google Scholar]
- 20. Knittel K., Losekann T., Boetius A., Kort R., Amann R. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71:467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kulm L. D., et al. 1986. Oregon subduction zone: venting, fauna, and carbonates. Science 231:561–566 [DOI] [PubMed] [Google Scholar]
- 22. Kundu N., Pal N., Sinha N., Budhiraja I. L. 2008. Paleo hydrate and its role in deep water Pliopleistocene gas reservoir in Krishna Godavari Basin, India. Proc. 6th Int. Conf. Gas Hydrates. https://circle.ubc.ca/handle/2429/1065
- 23. Lloyd K. G., Lapham L., Teske A. 2006. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl. Environ. Microbiol. 72:7218–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manheim F. T., Sayles F. L. 1974. Composition and origin of interstitial waters of marine sediments, based on deep sea drill cores, p. 527–568. In Goldberg E. D. (ed.), Marine chemistry. The sea: ideas and observations on progress in the study of the seas, vol. 5 John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 25. Michaelis W., et al. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013–1015 [DOI] [PubMed] [Google Scholar]
- 26. Milkov A. 2004. Global estimates of hydrate-bound gas in marine sediments: how much is really out there. Earth Sci. Rev. 66:183–197 [Google Scholar]
- 27. Nauhaus K., Treude T., Boetius A., Kruger M. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 7:98–106 [DOI] [PubMed] [Google Scholar]
- 28. Newberry C. J., et al. 2004. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program leg 190. Environ. Microbiol. 6:274–287 [DOI] [PubMed] [Google Scholar]
- 29. Niemann H., et al. 2005. Methane emission and consumption at the North Sea gas seep (Tommeliten area). Biogeoscience 2:335–351 [Google Scholar]
- 30. Niemann H., et al. 2006. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443:854–858 [DOI] [PubMed] [Google Scholar]
- 31. Orphan V. J., et al. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Orphan V. J., House C. H., Hinrichs K. U., McKeegan K. D., DeLong E. F. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U. S. A. 99:7663–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pachiadaki M. G., Lykousis V., Stefanou E. G., Kormas K. A. 2010. Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol. Ecol. 72:429–444 [DOI] [PubMed] [Google Scholar]
- 34. Phelps T. J., Pfiffner S. M., Sargent K. A., White D. C. 1994. Factors influencing the abundance and metabolic capacities of microorganisms in eastern coastal-plain sediments. Microb. Ecol. 28:351–364 [DOI] [PubMed] [Google Scholar]
- 35. Pohlman J. W., Ruppel C., Hutchinson D. R., Downer R., Coffin R. B. 2008. Assessing sulfate reduction and methane cycling in a high salinity pore water system in the northern Gulf of Mexico. Mar. Petrol. Geol. 25:942–951 [Google Scholar]
- 36. Price M. N., Dehal P. S., Arkin A. P. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rao D. P., et al. 1994. Analysis of multichannel seismic reflection and magnetic data along 130 N latitude across the Bay of Bengal. Mar. Geophys. Res. 16:225–236 [Google Scholar]
- 38. Reeburgh W. S. 1976. Methane consumption in Cariaco Trench waters and sediments. Earth Planet. Sci. Lett. 28:337–344 [Google Scholar]
- 39. Reeburgh W. S. 2007. Oceanic methane biogeochemistry. Chem. Rev. 107:486–513 [DOI] [PubMed] [Google Scholar]
- 40. Reitner J., Peckmann J., Reimer A., Schumann G., Thiel V. 2005. Methane-derived carbonate build-ups and associated microbial communities at cold seeps on the lower Crimean shelf (Black Sea). Facies 51:66–79 [Google Scholar]
- 41. Riedel M., et al. 2006. Geophysical and geochemical signatures associated with gas hydrate-related venting in the northern Cascadia margin. GSA Bull. 118:23–38 [Google Scholar]
- 42. Riedel M., Spence G. D., Chapman R. N., Hyndman R. D. 2002. Seismic investigations of a vent field associated with gas hydrates, offshore Vancouver Island. J. Geophys. Res. 107:2200 [Google Scholar]
- 43. Roussel E. G., et al. 2009. Archaeal communities associated with shallow to deep subseafloor sediments of the New Caledonia Basin. Environ. Microbiol. 11:2446–2462 [DOI] [PubMed] [Google Scholar]
- 44. Schloss P. D., et al. 2009. Introducing mothur: open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shipboard Scientific Party. 2003. Site 1251, p. 1–119. In Tréhu A. M., Bohrmann G., Rack F. R., Torres M. E., Bangs N. L., Barr S. R., Borowski W. S., Claypool G. E., Collett T. S., Delwiche M. E., Dickens G. R., Goldberg D. S., Gràcia E., Guèrin G., Holland M., Johnson J. E., Lee Y., Liu C., Long P. E., Milkov A. V., Riedel M., Schultheiss P., Su X., Teichert B., Tomaru H., Vanneste M., Watanabe M., Weinberger J. L., Proceedings of the Ocean Drilling Program. Initial report 204. Ocean Drilling Program, College Station, TX doi:10.2973/odp.proc.ir.204.110.2003 [Google Scholar]
- 46. Sivan O., Schrag D. P., Murray R. W. 2007. Rates of methanogenesis and methanotrophy in deep-sea sediments. Geobiology 5:141–151 [Google Scholar]
- 47. Suess E., et al. 1999. Gas hydrate destabilization: enhanced dewatering, benthic material turnover and large methane plumes at the Cascadia convergent margin. Earth Planet. Sci. Lett. 170:1–15 [Google Scholar]
- 48. Takai K., Komatsu T., Inagaki F., Horikoshi K. 2001. Distribution of archaea in a black smoker chimney structure. Appl. Environ. Microbiol. 67:3618–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takishita K., Kakizoe N., Yoshida T., Maruyama T. 2009. Molecular evidence that phylogenetically diverged ciliates are active in microbial mats of deep-sea cold-seep sediment. J. Eukaryot. Microbiol. 57:76–86 [DOI] [PubMed] [Google Scholar]
- 50. Teske A., et al. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trehu A. M., et al. 2004. Three-dimensional distribution of gas hydrate beneath southern Hydrate Ridge: constraints from ODP Leg 204. Earth Planet. Sci. Lett. 222:845–862 [Google Scholar]
- 52. van Loosdrecht M. C., Lyklema J., Norde W., Zehnder A. J. 1990. Influence of interfaces on microbial activity. Microbiol. Rev. 54:75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wegener G., Niemann H., Elvert M., Hinrichs K. U., Boetius A. 2008. Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ. Microbiol. 10:2287–2298 [DOI] [PubMed] [Google Scholar]
- 54. Weinberger J. L., Brown K. M. 2006. Fracture networks and hydrate distribution at Hydrate Ridge, Oregon. Earth Planet. Sci. Lett. 245:123–136 [Google Scholar]
- 55. Zyla T. A., Spence G., Riedel M., Whiticar M. J. 2010. Tracking and quantifying methane bubble plumes on the North Cascadia Margin, p. OS53A-1365 American Geophysical Union, San Francisco, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.