Abstract
The food-borne pathogen Listeria monocytogenes is a problem for food processors and consumers alike, as the organism is resistant to harsh environmental conditions and inimical barriers implemented to prevent the survival and/or growth of harmful bacteria. One mechanism by which listeriae mediate survival is through the accumulation of compatible solutes, such as proline, betaine and carnitine. In other bacteria, including Escherichia coli, the synthesis and accumulation of another compatible solute, trehalose, are known to aid in the survival of stressed cells. The objective of this research was to investigate trehalose metabolism in L. monocytogenes, where the sugar is thought to be transferred across the cytoplasmic membrane via a specific phosphoenolpyruvate phosphotransferase system and phosphorylation to trehalose-6-phosphate (T6P). The latter is subsequently broken down into glucose and glucose-6-phosphate by α,α-(1,1) phosphotrehalase, the putative product of the treA gene. Here we report on an isogenic treA mutant of L. monocytogenes 568 (568:ΔTreA) which, relative to the wild-type strain, displays increased tolerances to multiple stressors, including heat, high osmolarity, and desiccation. This is the first study to examine the putative trehalose operon in L. monocytogenes, and we demonstrate that lmo1254 (treA) in L. monocytogenes 568 indeed encodes a phosphotrehalase required for the hydrolysis of T6P. Disruption of the treA gene results in the accumulation of T6P which is subsequently dephosphorylated to trehalose in the cytosol, thereby contributing to the stress hardiness observed in the treA mutant. This study highlights the importance of compatible solutes for microbial survival in adverse environments.
INTRODUCTION
The disaccharide trehalose (α-d-glucopyranosyl-1,1-α-d-glucopyranoside) is widely distributed in nature and found in shellfish, insects, plants, mammals, bacteria, and fungi (2). It is a nonreducing sugar and compatible solute with unique physicochemical properties, which include acting as a molecular chaperone to prevent misfolding of proteins and stabilizing membranes through hydrogen bonding with phospholipids (46, 57). Trehalose also aids in the survival of many organisms during exposure to adverse conditions caused by temperature extremes, desiccation, high osmolarity, and oxidative stress (15).
Although, trehalose metabolism has been documented for Gram-negative bacteria such as Enterobacter sakazakii (Cronobacter) (8), Pseudomonas fluorescens (35), Rhizobium japonicum (52), and Salmonella enterica serovar Typhimurium (9), only that of Escherichia coli has received a thorough analysis (28, 32, 45). When subjected to high osmolarities, E. coli synthesizes large amounts of trehalose to act as a compatible solute. Perhaps the most interesting phenomenon surrounding this accumulation is the simultaneous high-osmolarity-dependent synthesis and degradation of the disaccharide (28). Spatial separation of these metabolic processes allows for their functional coexistence; the de novo synthesis is localized in the cytoplasm, whereas hydrolysis of trehalose is carried out in the periplasm (28). When the osmolarity of the external environment is low, E. coli uses exogenous trehalose as a carbon and energy source. This catabolic process involves the importation of the disaccharide via a trehalose-specific phosphotransferase system (PTS) resulting in its simultaneous phosphorylation to trehalose-6-phosphate (T6P), which in turn is cleaved by trehalose-6-phosphate hydrolase (α,α-phosphotrehalase) (45). Similar PTS uptake systems for trehalose catabolism are found in Gram-positive bacteria such as Bacillus subtilis which do not appear to accumulate the sugar for starvation or stress relief. Instead, trehalose is used solely as a carbon/energy source (47).
Listeria monocytogenes is an important Gram-positive food-borne pathogen that has received much attention due to its virulence and ability to survive and grow in foods and the food-processing environment. Information regarding trehalose metabolism in Listeria spp. is currently limited to inference based on the published genomic sequence of a putative trehalose operon (1, 21) and possible similarities to related Gram-positive bacteria (7, 14, 26, 47, 48). Also, growth studies have shown that generally Listeria spp. can utilize the disaccharide as their sole source of carbon and that utilization is repressed by the presence of glucose in the growth medium (19, 42, 49).
L. monocytogenes strain 568 is an environmental serotype 1/2a strain originally isolated from a shrimp-processing facility (25). In a previous study we reported on L. monocytogenes 568 transposon insertion mutants that displayed enhanced thermotolerance (18). In one of these mutants, the location of the Tn917 insertion was determined to be in the putative treA gene (lmo1254), the gene thought to encode phosphotrehalase, a phospho-(1,1)-glucosidase (21). The objective of the current study was to elucidate the reason for the observed thermal-resistant phenotype. Therefore, a treA gene deletion mutant of L. monocytogenes 568 was created to assess the impact of this gene on the biochemistry and physiology of the organism. Furthermore, the importance of the treA gene to a multitude of other food-relevant environmental stressors was examined. To the best of our knowledge, this is the first report involving experimental analysis of any of the three genes of the putative trehalose operon in L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. For long-term storage, all strains and transformants were kept at −80°C in broth media supplemented with glycerol (15%, wt/vol): brain heart infusion (BHI) for L. monocytogenes or Luria-Bertani (LB) for E. coli. Reconstitution of frozen stocks was done by streaking the cultures onto BHI or LB agar followed by incubation at 37°C. Recovery of survivors following stress treatments was carried out on tryptic soy agar supplemented with 0.6% yeast extract (TSA-YE) and 1% sodium pyruvate (4, 11). Survival studies for oxidative stress, acid stress, tolerance to ethanol, and osmotic stress were performed in BHI broth augmented according to the methods listed below. Trehalose utilization experiments were carried out in modified Welshimer's broth (MWB) (42). Antibiotic concentrations in selective media were as follows: for cloning treA into E. coli using TOPO pCR2.1, ampicillin (Amp), 100 μg/ml; for selection of E. coli carrying pAUL-A with the treA deletion insert (pAULA:ΔTreA), erythromycin (Erm), 250 μg/ml; for selection of pAULA:ΔTreA-transformed L. monocytogenes 568, Erm, 5 μg/ml; for complementation experiments with E. coli carrying pAM401, chloramphenicol (Crm), 50 μg/ml; and for selection of ΔTreA:pAM:TREOP-transformed L. monocytogenes, Crm, 10 μg/ml. All media and antibiotics were purchased from Oxoid Canada Inc. (Mississauga, ON, Canada) and Sigma-Aldrich Chemical Company (Oakville, ON, Canada), respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Species and strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| L. monocytogenes | ||
| 568 | Wild type; serotype 1/2a; from shrimp-processing plant | 25 |
| 32F11 | Tn917 mutant of 568; Ermr | 18 |
| 568:ΔTreA | 568 deletion mutant; TreA− | This study |
| ΔTreA:pAM:TREOP | pAM401:TreA complement of 568ΔT6P; Crmr Tets TreA+ | This study |
| 568/pAULAΔTreA | 568 carrying pAULA-ΔTreA with treA deletion insert | This study |
| E. coli | ||
| TCTOP:ΔTreA | TOP10 carrying pCR2.1 TOPO with 568 treA insert with deletion | This study |
| TCTOP:TREOP | TOP10 carrying pCR2.1 TOPO with intact 568 Tre operon | This study |
| DH5α | Cloning host; F−endA1 glnV44 thi-1 recA1 gyrA96 deoR nupG φ80lacZ ΔM15 Δ(lacZYA-argF)U169hsdR17 (rK− mK+) λ− | 24 |
| XL10-GOLD Kanr | Cloning host; endA1 glnV44 recA1 thi-1 gyrA96 relA1 lac hte Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 tetR F′[proAB lacIqZ ΔM15 Tn10(Tetr) Amy Tn5(Kanr)] | Stratagene |
| XLCVV1 | XL10-GOLD carrying pCVV1 expression vector | This study |
| TCE-pAULA:TreA | DH5α host carrying pAUL-A with treA deletion insert | This study |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR nupG recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 λ− | Invitrogen |
| Plasmids | ||
| pAUL-A | Gram-positive and Gram-negative shuttle vector; oriTS, LacZa′, Ermr | 10 |
| pAM401 | Gram-positive and Gram-negative shuttle vector; Crmr Tetr | 60 |
| pCR:ΔTreA | pCR2.1 TOPO with 568 treA insert with deletion | This study |
| pAULA:ΔTreA | pAUL-A with truncated 568 treA gene; Ermr | This study |
| pAM401:TREOP | pAM401 with intact 568 treA gene; Crmr, TetstreA+ | This study |
| pCVV1 | pCYB1-derived expression vector carrying B. subtilistreA; Tac promoter, Ampr | 56 |
| pCR2.1 TOPO | ColE1 Ampr KanrlacZα | Invitrogen |
DNA extractions, purification, and manipulations.
Large-scale plasmid DNA extractions of the vectors pAUL-A and pAM401 were carried out using the Qiagen Maxi-Prep kit (Qiagen Inc., Mississauga, ON, Canada) according to the manufacturer's instructions. Small-scale genomic DNA extractions were performed using the UltraClean microbial DNA isolation kit (MoBio Laboratories Inc., Carlsbad, CA). DNA-modifying enzymes (XbaI restriction endonuclease, T4 ligase, and calf intestinal alkaline phosphatase [CIAP]) were provided by New England BioLabs (Pickering, ON, Canada). DNA TOPO cloning kits were supplied by Invitrogen (Burlington, ON, Canada). KlenTaq and Taq DNA polymerases were supplied by Sigma-Aldrich.
Construction of a chromosomal ΔTreA deletion mutant.
A 462-bp deletion in the center of the treA gene, encoding phosphotrehalase, in L. monocytogenes 568 was created by allelic replacement by the gene splicing and overlap extension (SOE) method (29). Briefly, genomic DNA from an overnight culture of L. monocytogenes 568 was used as the template for two separate PCRs. Primers SOE-T6P-A (5′-ACGGTCTAGAACTGATTCTCTGGCGCTCAT-3′) and SOE-T6P-B (5′-GGATTGATAAAGGCGTGGAAGGCGCAACGATTCATTTTAT-3′) were used to generate a 591-bp amplicon at the 5′ end of the treA gene using KlenTaq polymerase. The second reaction produced a 491-bp amplicon at the 3′ end of the gene and employed primers SOE-T6P-C (5′-TTCCACGCCTTTATCAATCC-3′), and SOE-T6P-D (5′-CTGCTCTAGAGGGGATATTCCCGGTATCAC-3′). SOE-T6P-C was complementary to the 5′ tail of SOE-T6P-B. Sequences for the restriction endonuclease XbaI (underlined regions) were introduced into primers SOE-T6P-A and SOE-T6P-D to facilitate cloning. The PCR products were cleaned with the EZ-10 spin column PCR purification kit (Bio Basic, Inc., Markham, ON, Canada). Amplicons from each of the PCRs were diluted 1,000-fold, mixed in a 1:1 ratio, and used (1 μl) as the template for a new PCR using primers SOE-T6P-A and SOE-T6P-D. The 1,061-bp amplicon was run out on a 1% Tris-acetate-EDTA (TAE) agarose gel, excised, purified (GeneClean gel purification kit; Q-Biogene, CA), and TA cloned into E. coli TOP10 using the TOPO TA cloning kit. The presence of the insert was confirmed by digestion of the extracted plasmid with XbaI and fractionation on a TAE agarose gel. The ΔtreA fragment was excised, purified as before, and then ligated into the dephosphorylated temperature-sensitive shuttle vector pAUL-A with T4 DNA ligase. The resulting plasmid, designated pAULA:ΔTreA, was electroporated into E. coli DH5α (3, 36) and transformants selected on LB agar containing 250 μg/ml Erm.
Electroporation into L. monocytogenes 568 and selection of its isogenic treA deletion mutant.
A maxiprep of the pAULA:ΔTreA plasmid was prepared, and the DNA was electroporated into competent L. monocytogenes 568 cells (22). Selection of transformed Listeria was performed on BHI agar containing Erm (5 μg/ml) after 3 to 4 days at 28°C. The presence of the pAULA:ΔTreA plasmid was confirmed by plasmid extraction and restriction digestion of the DNA with XbaI. For the integration of pAULA:ΔTreA into the chromosome of L. monocytogenes 568, a single colony of a confirmed plasmid-carrying transformant was patched onto BHI agar with 5 μg/ml Erm, followed by incubation at 42°C for 2 days. The colony was transferred to a fresh plate and incubated under the same conditions. A total of three transfers/incubations were conducted. Merodiploid intermediates were confirmed by the presence of two amplicons (wild-type allele, 1,523 bp; deleted allele, 1,061 bp) by PCR using primers SOE-T6P-A and SOE-T6P-D. Spontaneous excision of the integrated plasmid via intramolecular homologous recombination of the deleted allele was achieved by 3 subsequent overnight incubations in BHI at 28°C in the absence of Erm followed by a final overnight incubation at 42°C in BHI. The culture was diluted in 0.1% peptone water, spiral plated onto BHI agar, and incubated at 35°C for 48 h. One hundred colonies were patched onto BHI and then replica plated onto MWB containing 0.5% trehalose as the sole source of carbon. The plates were incubated at 37°C for 48 h, and colonies displaying poor growth on the MWB-trehalose medium were subjected to PCR using primers SOE-T6P-A and SOE-T6P-D to select recombinants displaying a single amplicon of 1,061 bp. The in-frame deletion was confirmed by DNA sequencing, and the deletion mutant was named L. monocytogenes 568:ΔTreA.
Complementation of L. monocytogenes 568:ΔTreA.
Phosphotrehalase activity was restored in L. monocytogenes 568:ΔTreA by transforming the mutant with the shuttle vector pAM401 carrying an intact copy of the treA gene. Briefly, a full-length amplicon for the treA and treB genes, including the putative promoter region upstream of treB, was amplified from L. monocytogenes 568 using the primers TreOp-F (5′-CCTTCTAGAGGTCACCGGTTGGAT AAA TTCC-3′) and TreOp-R (5′-AATCTAGA ACCGGCAATGTCCAATAATTT-3′). The underlined bases denote XbaI restriction sites to facilitate cloning. The amplicon was TA cloned into TOPO pCR2.1, and transformants were selected on LB with Amp (100 μg/ml). The plasmid carrying the insert was isolated, purified, and then digested with XbaI. The insert was gel purified and ligated to pAM401 linearized with XbaI and dephosphorylated with CIAP. The resulting construct, pAM401:TREOP, was electroporated into competent cells of the deletion mutant L. monocytogenes 568:ΔTreA as described above. Also, pAM401 containing no insert was electroporated into the wild-type L. monocytogenes 568 to create L. monocytogenes 568/pAM401. This strain was used as a control in later experiments to account for possible fitness losses due to the presence of the plasmid in the complemented mutant. The transformants were selected on BHI containing Crm (10 μg/ml) and verified by PCR using the TreOp-F and TreOp-R primers. Restoration of treA activity was confirmed by growth on MWB-trehalose and measurements of phosphotrehalase activity (see below).
Preparation of crude cell extracts for enzyme assays.
Extracts from the mutant and wild-type L. monocytogenes 568 were prepared according to the method of Helfert et al. (26) with modifications. The cultures were grown in 50 ml of BHI with 1% trehalose for 16 h at 30°C and 200 rpm in an orbital shaker. The cells were harvested by centrifugation (6000 × g, 15 min, 0°C). Cell pellets were washed twice by resuspending them in 40 ml of bis-Tris sonication buffer (50 mM bis-Tris [pH 7.0], 10 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 2 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and repeating the centrifugation. The pellets were then resuspended in 5 ml of sonication buffer and sonicated on ice at 60% power for 20 s with a 30-s rest between pulses (the total pulse time was 5 min) with a Virsonic 600 cell disrupter (Virtis, Gardiner, NY). Cellular debris was removed by centrifugation (10,000 × g, 30 min, 0°C). The quantity of total protein in the supernatant was determined using an Ettan 2D-Quant kit (Amersham Biosciences, Inc., Baie d'Urfé, PQ, Canada).
Assay for α,α-(1,1)-phosphotrehalase activity.
The phosphotrehalase activity in crude cell extracts of the mutant and parent strains was measured as the amount of glucose liberated from the hydrolysis of T6P using a commercial glucose hexokinase (HK) kit (Sigma-Aldrich). A 500-μl aliquot of crude cell extract was incubated with 10 mM T6P for 2 h at 37°C. Control samples contained no added T6P. Following incubation, sample aliquots (200 μl) were mixed with 1.0 ml of glucose assay reagent. Negative control samples were combined with 1.0 ml sterile double-distilled water (ddH2O). Results were reported as liberated glucose per mg cell protein.
Effect of glucose on expression of treA.
In order to assay the repression of TreA activity by glucose, L. monocytogenes 568 was grown overnight at 37°C in MWB containing 10 mM trehalose. The overnight culture (10 μl) was used to seed 50 ml of fresh MWB containing 10 mM trehalose and 0, 1, 5, and 10 mM glucose. The cultures were incubated at 37°C at 200 rpm for 20 h and then harvested by centrifugation. The treA activity was measured as described above.
Extraction of intracellular listerial trehalose-6-phosphate.
The intracellular accumulation of T6P was determined in L. monocytogenes 568 and its treA mutant after growth (18 h, 30°C, 50 ml) in 5 different media: tryptic soy broth (TSB), TSB-YE, TSB with 1% trehalose (TSB-Tre), BHI, and MWB with 1% glucose. Cells were harvested by centrifugation (6,000 × g for 10 min at 0°C), resuspended in 1 ml of PBS (pH 7.0), and sonicated as before. Subsequently, an equal volume of ice-cold 2 M HClO4 was added along with glass beads, and tubes were shaken for 10 min. The cellular debris was removed by centrifugation at 20,000 × g (30 min, 0°C), and the samples were then neutralized with 3 M K2CO3 before being passed through LC-NH2 solid-phase ion-exchange columns (Supelco, Bellefonte, PA). Elution of T6P was carried out with 2 ml of 2.5 M ammonium hydroxide (pH 12.5). The ammonia was removed by freeze-drying followed by redissolving the sample in 50 mM bis-Tris buffer (pH 7.0).
Determination of T6P levels in cell extracts.
Intracellular T6P in L. monocytogenes 568 and 568:ΔTreA was determined using the method of Van Vaeck et al. (56). The inducible expression vector pCVV1 containing the intact treA gene from Bacillus subtilis (a gift from C. Van Vaeck and J. M. Thevelein, Katholieke Universiteit, Leuven, Flanders, Belgium) was electroporated into competent E. coli XL10-Gold (Stratagene, La Jolla, CA). Transformants were recovered on LB containing 100 μg/ml Amp. Isolation and purification of the overexpressed enzyme was carried out following the methods of Gotsche and Dahl (23) and Van Vaeck et al. (56) with modifications. In brief, 10 ml of an overnight culture of E. coli XL10-Gold carrying pCVV1 was used to seed 1 liter of LB broth containing 0.5 mM IPTG (isopropyl-β-d-thiogalactoside) and Amp (100 μg/ml). The culture was grown for 5 h at 37°C and 200 rpm. The cells were harvested at 6.000 × g for 10 min at 4°C. Washing and lysis of the pellets by sonication were carried out as described above, followed by removal of cellular debris by ultracentrifugation at 40,000 × g for 30 min at 4°C (Beckman SW40 rotor). Proteins in the supernatant were precipitated by stepwise addition of ammonium sulfate to 80% saturation. The solution was stirred overnight at 4°C, and the precipitate was collected by centrifugation (15,000 × g, 4°C, 20 min). The pellet was dissolved in 5 ml of extraction buffer and then dialyzed against several changes of bis-Tris buffer. Proteins were first fractionated by passage through a DEAE Sephadex A-50 anion-exchange column and elution in 5-ml fractions with an NaCl gradient. The presence of the 64-kDa phosphotrehalase enzyme in the fractions was determined on a 12% SDS-polyacrylamide gel by the method of Laemmli (33). These fractions were pooled, precipitated with ammonium sulfate, centrifuged, and redissolved in bis-Tris buffer. For size fractionation, the sample was loaded on a Superdex 75 gel filtration column equilibrated with bis-Tris buffer. Elution of 2-ml fractions was carried out with the same buffer. After analysis on a 12% SDS-polyacrylamide gel, fractions with a prominent 64-kDa band were pooled and analyzed for enzyme activity using pure T6P as a substrate (see above). In addition, the activity of the purified enzyme was quantified using p-nitrophenyl-α-d-glucopyranoside (PNPG) as a substrate, where 1 unit of phosphotrehalase will hydrolyze 1 nmol of PNPG/min at 37°C. The specific activity was calculated based on units/mg of protein as determined with the Ettan 2D Quant protein kit.
For the TP6 assay, various quantities of the intracellular cell extracts were mixed with 2.0 U of purified phosphotrehalase, and the total reaction volume was brought to 200 μl. The mixture was incubated at 37°C for 2 h. The reactions were run in duplicate. The amount of glucose liberated in the reaction was measured using the glucose HK assay.
Extraction and assay of trehalose from bacterial cultures.
Overnight TSB cultures were grown at 30°C and used to inoculate fresh 100-ml cultures in TSB, BHI, and TSB-Tre. The cultures were grown for 16 h at 30°C at 200 rpm and harvested by centrifugation (6,000 × g, 15 min, 2°C). The pellets were washed twice in phosphate-buffered saline (PBS) (pH 7.0), followed by mixing with 4 ml of sterile ultrapure water tempered to 2°C and sonication on ice. Cellular debris was removed by centrifugation (10,000 × g for 20 min) and saved for protein analysis. Precipitation of proteins in the supernatant was achieved by addition of acetic acid to pH 2.0, incubation on ice for 1 h, and centrifugation as before. The pH of each extract was adjusted to 6.0 with KOH (1 M), followed by storage at −80°C until analysis. The pellet from the acid precipitation was combined with the original pellet and solubilized in Laemmli buffer (33). The amount of extracted protein in the pellet was determined using the Ettan 2D-Quant kit according to the manufacturer's instructions (Amersham Biosciences).
Since trehalose consists of two glucose molecules, the glucose HK assay was used to measure the amount of glucose liberated following the hydrolysis of trehalose with purified porcine kidney trehalase (Sigma-Aldrich). Two hundred microliters of the deproteinized cell extracts was incubated with 0.05 U of trehalase at 37°C for 8 h. In order to compensate for endogenous glucose in the samples, controls with no trehalase were incubated along with each trehalase-treated sample. Also, standards containing pure trehalose were analyzed. Following incubation, the glucose content in all samples were determined by the glucose HK method, and intracellular the trehalose content {0.5 × ([glucosetrehalase] − [glucoseno trehalase])} was reported as μg/mg cell protein.
Assessment of stress tolerance.
L. monocytogenes 568:ΔTreA and wild-type L. monocytogenes 568 were subjected to several stress conditions. For heat treatments, overnight cultures in BHI, TSB, TSB-YE, and TSB-Tre were grown at 30°C. Aliquots (10 μl) from each culture were used to inoculate 5 ml of the same medium, followed by incubation for 16 h at 30°C. For cold-temperature growth curves, the cells were first cultured in BHI for 5 days at 4°C. For the remaining stress studies, the cells were cultured in BHI and then grown in 5 ml of BHI with 1% trehalose for 16 h at 30°C. Harvest of cultures was by centrifugation at 6,000 × g for 10 min at room temperature followed by washing once with 0.1% peptone water and resuspension in the appropriate test medium (see below).
Heat treatments.
The cell pellets were washed twice in 0.1% peptone water and then diluted in the original medium to obtain initial concentrations of 107 to 108 CFU/ml. Samples (100 μl) from each treatment were distributed into 16 thin-walled 0.2-ml PCR tubes. The tubes were placed in a Biometra Tgradient thermocycler (Whatman Biometra, Göttingen, Germany) and heat treated at 52°C for 105 min. Duplicate samples were removed at 15-min intervals and cooled in an ice-water bath for 1 min. The samples were diluted in sterile 0.1% peptone water and spiral plated onto TSA-YE containing 1% sodium pyruvate. CFU were enumerated following 72 h of incubation at 35°C, and the log values were plotted against time to obtain thermal death time curves. The entire experiment was repeated 3 times.
Cold growth.
Duplicate tubes containing 5 ml of BHI tempered to 4°C were inoculated with 10 μl of the 5-day culture. The tubes were incubated at 4°C without shaking, and aliquots were removed periodically for absorbance measurements and colony enumerations. Absorbance measurements were taken at 600 nm, and cell enumerations were performed by spiral plating on TSA-YE (48 h at 35°C). The experiment was repeated twice.
Resistance to freeze-thaw cycles.
The cell pellets were washed in sterile phosphate-buffered saline (PBS) (pH 7.0) then resuspended in 10 ml of PBS in 50-ml polypropylene centrifuge tubes to obtain cell densities of 107 to 108 CFU/ml. The initial cell numbers were determined by plating on TSA-YE. The tubes were frozen to −80°C in an ultra-low-temperature freezer. After 2 h, the suspensions were thawed in a water bath (37°C) and an aliquot was diluted and spiral plated. After 30 min at 37°C, the tubes were placed in the freezer, and they were thawed again after 24 h to determine the viable CFU. The freeze-thaw cycles were continued for 9 days, with samples being taken every 24 h. Colony enumerations were performed after 72 h at 35°C. The experiment was repeated 3 times.
Resistance to desiccation.
The washed pellets from each growth medium were resuspended in 0.1% peptone water to cell densities of ca. 108 CFU/ml. Ten microliters of each suspension (i.e., 106 CFU) was aseptically transferred to 3 wells of a 12-well flat-bottomed tissue culture plate, followed by drying for 2 h in a biosafety cabinet (Baker Co., Sanford, ME) with lids removed. Subsequently, the plates were stored in a vacuum jar containing 10- to 20-mesh Drierite desiccant (WA Hammond Drierite Co., Xenia, OH) at ambient temperature (20°C) for 14 days. A Hobo Pro v2 temperature/relative humidity (RH) data logger was also placed inside the jar to record the RH. Following the desiccation period, BHI (5 ml) was added to the wells of each culture plate, which were placed in an incubator at 30°C and monitored for growth (A600). Absorbance readings were taken until the cells reached stationary phase. CFU were determined by extrapolating values from a standard curve relating CFU to absorbance at 600 nm (not shown), and the Microsoft Excel add-in program DMFit (version 2.1; http://www.ifr.ac.uk/safety/dmfit/) was used to determine lag times and growth rates. Three replicates of this experiment were carried out.
Survival in acid, ethanol, hydrogen peroxide, and sodium chloride.
The pellets were resuspended to 107 to 108 CFU/ml in BHI adjusted to achieve one of the following stress conditions: pH 3.5, adjusted with HCl; 18% (vol/vol) ethanol; 0.1% (vol/vol) H2O2; and 20% (wt/vol) NaCl. For each sample treatment, a 100-μl aliquot was removed immediately following mixing, diluted in peptone water, and spiral plated on TSA-YE. Samples were removed and spiral plated at specified time intervals for each sample treatment. All plates were incubated at 35°C for 72 h before colony enumerations. These experiments were replicated at least twice.
Statistical analysis.
Stress test data were subjected to analysis of variance (ANOVA) using SYSTAT software, and pairwise comparisons were assessed by the Tukey method at the 95% confidence level. Critical parameters for analysis of thermal death curves were obtained using the Pruitt-Kamau model (equations 1 and 2) (43). Survivor curves were modeled based on a modification of the Fermi distribution model of Whiting (59) for curves not exhibiting a tailing phenomenon:
where N is the number of survivors at any time (t), N0 is the initial population at t = 0, β is the maximum specific death rate, and tcm is the lag period. For comparison of the overall thermal resistance for the wild type and its mutant, a logistic decimal reduction time (DL) was calculated as previously described by Pruitt and Kamau (43):
Where appropriate, Student's t test (α = 0.05) was used for direct comparison of the means obtained for trehalose or T6P accumulation in the mutant versus the wild type.
RESULTS
Growth kinetics of the deletion mutant.
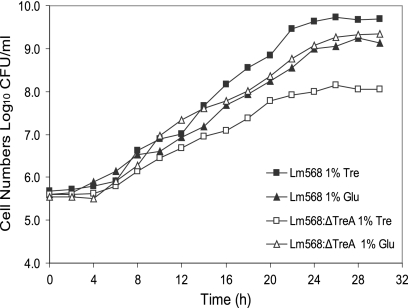
DNA sequencing demonstrated that the L. monocytogenes 568:ΔTreA mutant contained a 462-bp in-frame deletion in the central region of the putative treA gene. The colony morphology of the deletion mutant was not different from that of the wild-type L. monocytogenes 568 when grown on TSA-YE or BHI agar. The growth kinetics in BHI, TSB (data not shown), and MWB with glucose (Fig. 1) were also similar. Interestingly, L. monocytogenes 568:ΔTreA grew in MWB with 1.0% trehalose as the sole carbon source, although the growth rate was significantly (P < 0.05) decreased and a lower maximum cell density was obtained (Fig. 1). To confirm that the mutant was indeed using trehalose and not another constituent in the medium, both the wild-type and the mutant were inoculated into MWB with no added carbon source. Neither culture grew after 48 h at 37°C. Also investigated was the possibility that the apparent utilization of trehalose by the mutant was due to natural degradation of the disaccharide into glucose during incubation at 37°C. However, no glucose could be detected in the sterile MWB with 1% trehalose following 48 h at 37°C using either the glucose HK assay or high-pressure liquid chromatography (HPLC) analysis (data not shown).
Fig. 1.
Growth curves for L. monocytogenes 568 and its treA mutant 568:ΔTreA in modified Welshimer's broth (42) at 37°C and 150 rpm. The values displayed for each data point are the means ± standard deviations (n − 1) for four samples (two replicates from two independent experiments).
Enzymatic activity of crude cell extracts.
According to the published genome map of L. monocytogenes EGD-e (GenBank accession number AL591824), the only putative transport mechanism for the utilization of trehalose is through the enzyme IIBC trehalose-specific PTS (treB) and phosphotrehalase (treA). In order to verify the absence of trehalase activity, the glucose hexokinase assay was used to determine whether glucose is liberated from pure trehalose during incubation for 4 h at 37°C with crude cell proteins extracted from the mutant and the wild type. In neither case were increases in glucose detected in comparison to the control extracts without trehalose.
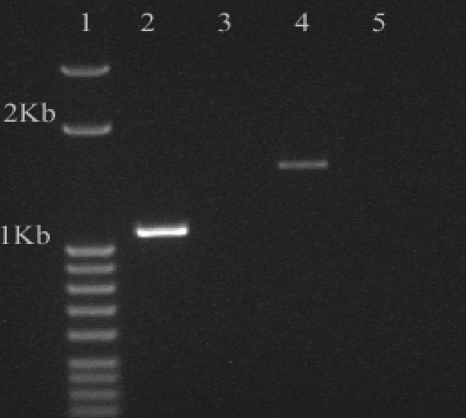
Reverse transcription-PCR (RT-PCR) demonstrated that a truncated phosphotrehalase transcript was produced by the deletion mutant (Fig. 2); however, the enzyme was not active, as free glucose was not produced from T6P during incubation of crude cell extracts from the mutant for 1, 4, and 8 h at 37°C. Conversely, extracts from the wild-type strain grown in MWB (with trehalose) contained high levels of phosphotrehalase activity, since 10 μl of cell extract liberated large quantities of glucose from T6P after only 1 h at 37°C (Fig. 3).
Fig. 2.
Transcriptional analysis by RT-PCR of the treA genes in Listeria monocytogenes 568 and mutant 568:ΔTreA. Lane 1, DNA ladder; lane 2, L. monocytogenes 568:ΔTreA amplicon from cDNA; lane 3, control for L. monocytogenes 568:ΔTreA with no RT added to the reaction mixture; lane 4, L. monocytogenes 568 amplicon from cDNA; lane 5, control for L. monocytogenes 568 with no RT added to the reaction mixture.
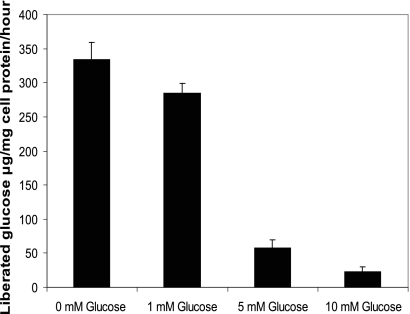
Fig. 3.
Repression of phosphotrehalase activity in Listeria monocytogenes 568 by the presence of glucose in modified Welshimer's broth (MWB) supplemented with 10 mM trehalose and various concentrations of glucose. The results show the means ± standard deviations for four samples (two replicates from two independent experiments).
Repression of treA by glucose.
We examined the effect of the presence of glucose on the treA activity in early-stationary-phase cells of L. monocytogenes 568. Crude extracts from cells grown in different concentrations of glucose demonstrated that the transcription of treA or enzyme activity was repressed by the presence of glucose (Fig. 3). Cells grown in the presence of 10 mM trehalose as the sole carbon source exhibited the highest level of phosphotrehalase activity, with more than 80 μg of glucose/mg cell protein being liberated from T6P after 1 h at 37°C. The addition of glucose (5 and 10 mM) to MWB cultures resulted in only 29 and 12 μg of glucose/mg protein being released from T6P, respectively. At 1 mM glucose, catabolite repression was not as strict, since crude cell extracts liberated 71 μg of glucose/mg cell protein.
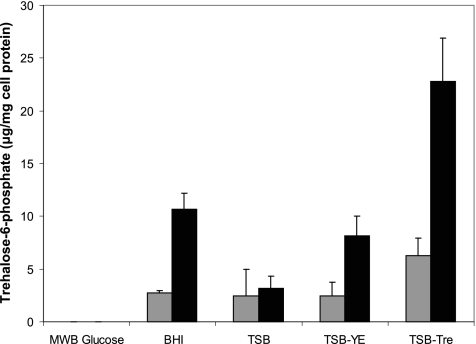
Accumulation of intracellular trehalose-6-phosphate.
L. monocytogenes 568 and 568:ΔTreA were assessed for intracellular levels of T6P after growth in different substrates. The growth medium affected the concentration of T6P in both the wild-type and the mutant. This could be correlated to the amount of trehalose present in the medium (Fig. 4). As such, neither strain displayed detectable levels of T6P after growth in MWB with glucose as the carbon source. However, where low levels of trehalose were detected in TSB (72 mg/liter), the phosphorylated sugar was present in cell extracts in small amounts after growth in this medium: 2.4 and 3.2 μg/mg cell protein for L. monocytogenes 568 and 568:ΔTreA, respectively. Supplementing TSB with yeast extract (0.6%, wt/vol) resulted in a significant increase (P < 0.001) in the intracellular pool of T6P for the mutant (8.2 μg/mg cell protein) but not for the wild-type strain (2.5 μg/mg cell protein), in comparison to growth in the unmodified medium. Yeast extract was found to contain substantial levels of trehalose, as a 0.6% (wt/vol) solution contained approximately 730 mg/liter. The addition of 1% trehalose to TSB resulted in more than twice as much T6P in the wild type (6.3 μg/mg cell protein) as for cells grown in TSB without trehalose, while growth in BHI did not significantly (P > 0.05) raise T6P levels (2.7 μg/mg cell protein), although BHI was found to naturally contain approximately 300 mg/liter of trehalose. In contrast, L. monocytogenes 568:ΔTreA showed a gradient of increasing quantities of T6P depending on the type of medium. Although T6P levels for cells grown in TSB were not significantly (P > 0.05) greater than those for the wild type, the addition of yeast extract and 1% trehalose increased intracellular T6P from levels of 3.2 to 8.2 and 22.8 μg/mg cell protein, respectively. Moreover, free trehalose in BHI was effectively transported into the L. monocytogenes 568:ΔTreA mutant, since cells grown in this medium accumulated the second-highest levels of T6P (10.7 μg/mg cell protein).
Fig. 4.
Intracellular accumulation of trehalose-6-phosphate by L. monocytogenes 568 (gray bars) and L. monocytogenes 568:ΔTreA (black bars) after growth in five different media (MWB plus Glu, TSB, TSB-YE, TSB-Tre, and BHI). The results show the means ± standard deviations for four samples (two replicates from two independent experiments).
Quantification of trehalose in cell extracts and phosphatase activity.
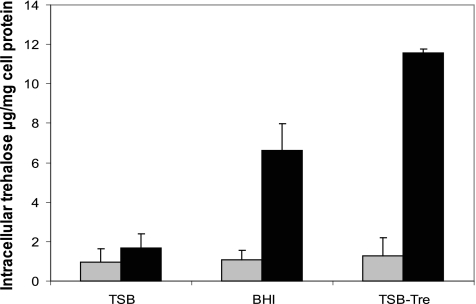
Intracellular trehalose was detected in all cell extracts regardless of treatment or strain (Fig. 5). Trehalose levels for the wild type did not depend (P > 0.05) on the trehalose content in the growth medium, as concentrations ranged from 0.95 to 1.3 μg/mg cell protein. Levels of trehalose in L. monocytogenes 568:ΔTreA extracts from the same treatment were consistently significantly greater (P < 0.05) than those for L. monocytogenes 568; e.g., mutant cells grown in TSB contained twice the trehalose levels found in cells of L. monocytogenes 568 grown in the same medium. In BHI (300 mg/liter trehalose), mutant trehalose concentrations increased significantly (P < 0.05) to approximately 6.6 μg/mg protein, and the level was even higher (11.6 μg/mg protein) when the mutant was grown in TSB-Tre (10,070 mg/liter trehalose). Current knowledge suggests that trehalose is imported into Listeria only via the PTS system as T6P. Therefore, the formation of trehalose in the mutant due to dephosphorylation of T6P was investigated by incubating crude extracts from L. monocytogenes 568:ΔTreA grown in MWB plus 1% glucose with T6P (10 mM) for 4 h at 37°C. After removal of phosphorylated sugars using ion exchange as before, sample trehalose was digested with porcine trehalase for 2 h to release 0.89 ± 0.21 μg glucose/mg protein. Levels of glucose in control extracts not treated with trehalase remained unchanged. This indicates that the crude cell extracts contain phosphatase activity capable of cleaving the phosphate group from T6P.
Fig. 5.
Intracellular trehalose contents in Listeria monocytogenes 568 (gray bars) and its treA mutant L. monocytogenes 568:ΔTreA (black bars) after growth in either TSB, BHI, or TSB-Tre (supplemented with 1% trehalose) for 16 h at 37°C. The results show the means ± standard deviations for four samples (two replicates from two independent experiments).
Heat tolerance as affected by growth medium.
All thermal death curves from the heat treatment (52°C) of early stationary-phase L. monocytogenes 568 and 568:ΔTreA cells from four different broth media displayed a lag period prior to entering a log-linear region of maximum inactivation (not shown). The results for the model parameters are given in Table 2.
Table 2.
Thermal inactivation kinetics parameters for Listeria monocytogenes strains 568 and 568:ΔTreA at 52°C following growth on different mediaa
| Strain and growth medium (trehalose content, mg/liter) | βb | ASEc (β) | tcmd | ASE (tcm) | Logistic DL valuee (min) |
|---|---|---|---|---|---|
| 568 | |||||
| TSB (70) | 0.235 | 0.009 | 12.86 | 1.70 | 22.57 |
| TSB-YE (730) | 0.228 | 0.010 | 12.09 | 1.63 | 22.11 |
| TSB-Tre (10,070) | 0.202 | 0.019 | 15.62 | 3.26 | 26.35 |
| BHI (310) | 0.196 | 0.013 | 18.29 | 2.27 | 29.95 |
| 568:ΔTreA | |||||
| TSB (70) | 0.203 | 0.010 | 11.80 | 2.27 | 23.05 |
| TSB-YE (730) | 0.176* | 0.012 | 14.31 | 3.13 | 27.14 |
| TSB-Tre (10,070) | 0.161* | 0.005 | 19.21* | 1.43 | 33.31* |
| BHI (310) | 0.152* | 0.004 | 29.03* | 1.10 | 44.09* |
The inactivation kinetics was modeled using the modified Fermi distribution function (43). *, significantly different from value for the wild-type 568 for the same treatment (P < 0.05).
β, inactivation rate parameter for the log-linear region of the death curve.
ASE, asymptotic standard error.
tcm, lag parameter prior to the region of maximum inactivation rate.
DL, time for the initial log decline when a lag phase exists (equation 2).
The DL (ca. 30 min) for the wild-type strain in BHI was significantly longer (P < 0.05) than DL values obtained in TSB or TSB-YE (22.6 and 22.1 min, respectively). However, there was no significant difference (P > 0.05) between L. monocytogenes 568 cells grown and heated in BHI or TSB-Tre. This indicated that BHI (trehalose content, 310 mg/liter) in general induces a higher level of heat resistance in L. monocytogenes 568 than the other media used in this study. Growth in TSB (trehalose content, 70 mg/liter) resulted in similar thermal resistance for the mutant and the parent strain, as DL values were not significantly different (P > 0.05) from each other (Table 2). Supplementation of TSB with either 0.6% yeast extract (trehalose content, 730 mg/liter) or 1% trehalose (10,070 mg/liter) resulted in significant increases (P < 0.05) in heat resistance for the mutant, with TSB-Tre cultures showing the highest level of thermotolerance (DL = 33.3 min) among the TSB-based media (Table 2). This progression in thermal resistance correlated with increases in both intracellular T6P accumulated from the uptake of trehalose (Fig. 4) and high intracellular trehalose levels (Fig. 5). Interestingly, L. monocytogenes 568:ΔTreA cells grown in BHI were by far the most thermal-resistant cells observed in this study (DL = 44.1 min) despite the fact that lower T6P and/or trehalose levels were found in these cells than in cells grown in TSB-Tre (Fig. 4 and 5) or TSB-YE. This indicates that other factors in BHI apart from an endogenous content of trehalose increased the thermal resistance of L. monocytogenes 568:ΔTreA. The enhanced thermal resistance of the mutant seen in BHI and TSB-Tre appears to be due mainly to an enhancement of the lag period (tcm values) (Table 2).
Tolerance to other environmental stresses.
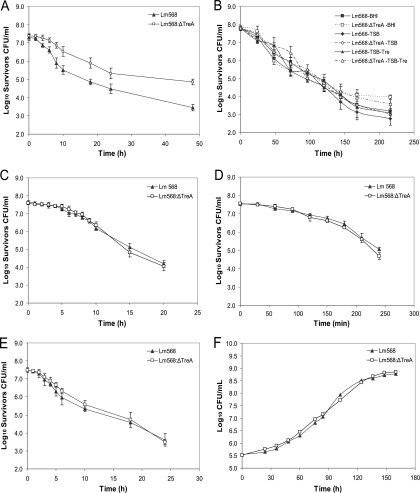
In addition to heat stress, several other stress factors were examined to evaluate the phosphotrehalase mutant's stress hardiness. The L. monocytogenes 568:ΔTreA mutant demonstrated an increased capacity to survive high osmolarity (20% NaCl), repeated freeze-thaw cycles, and desiccation. After 6 h in BHI with 20% NaCl, survivor counts for the phosphotrehalase mutant were significantly higher (P < 0.05) than those for L. monocytogenes 568 at all subsequent sampling times, resulting in 1.5 log more L. monocytogenes 568:ΔTreA survivors than wild-type survivors after 48 h (Fig. 6A). Less dramatic but noteworthy was the result of the exposure to repeated freeze-thaw cycles. Although, the mutant generally did not survive significantly (P > 0.05) better than the wild type for the majority of sampling times, the number of L. monocytogenes 568:ΔTreA survivors in BHI were 0.8 log greater than that of L. monocytogenes 568 survivors in the same medium (Fig. 6B). Moreover, L. monocytogenes 568:ΔTreA survived desiccation (RH of 16%) better than L. monocytogenes 568, as desiccated 568:ΔTreA cells exhibited a significantly (P < 0.05) shorter lag time (23.2 h) than L. monocytogenes 568 cells (27.6 h) prior to the onset of exponential growth following the treatment (Table 3). On the other hand, there were no significant differences (P > 0.05) between the survival and growth of the wild type and L. monocytogenes 568:ΔTreA when they were subjected to the other stress factors (ethanol, H2O2, low pH, and low temperature) examined in this study (Fig. 6C to F).
Fig. 6.
Effects of various stressors on the survival or growth of Listeria monocytogenes 568 and mutant L. monocytogenes 568:ΔTreA. Cells were subjected to high osmolarity (20% NaCl in BHI) (A), repeated freeze-thaw cycling (−80 and 37°C every 24 h in BHI, TSB, and TSB-Tre) (B), ethanol (18%, vol/vol) (C), hydrogen peroxide (0.1%, vol/vol) (D), high acidity (pH 3.5) (E), and low temperature (4°C) (F). The results are the mean populations for three independent experiments with corresponding standard errors.
Table 3.
Desiccation tolerances of Listeria monocytogenes strain 568 and its phosphotrehalase mutant 568:ΔTreAa
| Treatment | 568 |
568:ΔTreA |
||
|---|---|---|---|---|
| Lag time (h) | Specific growth rate (h−1) | Lag time (h) | Specific growth rate (h−1) | |
| Untreated | 4.36 ± 0.64 | 0.882 ± 0.039 | 4.56 ± 0.41 | 0.855 ± 0.029 |
| Desiccated | 27.57 ± 0.90* | 0.861 ± 0.018 | 23.24 ± 0.75* | 0.878 ± 0.022 |
Cells (∼106 CFU) were air dried for 2 h in the bottom of 12-well tissue culture plates and then desiccated for 14 days at 16% RH and 20°C. Following desiccation, BHI was added to the wells and growth of untreated (control) and desiccated cells at 30°C monitored by measuring absorbance at 30-min intervals. Analysis of growth curves was done using DMFit (version 2.1; http://www.ifr.ac.uk/safety/dmfit/). *, significant difference (P < 0.05) between the means from triplicate experiments as determined by Student's t test.
Complementation of 568:ΔT6P.
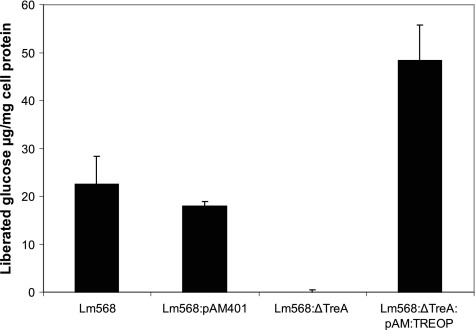
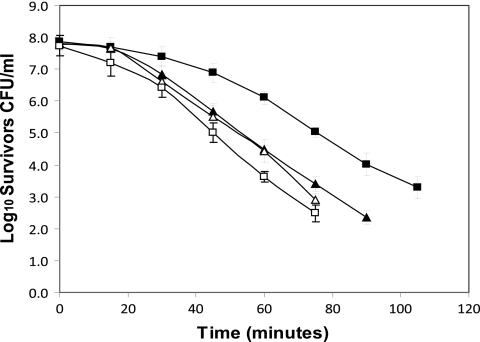
In order to confirm that the deletion in the treA gene was responsible for the observed phenotype, L. monocytogenes 568:ΔTreA was back complemented with a vector containing the intact treA gene. As the promoter for the tre operon resides upstream from the treB gene (encoding the trehalose-specific PTS enzyme IIBC), it was decided to clone the entire treA/treB region into the XbaI site of the Gram-positive shuttle vector pAM401 to create pAM:TREOP. The construct was electroporated into mutant L. monocytogenes 568:ΔTreA and the resultant complemented strain named ΔTreA:pAM:TREOP. The phosphotrehalase activity in ΔTreA:pAM:TREOP was restored, as crude cell extracts liberated nearly 50 μg of glucose/mg cell protein from 10 mM T6P after 1 h at 37°C. This indicated an overexpression of treA, since extracts from L. monocytogenes 568 liberated less than half this amount (22.5 μg/mg cell protein) of glucose under the same conditions (Fig. 7). Moreover, the thermal death profile for the complemented mutant cells demonstrated a marked reduction in thermotolerance relative to mutant L. monocytogenes 568:ΔTreA (Fig. 8). The calculated DL values for L. monocytogenes 568 and ΔTreA:pAM:TREOP were 30.5 and 23.1 min, respectively. However, the DL value of 45.9 min for mutant L. monocytogenes 568:ΔTreA was significantly greater (P < 0.05) (Table 4). It should be noted that the resulting phenotype was the result of the activation of the treA gene and not due to the presence of the pAM401 vector, since the phosphotrehalase activity and heat resistance of L. monocytogenes 568/pAM401 (DL value = 28.5 min) did not vary significantly (P > 0.05) from those of wild-type L. monocytogenes 568.
Fig. 7.
Phosphotrehalase activity in crude cell extracts from L. monocytogenes 568 (wild type), L. monocytogenes 568/pAM401 (wild type with pAM401 shuttle vector), L. monocytogenes 568:ΔTreA (treA deletion mutant of L. monocytogenes 568) and its trans complement L. monocytogenes 568:ΔTreA:pAM:TREOP (the entire tre operon carried on the pAM401 shuttle vector) as measured by the glucose hexokinase assay. The results show the means ± standard deviations (n − 1) for four samples (two replicates from two independent trials).
Fig. 8.
Heat resistance of L. monocytogenes 568, the treA mutant, and the back-complemented treA mutant. Wild-type strain L. monocytogenes 568 (▴), L. monocytogenes 568/pAM401 (▵), deletion mutant L. monocytogenes 568:ΔTreA (■), and pAM401:TREOP-transformed treA mutant ΔTreA:pAM:TREOP (□), were grown for 16 h at 37°C in BHI broth and subjected to a heat treatment of 52°C. The plot represents the mean survivors from three independent heat trials ± standard errors.
Table 4.
Thermal inactivation kinetics parameters at 52°C for Listeria monocytogenes 568, 568:ΔTreA, complemented mutant ΔTreA:pAM:TREOP, and 568/pAM401a
| Strain | βb | ASEc (β) | tcmd | ASE (tcm) | Logistic DL valuee (min) |
|---|---|---|---|---|---|
| 568 | 0.175 | 0.006 | 17.47 | 1.76 | 30.54 |
| 568:ΔTreA | 0.141* | 0.005 | 29.64* | 1.79 | 45.88* |
| ΔTreA:pAM:TREOP | 0.186 | 0.008 | 10.79* | 2.12 | 23.08 |
| 568/pAM401 | 0.174 | 0.012 | 15.35 | 3.66 | 28.49 |
The inactivation kinetics was modeled using the modified Fermi distribution function (43). *, significantly different from value for the wild-type 568 for the same treatment (P < 0.05).
β, inactivation rate parameter for the log-linear region of the death curve.
ASE, asymptotic standard error.
tcm, lag parameter prior to the region of maximum inactivation rate.
DL, time for the initial log decline when a lag phase exists (equation 2).
DISCUSSION
In this study, we have demonstrated that a 462-bp deletion in the treA gene (lmo1254 for strain EGD-e) of L. monocytogenes strain 568 results in a thermo- and osmotolerant phenotype when the cells are grown in complex media containing trehalose. To our knowledge this represents the first study investigating the putative trehalose operon of Listeria. Based on DNA sequence, the predicted protein encoded by treA in L. monocytogenes EGD-e is highly similar to α,α-(1,1)-phosphotrehalase in Bacillus subtilis (12, 23, 47, 48) and to T6P hydrolase, the product of the treC gene, of E. coli (45). The predicted trehalose operon in Listeria was also shown to be similar in organization to the catabolic operons of other closely related bacteria such as Lactococcus lactis and Lactobacillus plantarum (1). In addition to treA, this operon also includes treB, which encodes a trehalose-specific IIBC transport protein complex required for the phosphorylation of trehalose via the phosphoenolpyruvate PTS (PEP-PTS) (47). The operon is regulated by the product of treR, a protein highly similar to the GntR-like family of regulators. The deletion of this gene in Bacillus subtilis strain 168 results in the constitutive expression of the operon (48).
In B. subtilis, T6P is formed during the translocation of trehalose across the cytoplasmic membrane via the PEP-PTS, and this in turn is hydrolyzed by phosphotrehalase to glucose and glucose-6-phosphate. We have demonstrated that the product of treA is indeed required for the cleavage of T6P in L. monocytogenes 568. Crude cell lysates from the wild-type strain efficiently hydrolyzed pure T6P, while L. monocytogenes 568:ΔTreA lysate was devoid of this activity. According to the genomic map of L. monocytogenes strain EGD-e, there does not appear to be another putative mechanism for the passage of trehalose into the cytoplasm. Also, L. monocytogenes appears not to produce exogenous trehalases, as free glucose was not detected in MWB following growth of L. monocytogenes 568 with trehalose as the sole carbon source. Therefore, trehalose must first pass through a trehalose-specific or semispecific channel in the cytoplasmic membrane, requiring phosphorylation by a trehalose-specific enzyme IIBC complex (40) followed by cleavage of T6P by phosphotrehalase in the cytosol.
Depending on the trehalose concentrations in the growth media, our treA mutant accumulated high levels of T6P. In B. subtilis, trehalose is the external molecular inducer of the operon, whereby, in its absence, the TreR repressor turns down the transcription of treA (48). Moreover, T6P acts as the internal inducer for treA, where the phosphorylated sugar interferes with the repressor/tre-operator interaction, thus allowing for the transcription of the treA gene to proceed. Additionally, a cis-acting catabolite responsive element (CRE) has been identified in the promoter region of the operon just upstream of the open reading frame (ORF) for treB (37). Catabolites such as glucose, fructose, or mannitol repress the uptake of trehalose (12, 19, 26). Here, glucose was shown to act as a repressor in a concentration-dependent manner, as decreasing phosphotrehalase activities were observed in extracts from L. monocytogenes 568 cells cultured in MWB with trehalose and increasing levels of glucose.
Studies conducted with yeasts have indicated that a buildup of T6P is toxic and results in increased sensitivity to environmental stresses (13, 17, 20, 55, 56). Duong et al. (14) also found that a Lactobacillus acidophilus treB (phosphotrehalase) deletion mutant was intolerant to stresses normally endured by the wild-type strain. In our study, elevated concentrations of intracellular T6P in L. monocytogenes 568:ΔTreA came with a significant increase in the thermo- and osmotolerance relative to those of the parent L. monocytogenes 568. As the phosphorylated version of the disaccharide does not provide the same protection as trehalose (30, 39), this cannot explain the increase in stress hardiness. However, the increase in T6P came together with an accumulation of trehalose, reaching levels corresponding to ca. 50% of the T6P levels. Moreover, as no trehalase activity was observed in these extracts, trehalose levels remained stable. To explain the presence of trehalose in the mutant, dephosphorylation of T6P in crude mutant cell extracts was demonstrated in repeated experiments. This dephosphorylation of T6P and conversion into the compatible solute, trehalose, may be through the action of nonspecific phosphatases (15), which could explain why high levels of T6P are not causing the stress sensitivity. The same benefit may not be seen in phosphotrehalase mutants of other microorganisms such as E. coli, which possesses a separate mechanism for trehalose synthesis/degradation, including a trehalase that would directly cleave the trehalose molecule, thus preventing its simultaneous accumulation (53). Also, the facts that L. monocytogenes 568:ΔTreA grew at a highly reduced rate in MWB with trehalose as the sole carbon source and that a truncated transcript from treA was detected may suggest some residual enzyme activity below the sensitivity of our assay.
When exposed to environmental stresses, intracellular trehalose can provide protection to cells of both bacteria and yeasts. In Saccharomyces cerevisiae, biosynthesis of the disaccharide is initiated by the same stimuli that induce a heat shock response (16, 17, 30, 38). In E. coli, the putative stationary-phase sigma factor σs controls the expression of otsA and otsB, the genes encoding T6P synthase and T6P phosphatase, respectively. However, this is only for stationary-phase-associated thermotolerance, as trehalose biosynthesis is not needed for the development of adaptive thermotolerance in logarithmic-phase cells (27). Back complementation with treA (and treB) removed the intracellular trehalose and decreased the thermotolerance at 52°C in comparison to that of L. monocytogenes 568:ΔTreA; however, the ΔTreA:pAM:TREOP complemented strain was also slightly more heat sensitive than wild-type L. monocytogenes 568. This may be explained by the fact that the trans-complemented mutant overexpresses phosphotrehalase activity. Normally, the treA gene is induced by the presence of T6P reaching a threshold level. Therefore, in the presence of extracellular trehalose, a basal intracellular T6P level exists. However, if treA is constitutively overexpressed, accumulation of T6P would cease and therefore no dephosphorylation of T6P could occur in ΔTreA:pAM:TREOP; hence, trehalose would be absent in the cytoplasm. The mechanism by which trehalose confers thermotolerance is thought to be through its interaction with cellular proteins, preventing their denaturation and aggregation. Therefore, trehalose may act as a molecular chaperone working synergistically with heat shock proteins at elevated temperatures (31, 57).
In addition to having elevated heat resistance, the treA mutant was also more resistant than L. monocytogenes 568 to high osmolarity and desiccation. A 1.5-log increase in survivors was observed for the mutant over L. monocytogenes 568 after 48 h in the presence of 20% NaCl. Since desiccation can be viewed as an extreme case of osmotic stress (8, 41), further evidence for increased resistance to osmotic stress and membrane protection is provided by the finding that L. monocytogenes 568:ΔTreA had shorter recovery times than L. monocytogenes 568 when reconstituted in BHI after desiccation. This observation was presumably due to the presence of more surviving and/or undamaged cells. Listeria spp. normally deal with osmotic stress by accumulating the compatible solutes glycine betaine, carnitine, and proline (51). However, other bacteria display an osmotically induced accumulation of trehalose (44). For example, E. coli will accumulate trehalose under high osmolarity if the preferred osmolyte, glycine betaine, is not available (53). The preference for glycine betaine over trehalose under desiccation is interesting, since in E. coli it gives no survival advantage, yet accumulated endogenous trehalose provides significant protection for these cells (58). This indicates that for E. coli the role of these compatible solutes differs during conditions of drying and those of milder osmotic stress.
In other microorganisms, such as E. coli and Saccharomyces cerevisiae, intracellular trehalose accumulation also gives protection against high alcohol concentrations, oxidative stress, and cold temperatures (6, 31, 50). However, enhanced survivability was not observed when L. monocytogenes 568:ΔTreA was subjected to these stressors. L. monocytogenes 568 appears to have a naturally high resistance to ethanol and H2O2, relative to other L. monocytogenes strains (34). Therefore, the margin of protective capacity may be too narrow to expect increased survivability for the mutant. Also, since listeriae are known psychrotrophs (5, 54), it is not surprising that the accumulation of trehalose by the mutant, did not affect its growth profile at 4°C. However, in the case of mesophiles, such as E. coli, the accumulation of trehalose would be highly advantageous for survival at 4°C (31).
To conclude, this is the first study reporting on any of the genes of the putative trehalose operon in L. monocytogenes. We have demonstrated that lmo1254 in L. monocytogenes 568 indeed encodes a phosphotrehalase that is required for the hydrolysis of T6P. Disruption of the treA gene results in the accumulation of T6P which is subsequently dephosphorylated, resulting in a buildup of the compatible solute trehalose. Although listeriae do not have a natural mechanism for the synthesis of trehalose as seen in other bacteria (e.g., E. coli), we have shown that the subsequent accumulation of intracellular trehalose in an L. monocytogenes strain 568 treA mutant leads to a phenotype of enhanced resistance against heat, high osmolarity, desiccation, and freeze-thaw cycling stresses. Therefore, in the artificially induced mutant, trehalose provides benefits similar to those observed in bacteria naturally synthesizing this molecule as a protectant against harsh conditions. This mutant may be used as a model system to gain a better understanding of the protective capacity of organic osmolytes in Listeria monocytogenes and other closely related bacteria.
ACKNOWLEDGMENTS
We thank Michael Jordan and Kathleen Munro Pennell for their technical support with HPLC.
Funding of this research was through the support of Agriculture and Agri-Food Canada (AAFC) and the Natural Sciences and Engineering Research Council of Canada (NSERC).
Footnotes
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Andersson U., Molenaar D., Rådström P., de Vos W. M. 2005. Unity in organization and regulation of catabolic operons in Lactobacillus plantarum, Lactococcus lactis and Listeria monocytogenes. Syst. Appl. Microbiol. 28:187–195 [DOI] [PubMed] [Google Scholar]
- 2. Argüelles J. C. 2000. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol. 174:217–224 [DOI] [PubMed] [Google Scholar]
- 3. Ausubel F. A., et al. 1990. Current protocols in molecular biology. Wiley-Interscience, New York, NY [Google Scholar]
- 4. Baird-Parker A. C., Davenport E. 1965. The effect of recover medium on isolation of Staphylococcus aureus after heat treatment and after storage of frozen or dried cells. J. Appl. Bacteriol. 28:390–402 [DOI] [PubMed] [Google Scholar]
- 5. Bayles D. O., Annous B. A., Wilkinson B. J. 1996. Cold stress proteins induced in Listeria monocytogenes in response to temperature downshift and growth at low temperatures. Appl. Environ. Microbiol. 62:1116–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benaroudj N., Lee D. H., Goldberg A. L. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276:24261–24267 [DOI] [PubMed] [Google Scholar]
- 7. Bhumiratana A., Anderson R. L., Costilow R. N. 1974. Trehalose metabolism by Bacillus popillae. J. Bacteriol. 119:484–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breeuwer P., Lardeau A., Peterz M., Joosten H. M. 2003. Desiccation and heat tolerance of Enterobacter sakazakii. J. Appl. Microbiol. 95:967–973 [DOI] [PubMed] [Google Scholar]
- 9. Cánovas D., Fletcher S. A., Hayashi M., Csonka L. N. 2001. Role of trehalose in growth at high temperature of Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:3365–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakraborty T., et al. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clark C. W., Witter L. O., Ordal Z. J. 1968. Thermal injury and recovery of Streptococcus faecalis. Appl. Microbiol. 16:1764–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dahl M. K. 1997. Enzyme IIGlc contributes to trehalose metabolism in Bacilus subtilis. FEMS Microbiol. Lett. 148:233–238 [Google Scholar]
- 13. De Virgilio C., et al. 1993. Disruption of TPS2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur. J. Biochem. 212:315–323 [DOI] [PubMed] [Google Scholar]
- 14. Duong T., Barrangou R., Russell W. M., Klaenhammer T. R. 2006. Characterization of the tre locus and analysis of trehalose cryoprotection in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 72:1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elbein A. D., Pan Y. T., Pastuszak I., Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R–27R [DOI] [PubMed] [Google Scholar]
- 16. Eleutherio E. C. A., Araujo P. S., Panek A. D. 1993. Protective role of trehalose during heat stress in Saccharomyces cerevisiae. Cryobiology 30:591–596 [DOI] [PubMed] [Google Scholar]
- 17. Elliott B., Haltiwanger R. S., Futcher B. 1996. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics 144:923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ells T. C., Speers R. A., Truelstrup Hansen L. 2009. Insertional mutagenesis of Listeria monocytogenes 568 reveals genes that contribute to enhanced thermal resistance. Int. J. Food Microbiol. 136:1–9 [DOI] [PubMed] [Google Scholar]
- 19. Evans Gilbreth S., Benson A. K., Hutkins R. W. 2004. Catabolite repression and virulence gene expression in Listeria monocytogenes. Curr. Microbiol. 49:95–98 [DOI] [PubMed] [Google Scholar]
- 20. Franco A., et al. 2000, Characterization of Tpp1+ as encoding a main trehalose-6P phosphatase in the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 182:5880–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glaser P., et al. 2001. Comparative genomics of Listeria species. Science 294:849–852 [DOI] [PubMed] [Google Scholar]
- 22. Gorski L., Palumbo J. D., Nguyen K. D. 2004. Strain-specific attachment of Listeria monocytogenes to alfalfa sprouts. J. Food Prot. 67:2488–2495 [DOI] [PubMed] [Google Scholar]
- 23. Gotsche S., Dahl M. K. 1995. Purification and characterization of the phosphor-α-(1,1)-glucosidase (TreA) of Bacillus subtilis 168. J. Bacteriol. 177:2721–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanahan D. 1985. Techniques for transformation of E. coli, p. 109–135In Glover D. M. (ed.), DNA cloning: a practical approach, vol. 1 IRL Press, McLean, VA [Google Scholar]
- 25. Hefford M. A., et al. 2005. Proteomic and microscopic analysis of biofilms formed by Listeria monocytogenes 568. Can. J. Microbiol. 51:197–208 [DOI] [PubMed] [Google Scholar]
- 26. Helfert C., Gotsche S., Dahl M. K. 1995. Cleavage of trehalose-phosphate in Bacillus subtilis is catalyzed by a phospho-α-(1-1)-glucosidase encoded by the treA gene. Mol. Microbiol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 27. Hengge-Aronis R., Klein W., Lange R., Rimmele M., Boos W. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horlacher R., Boos W. 1997. Characterization of TreR, the major regulator of the Escherichia coli trehalose system. J. Biol. Chem. 272:13026–13032 [DOI] [PubMed] [Google Scholar]
- 29. Horton R. M., Cai Z., Ho S. N., Pease L. R. 1990. Gene splicing by overlap extension: tailor made genes using the polymerase chain reaction. Biotechniques 8:528–535 [PubMed] [Google Scholar]
- 30. Hottiger T., De Virgilio C., Hall M. N., Boller T., Wiemken A. 1994. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur. J. Biochem. 219:187–193 [DOI] [PubMed] [Google Scholar]
- 31. Kandror O., Deleon A., Goldberg A. L. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. U. S. A. 99:9727–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klein W., Horlacher R., Boos W. 1995. Molecular analysis of treB encoding the Escherichia coli enzyme II specific for trehalose. J. Bacteriol. 177:4043–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 34. Lin T.-D., Chou C.-C. 2004. Effect of heat shock on the thermal tolerance and susceptibility of Listeria monocytogenes to other environmental stresses. Food Microbiol. 21:605–610 [Google Scholar]
- 35. Matthijs S., Koedam N., Cornelis P., De Greve H. 2000. The trehalose operon of Pseudomonas fluorescens ATCC 17400. Res. Microbiol. 151:845–851 [DOI] [PubMed] [Google Scholar]
- 36. Miller E. M., Nickoloff J. A. 1995. Escherichia coli electrotransformation, p. 105–114In Nickoloff J. A. (ed.), Electroporation protocols for microorganisms. Humana Press, Totowa, NJ [Google Scholar]
- 37. Miwa Y., Nakata A., Ogiwara A., Yamamoto M., Fujita Y. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acid Res. 28:1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neves M.-J., François J. 1992. On the mechanism by which a heat shock induces trehalose accumulation in Saccharomyces cerevisiae. Biochem. J. 288:859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Byrne C. P., Booth I. R. 2002. Osmoregulation and its importance to foodborne microorganisms. Int. J. Food Microbiol. 74:203–216 [DOI] [PubMed] [Google Scholar]
- 40. Postma P., Lengeler J. W., Jacobson G. R. 1993. Phosphoenolpyruvate:carbon phosphotransferase systems of bacteria. Microbiol. Rev. 57:543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Premaratne R. J., Lin W.-J., Johnson E. A. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pruitt K. M., Kamau D. N. 1993. Mathematical models of bacterial growth, inhibition and death under combined stress conditions. J. Ind. Microbiol. 12:221–231 [Google Scholar]
- 44. Purvis J. E., Yomano L. P., Ingram L. O. 2005. Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl. Environ. Microbiol. 71:3761–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rimmele M., Boos W. 1994. Trehalose-6-phosphate hydrolase of Escherichia coli. J. Bacteriol. 176:5654–5664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rudolph A. S., Crowe J. H., Crowe L. M. 1986. Effects of three stabilizing agents—proline, betaine, and trehalose—on membrane phospholipids. Arch. Biochem. Biophys. 15:134–143 [DOI] [PubMed] [Google Scholar]
- 47. Schöck F., Dahl M. K. 1996. Analysis of DNA flanking the treA gene of Bacillus subtilis reveals genes encoding a putative specific enzyme IITre and a potential regulator of the trehalose operon. Gene 175:59–63 [DOI] [PubMed] [Google Scholar]
- 48. Schöck F., Dahl M. K. 1996. Expression of the tre operon of Bacillus subtilis 168 is regulated by the repressor TreR. J. Bacteriol. 178:4576–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seeliger H. P. R., Jones D. 1986. Genus Listeria, p. 1235–1245In Sneath P. H. A., Mair N. S., Sharpe M. E., Holt J. G. (ed.), Bergey's manual of systematic bacteriology, vol. 2 Williams and Wilkins, Baltimore, MD [Google Scholar]
- 50. Sharma S. C. 1997. A possible role of trehalose in osmotolerance and ethanol tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 152:11–15 [DOI] [PubMed] [Google Scholar]
- 51. Sleator R. D., Gahan C. G. M., Hill C. 2003. A postgenomic appraisal of osmotolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 69:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Streeter J. G. 1985. Accumulation of α,α-trehalose by Rhizobium bacteria and bacteroids. J. Bacteriol. 164:78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ström A. R., Kaasen I. 1993. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol. Microbiol. 8:205–210 [DOI] [PubMed] [Google Scholar]
- 54. Tasara T., Stephan R. 2006. Cold stress tolerance of Listeria monocytogenes: a review of molecular adaptive mechanisms and food safety implications. J. Food Prot. 69:1473–1484 [DOI] [PubMed] [Google Scholar]
- 55. Van Dijck P., De Rop L., Szlufcik K., Van Ael E., Thevelein J. M. 2002. Disruption of the Candida albicans TPS2 gene encoding trehalose-6-phosphate phosphatase decreases infectivity without affecting hypha formation. Infect. Immun. 70:1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Van Vaeck C., Wera S., Van Dijck P., Thevelein J. M. 2001. Analysis and modification of trehalose 6-phosphate levels in the yeast Saccharomyces cerevisiae with the use of Bacillus subtilis phosphotrehalase. Biochem. J. 353:157–162 [PMC free article] [PubMed] [Google Scholar]
- 57. Welch W. J., Brown C. R. 1996. Influence of molecular and chemical chaperones on protein folding. Cell Stress Chaperones 1:109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Welsh D. T., Herbert R. A. 1999. Osmotically induced intracellular trehalose, but not glycine betaine accumulation promotes desiccation tolerance in Escherichia coli. FEMS Microbiol. Lett. 174:57–63 [DOI] [PubMed] [Google Scholar]
- 59. Whiting R. C. 1993. Modeling bacterial survival in unfavourable environments. J. Ind. Microbiol. 12:240–246 [Google Scholar]
- 60. Wirth R., An F. Y., Clewell D. B. 1986. Highly efficient protoplast transformation system with Streptococcus faecalis and a new Escherichia coli-Streptococcus faecalis shuttle vector. J. Bacteriol. 165:831–836 [DOI] [PMC free article] [PubMed] [Google Scholar]