Abstract
Polyene macrolides are important antibiotics used to treat fungal infections in humans. In this work, acyltransferase (AT) domain swaps, mutagenesis, and cross-complementation with heterologous polyketide synthase domain (PKS) loading modules were performed in order to facilitate production of new analogues of the polyene macrolide nystatin. Replacement of AT0 in the nystatin PKS loading module NysA with the propionate-specific AT1 from the nystatin PKS NysB, construction of hybrids between NysA and the loading module of rimocidin PKS RimA, and stepwise exchange of specific amino acids in the AT0 domain by site-directed mutagenesis were accomplished. However, none of the NysA mutants constructed was able to initiate production of new nystatin analogues. Nevertheless, many NysA mutants and hybrids were functional, providing for different levels of nystatin biosynthesis. An interplay between certain residues in AT0 and an active site residue in the ketosynthase (KS)-like domain of NysA in initiation of nystatin biosynthesis was revealed. Some hybrids between the NysA and RimA loading modules carrying the NysA AT0 domain were able to prime rimocidin PKS with both acetate and butyrate units upon complementation of a rimA-deficient mutant of the rimocidin/CE-108 producer Streptomyces diastaticus. Expression of the PimS0 loading module from the pimaricin producer in the same host, however, resulted in production of CE-108 only. Taken together, these data indicate relaxed substrate specificity of NysA AT0 domain, which is counteracted by a strict specificity of the first extender module KS domain in the nystatin PKS of Streptomyces noursei.
INTRODUCTION
Biosynthesis of polyketides by bacteria has received much attention during the last 2 decades due to the high commercial potential of the latter compounds as biologically active molecules for the treatment of diverse human diseases. Macrolides represent a large group of polyketides currently used as antibacterial, antifungal, antitumor, and immunosuppressive agents, structurally characterized by the macrolactone ring, which represents a cyclized polyketide chain. The latter is biosynthetically assembled by large enzymes, type I polyketide synthases (PKS), in a manner similar to fatty acid biosynthesis (34). PKS responsible for the macrolactone ring assembly are organized in modules, each performing decarboxylative condensation of the carboxylic acid units such as acetate, propionate, or butyrate onto the growing polyketide chain. Each ketide unit incorporated into the chain can undergo in-synthesis modification by the reductive domains present in the next extension module, thus providing chemical modification that will be reflected in the final product. The type I PKS are thus considered nature's assembly lines for the biosynthesis of macrolides, and a number of alterations have been introduced into these systems, resulting in biosynthesis of “unnatural” polyketides with properties distinctly different from those of the original molecules (20). The success of the type I PKS manipulation in terms of obtaining desired product depends on many factors, and in certain cases failure could not be easily explained. Such factors as the availability of starter and extender units, the structural integrity of the PKS proteins (including both enzymatic domains and linkers between them), and the ability of the next-in-line module to accept and process altered substrate all seem to have a profound impact on the functionality of these proteins (34).
Polyketide chain assembly by the type I PKS begins with the loading of a starter unit, in most cases acetate or propionate, onto the first PKS extender module (13). This process is accomplished by a loading module, which can be either fused to the first extender module or represented by a separate polypeptide, such as in the case of polyene macrolide PKS (2). The mechanism of polyketide biosynthesis initiation by the loading module depends on the composition of this module. The loading modules containing ketosynthase (KS)-like domains presumably recruit malonyl coenzyme A (malonyl-CoA) or methylmalonyl-CoA, which are decarboxylated to yield acetate or propionate starter units, respectively. The KS-like domains in such loading modules usually contain Glu instead of Cys in their active sites, and this residue has been assumed to be important for decarboxylase activity of the KS-like domains (4). In certain cases, the KS-like domain is absent from the loading modules, suggesting that they utilize acetyl- or propyonyl-CoA to initiate polyketide synthesis. In any case, the choice of the starter unit is ensured by the acyltransferase (AT) domain in the loading module, which can have either strict or relaxed specificity (19, 25, 34). Since changing the specificity of the AT domain can provide new engineered polyketides, substantial efforts in dissecting the specificity issues have been made recently. Earlier, it had been noticed that the specificity of the AT domains toward different substrates is reflected in the amino acid sequences of these domains (10). Since then, the specificities of several PKS modules toward a starter or extender unit have successfully been changed either by replacing the whole AT domains or by introducing specific mutations (9, 18, 19, 22).
Nystatin A1 (Fig. 1) is a medically important antifungal antibiotic, and it was recently shown that nystatin analogues with high antifungal activity and lower toxicity can be obtained via manipulation of the nystatin biosynthetic genes (7). Among the envisaged analogues that would be interesting to produce and analyze are those that would contain an extended or modified C-37 side chain resulting from the initiation of biosynthesis with a starting unit other than acetate. Earlier studies on the loading module NysA of the nystatin PKS, which contains a KS-like domain with a Ser instead of Cys residue in its active site, revealed that both malonyl-CoA and acetyl-CoA can be used for initiation of nystatin biosynthesis (6) (Fig. 2). In the current work, we attempted to change the specificity of the nystatin PKS loading module in order to prime nystatin biosynthesis with propionate or butyrate rather than acetate. We used the RimA PKS loading module for rimocidin (Fig. 1) from S. diastaticus variant 108; RimA PKS is capable of loading the extender module with either acetate or butyrate (21, 26) (Fig. 2). We additionally used the AT1 from NysB with specificity for methylmalonyl-CoA. Although our attempts, which included module swaps, hybrid construction, and site-specific mutagenesis, were unsuccessful in terms of producing new nystatin analogues, important information regarding the initiation of polyene macrolide biosynthesis, as well as the interplay between certain amino acid residues in the loading module, was obtained. Heterologous expression of certain NysA-RimA hybrids revealed relaxed substrate specificity of the NysA AT domain, which, keeping in mind our previous attempts of loading module engineering (6), implies a gate-keeping function of the first extender module in the nystatin PKS. These data will provide important support for rational design of future PKS manipulations aimed at the production of new macrolides.
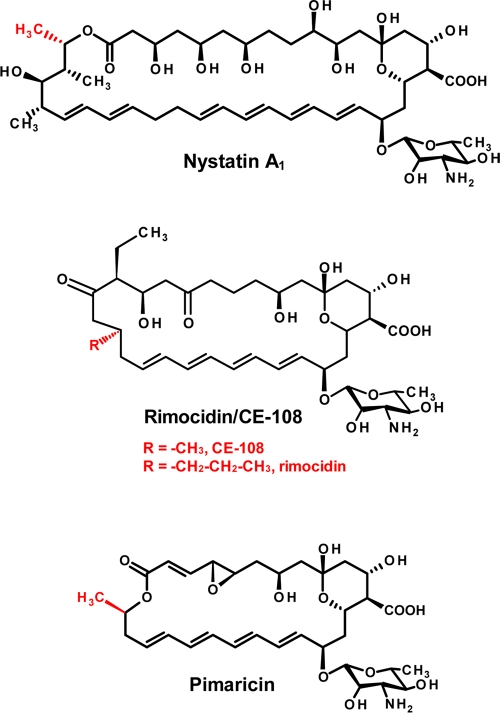
Fig. 1.
Chemical structures of nystatin A1 (acetate starter), rimocidin/CE-108 (acetate/butyrate starter), and pimaricin (acetate starter).
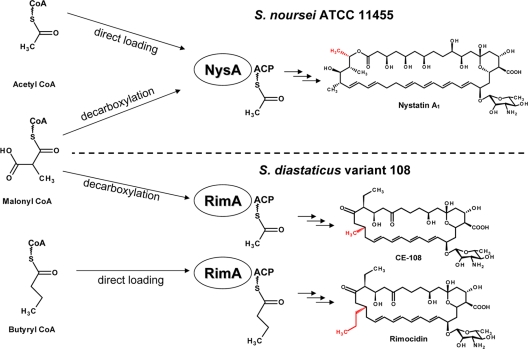
Fig. 2.
Current models for initiation of polyene macrolide biosynthesis in S. noursei (nystatin producer) and S. diastaticus variant 108 (rimocidin/CE-108 producer).
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bacterial strains and plasmids used and constructed during this study are listed in Table S1 in the supplemental material. S. noursei strains were maintained on ISP2 agar medium (Difco), S. diastaticus variant 108 strains were grown in SYM2 medium (28), and Escherichia coli strains were grown in L broth or L agar (24). S. diastaticus variant 108 strains were manipulated by protoplasting, transfection, and transformation as described elsewhere (15). Intraspecific conjugation was carried out as described previously (27).
Conjugal plasmid transfer from E. coli ET 12567(pUZ8002) and the gene replacement procedure in S. noursei were performed as described elsewhere (29). Transformation of E. coli was performed as described in the work of Sambrook et al. (24). Agar and liquid media, where appropriate, were supplemented with antibiotics using the following concentrations: chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; thiostrepton, 50 μg/ml; erythromycin, 100 μg/ml; ampicillin, 100 μg/ml; and apramycin, 50 μg/ml. Analysis of the production of polyene macrolides by S. noursei strains was done by liquid chromatography-mass spectroscopy (LC-TOF) of dimethyl sulfoxide (DMSO) extracts of the 5-day-old cultures from well plate cultivations according to the work of Bruheim et al. (8). The well plate cultivation was performed in semidefined 0.5× SAO-23 medium according to the protocol described previously (31). For feeding experiments with either crotonic or butyric acids, stock solutions (1 M, pH 7.0) were added in equal portions at 48, 60, and 72 h during fermentations to a final concentration of 15 mM. Production of polyene macrolides from S. diastaticus variant 108 was tested by high-performance liquid chromatography (HPLC) analysis of methanol extracts from the 5-day-old cultures grown on the SYM2 plates as previously described (28).
DNA manipulations, DNA sequencing, and PCR.
General DNA manipulations were performed as described in the work of Sambrook et al. (24). DNA fragments from agarose gels were purified using the Qiaex II kit (Qiagen, Germany) or PerfectPrep Gl cleanup kit (Eppendorf, Germany). Oligonucleotide primers were purchased from Qiagen (Germany), and primers used for QuikChange (site-directed mutagenesis) were PAGE purified. DNA sequencing was performed using the ABI Prism BigDye Terminator cycle sequencing kit and the Genetic Analyzer 3100 (Applied Biosystems, Inc.). The PCRs were performed with the Expand high-fidelity PCR system (Roche Molecular Biochemicals) using the Eppendorf Mastercycler (Eppendorf, Germany) as described previously (6). The DNA sequence encompassing the regions used in this study has been reported previously by Brautaset et al. (5).
Construction of the RimA expression vector.
The rimA gene was excised from the pSM736 plasmid as a 5.9-kb BglII fragment, treated with the Klenow fragment of DNA polymerase, and cloned into the HincII site of pGEM3Zf(−). After verification of the correct orientation, the 5.9-kb XbaI-HindIII rimA-containing fragment from the resulting construct was cloned, together with the HindIII-XbaI fragment containing the nysAp promoter (30), into the pSOK804 vector. The resulting plasmid, pRIM1, was used in complementation experiments.
Replacement of AT0 with AT1 in NysA using gene SOEing.
Replacements of the AT0 in NysA with the methylmalonyl-specific AT1 from the nystatin PKS NysB were conducted both with and without the interdomain linker region from module 1 of NysB by using the gene splicing by overlap extension (SOEing) technique (12). The acetyltransferase AT1 coding region in nysB was fused with nysA by using PCR. The fusion points used for the assembly procedure were in all cases positioned in the interdomain linker regions, and the fusions were done within overlapping DNA fragments, ensuring that there were no gaps in the amino acid alignments. This resulted in seamless hybrids, thereby avoiding any unwarranted structural effects on the resulting hybrid proteins. The plasmid pl76B11 (5) served as a template for the amplified fragments. Fusion primers and flanking primers with restriction sites are given in Table S2 in the supplemental material. All the resulting constructs were verified by DNA sequencing.
(i) Without linkers.
As a primary step, the AT1 coding region and the KSS and DH0 (dehydratase domain) coding regions with C-terminal and N-terminal linkers, respectively, were amplified separately. A region containing the KSS coding region with the C-terminal linker was amplified with flanking primer mluI-F, containing an MluI site located upstream of ks0, and fusion primer ks0link-R, yielding the ks0linkup DNA fragment (0.73 kb). The AT1 coding region was amplified as two parts with hybrid primers at1-F and at1-R, and primers on both sides of NruI, nruI-F and nruI-R, yielding DNA fragments at1casup (0.90 kb) and at1casdown (0.11 kb), respectively. The DNA fragment encoding the DH0 domain with N-terminal linker was amplified with a fusion primer, dh0-F, and a flanking primer, pstI-R, containing a PstI recognition site located downstream of the DH0 coding sequence, yielding the dh0link fragment (0.82 kb). A secondary PCR fused ks0linkup with at1casup and at1casdown with dh0link, resulting in at1casup (1.6 kb) and at1casdown (0.90 kb), respectively. These fragments were respectively cloned into pLIT38NruI(−) MluI-NruI and pLIT38NruI(−) NruI-PstI, resulting in pAT1casup and pAT1casdown plasmids. The MluI-NruI and NruI-PstI fragments from the latter constructs were excised and ligated into pLITMUS38 MluI-PstI, yielding pAT1cassette. Subsequently, the MluI-NcoI fragment (2.42 kb) from pAT1cassette was ligated into the pl76ES4.6 MluI-NcoI fragment (5.40 kb), resulting in pNysA-AT1cas. Finally, the 4.67-kb HindIII-EcoRI fragment from pNysA-AT1cas was excised and ligated into the HindIII-EcoRI sites of vector pSOK804, resulting in the integrative vector pSOKAT1cassette for expression of this hybrid.
(ii) With linkers.
A similar procedure was employed for replacing the intrinsic AT0 domain of NysA with N- and C-terminal linkers by the corresponding part of the NysB protein encompassing the AT1 domain and linkers. First, the ks0up DNA fragment (0.44 kb) was amplified using primers mluI-F and ks0-R. The AT1 coding region with interdomain linker regions was amplified first as two different fragments with hybrid primers ks1link-F and dh1link-R and primers on both sides of NruI, nruI-R and nruI-F, giving PCR fragments at1linkup (1.21 kb) and at1linkdown (0.16 kb), respectively. Next, the dh0down fragment (0.76 kb) was amplified using primers dh0link-F and pstI-R. A second PCR subsequently resulted in the at1briup (0.44 kb) and at1bridown (0.16 kb) fragments, which were cloned into the pLIT38NruI(−) MluI-NruI and pLIT38NruI(−) NruI-PstI, yielding the pAT1briup and pAT1bridown constructs, respectively. The latter vectors were processed in the same manner as the corresponding vectors in the previous section (see above), yielding the pAT1bridge plasmid. The 2.4-kb MluI-NcoI fragment from the latter vector was inserted into the pl76ES4.6 MluI-NcoI fragment (5.40-kb), resulting in pNysA-AT1bri. Finally, the 4.6-kb HindIII-EcoRI fragment from pNysA-AT1bri was excised and ligated into the HindIII-EcoRI sites of vector pSOK804, resulting in the integrative vector pSOKAT1bridge.
Construction of the NysA-RimA hybrids. (i) Fusion of nysAp promoter to rimA CoL.
DNA fragments of 250 bp and 239 bp were amplified by PCR using primers NR-F1-U (5′-CGGGATCCACCATGATTACGCCAAGCTATTTAGG-3′) and NR-F1-M-r (5′-GTGTGGACGGGCACCATGCCTTGCTCACCCCTG-3′) on template pL76ES4.6 and primers NR-F1-M (5′-CAGGGGTGAGCAAGGCATGGTGCCCGTCCACAC-3′) and NR-F1-D (5′-ACCGCTCGAGGACGCCCGATTCCTGGAG-3′) on template pRIM1, respectively. The latter plasmid harbors the rimA gene of S. diastaticus variant 108. These PCR products were purified and fused in a new PCR together with primers NR-F1-U and NR-F1-D to yield a PCR product of 456 bp, which was cloned as the BamHI-XhoI fragment into pGEM-7zf(+) to yield plasmid pF1BX. The latter construct was sequenced to confirm correct fusion.
(ii) Fusion of rimA acyl carrier protein (ACP) to nysA KS.
DNA fragments of 309 bp and 870 bp were amplified by PCR using primers NR-F3-U (5′-CAGGATCCCAGGTCCTCGGCAGCAG-3′) and NR-F3-M-r (5′-CGACGACCACCACCGGATCGGCCTCG-3′) on template pRIM1 and primers NR-F3-M (5′-CGAGGCCGATCCGGTGGTGGTCGTCG-3′) and NR-F3-D (5′-AGACCTCGAGTGACGCGTTCCTGGGC-3′) on template pL76ES4.6, respectively. These PCR products were purified and fused in a new PCR together with primers NR-F3-U and NR-F3-D to yield a PCR product of 1,153 bp, which was cloned into the vector pPCR-Script Amp SK(+) with the PCR-Script Amp cloning kit (Promega) to yield plasmid pF3BX′ and sequenced to confirm correct fusion.
(iii) Fusion of rimA AT to nysA DH.
DNA fragments of 208 bp and 227 bp were amplified by PCR using primers NR-F4-U (5′-ATGGATCCGGAGGAGCGCGTCACC-3′) and NR-F4-M-r (5′-CGGGCCAGTCGACGGGTGCG-3′) on template pRIM1 and primers NR-F4-M (5′-CGCACCCGTCGACTGGCCCG-3′) and NR-F4-D (5′-ACCACTCGAGGTGGGTGCGGAGGGAG-3′) on template pL76ES4.6, respectively. These PCR products were purified and fused in a new PCR together with primers NR-F4-U and NR-F4-D to yield a PCR product of 415 bp, which was cloned as the BamHI-XhoI fragment into pGEM-7zf(+) to yield plasmid pF4BX and sequenced to confirm correct fusion.
(iv) Fusion of rimA KS to nysA AT.
A DNA fragment of 1,164 bp was amplified from template pL76ES4.6 with primers NR-F5-U (5′-CGGGATCCGTCTTCTCCGGACAGGGCAGCC-3′) and NR-F4-D and cloned directly into dephosphorylated (calf intestinal phosphatase; New England BioLabs) vector pGEM-3zf(+) to yield plasmid pF5BX*, which was sequenced to confirm correct fusion.
(v) Construction of complementation plasmid for nysA-rimA hybrid C.
A 3,760-bp HindIII-BspEI fragment from pLit28-F1-RIM, a 1,037-bp BspEI-ApaI fragment from pF5BX*, a 1,845-bp ApaI-EcoRI fragment from pL76ES4.6, and pSOK804-cut EcoRI-HindIII were ligated to yield complementation plasmid pNR-C.
(vi) Construction of complementation plasmid for nysA-rimA hybrid D.
A 4,690-bp HindIII-NotI fragment from pLit28-F1-RIM, a 139-bp NotI-ApaI fragment from pF4BX, a 1,845-bp ApaI-EcoRI fragment from pL76ES4.6, and pSOK804-cut EcoRI-HindIII were ligated to yield complementation plasmid pNR-D.
(vii) Cloning of rimJ under the control of nysRIVp promoter into the plasmids for expression of nysA-rimA hybrids.
A 1,435-bp fragment containing rimJ from pSM765 was amplified using primers RimJ-f (5′-ATATTCTAGACGCCTTTTCCGGAGGCTC-3′) and RimJ-r (5′- ATAACACCGG TGAAGCTTGCGTCCCGCAAGGGC-3′). The use of these primers introduced an XbaI site at the 5′ end and HindIII and SgrAI restriction sites at the 3′ end of the rimJ PCR product. A 3.5-kb plasmid, pNR4pHX [containing an nysRIV promoter cloned as a HindIII-XbaI fragment in pGEM3Zf(−)] was linearized through digestion with XmaI and XbaI and ligated with the XbaI- and SgrAI-digested rimJ PCR product, resulting in pNR4rimJ. In the latter plasmid, rimJ appeared under the control of the nysRIVp promoter. A 1,751-bp HindIII cassette from pNR4rimJ was cloned into the HindIII site of the nysA-rimA hybrid expressing plasmids constructed as described above.
Site-directed mutagenesis of the nysA AT0 coding region.
To prepare a suitable template for site-directed mutagenesis, the 1.76-kb MluI-ApaI fragment from plasmid pl76ES4.6 was excised and ligated into pLITMUS38, resulting in plasmid plit38-AT0. Site-directed mutagenesis using QuikChange (Stratagene) was done on a plit38-AT0 template and, successively, on the mutants thereof (referred to below as plit38-mutX). Stepwise exchange of specific amino acids in AT0 was conducted by altering the codons for amino acids in the nystatin PKS loading AT0 domain as follows: T56V, G57D, W58V, V89Q, S184D, H185Y, F187S, and M117I. The primers used in the site-directed mutagenesis are given in Table S3 in the supplemental material.
The mutant plasmids plit38-mutX were isolated, and all mutations were verified by DNA sequencing. This was followed by isolation of the 1.76-kb MluI-ApaI fragment from plit38-mutX and ligation into the 6.04-kb MluI-ApaI fragment of pl76ES4.6, yielding pl76ES4.6AT0-mutX constructs. HindIII-EcoRI fragments (4.65 kb) from pl76ES4.6AT0_mutX was excised and ligated into the HindIII-EcoRI sites of vector pSOK804, resulting in the nysA integration vectors pAT0_mutX.
Combining mutation in AT0 with KSC and KSQ.
The mutations in the NysA KS-like domain's “active site” were constructed previously (6). These constructs were combined with some of the mutations in AT0 by using conventional cloning techniques. A 1.05-kb MluI-HindIII fragment from plasmids pKSCAT0 and pKSQAT0 was respectively excised and ligated into the 6.75-kb MluI-HindIII fragment from certain pl76ES4.6AT0_mutX constructs, resulting in pKSCAT0-mutX and pKSQAT0-mutX. The latter plasmids were used to complement S. noursei nysA mutant NDA59.
RESULTS
Expression of the rimocidin PKS loading module restores nystatin biosynthesis in the S. noursei nysA mutant.
The RimA PKS loading module of S. diastaticus variant 108 is capable of loading both acetate and butyrate onto the first rimocidin PKS extension module, resulting in production of CE-108 and rimocidin, respectively. We hypothesized that RimA might also initiate nystatin biosynthesis and is able to prime the NysB PKS with butyrate, which would result in the production of a new nystatin analogue. The rimA gene was placed under the control of the nysAp promoter, yielding the pRIM1 vector, which was introduced into nysA-deficient S. noursei NDA59 (6). Testing of the transconjugants for polyene macrolide production revealed nystatin as the final product, and no new nystatin analogues could be detected. Keeping in mind that precursor supply might be a limiting factor for the biosynthesis of new analogues, feeding experiments were performed according to the work of Liu and Reynolds (17). However, supplementation of the fermentation medium with either crotonic or butyric acid did not result in the production of new nystatin analogues. Thus, despite that the rimA gene seemed to be functional in complementing the nysA mutation, the biosynthetic pathway for nystatin could successfully proceed only when primed with acetate.
Complementation of the S. diastaticus variant 108 rimA disruption mutant with the nysA gene was also attempted. For that purpose, nysA was cloned under the control of the xysA promoter (xysAp) of the xylanase gene from S. halstedii JM8 (23). A BstEII-BglII fragment carrying the nysA gene was fused to the xysAp promoter and cloned in several steps as described in Table S1 in the supplemental material. The resulting recombinant plasmid, pSM767, was introduced into S. diastaticus variant 108 (wild type) and then transferred by intraspecific conjugation into the rimA-deficient mutant PM1-500. No tetraene production was observed in the fermentation broth of the latter recombinant strain, suggesting the inability of NysA to prime rimocidin biosynthesis, most probably due to the lack of a proper interaction with RimB. Together, these results imply that NysB has a more flexible docking domain than RimB, allowing it to interact with both NysA and RimA.
Replacements of the AT0 domain in NysA with AT1 of NysB by using gene SOEing.
Next, as an attempt to change the specificity of NysA toward methylmalonyl-CoA, we replaced AT0 with AT1 from NysB (providing the first extender unit) using restriction sites created through PCR. The AT1 domain of NysB has a clear specificity toward methylmalonyl-CoA, as it ensures incorporation of a propionate extender during the nystatin biosynthesis (5). Such hybrids proved to be nonfunctional, as they could not initiate biosynthesis of nystatin or any other nystatin-related polyene macrolides (6). This could be attributed to the changes in amino acid sequences within the KSS-AT0 and AT0-DH0 linkers in NysA, more specifically introduction of sequences coding for additional amino acid residues during the hybrid assembly. Relatively little is known about the linker regions, and it can be anticipated that these regions affect how the domains are functioning within the module (34). Keeping this in mind, we employed a method of gene SOEing that ensures the accurate fusion of virtually any DNA sequences at any nucleotide by using a PCR-based technique (12). The strategy for gene SOEing was chosen for precise splicing of the AT1 domain into the NysA either with or without its original KS1-AT1 and AT1-DH1 linkers. The pSOK804-based plasmids pSOKAT1cassette and pSOKAT1bridge expressing recombinant NysA containing AT1 with or without its original linkers (Fig. 3), respectively, were constructed. Both plasmids were introduced by conjugation into the nysA-deficient S. noursei NDA59 mutant, which was unable to produce nystatin. It has previously been shown (6) that complementation of this strain with nysA expressed in trans restores nystatin production almost to wild-type levels. Analysis of metabolites produced by the above-described pSOKAT1cassette- and pSOKAT1bridge-containing recombinant strains revealed that neither of the constructs could restore nystatin production or initiate synthesis of novel nystatin analogues in the NDA59 mutant.
Fig. 3.
Schematic representation of the NysA and RimA loading modules and their hybrids. Solid boxes and thin lines represent RimA enzymatic domains and interdomain linkers, respectively.
Certain hybrids between NysA and rimocidin PKS loading module RimA are able to initiate nystatin biosynthesis.
In the next attempt to change the starter unit specificity of the NysA, the gene SOEing technique was applied for the construction of hybrid loading modules between NysA and the initiator protein of the rimocidin/CE-108 PKS RimA. The latter protein is composed of two modules, one containing carboxylic acid-CoA ligase (CoL) and ACP, and another containing KSS, AT, and ACP domains (26). RimA was shown to be responsible for the choice of starting unit in rimocidin and CE-108 biosynthesis, incorporating either acetate (CE-108) or butyrate (rimocidin) (26). The exact mechanism by which RimA selects the starter unit is not presently clear, and it was not possible to predict whether the CoL or AT domain in RimA could be responsible for the incorporation of butyrate. Although the AT domain in RimA was originally proposed to be acetate specific, reexamination of the sequence revealed that two residues critical for specificity, His185 and Phe187, highly conserved in acetate-specific domains (10), are replaced with Val and Ala, respectively.
Four hybrid loading module genes were constructed based on rimA and nysA by using gene SOEing (Fig. 3) and placed under the control of the nysAp promoter, which was shown to function efficiently during previous complementation experiments (6, 30). The C-terminal part of the hybrids was always represented by the NysA counterpart in order to ensure proper interaction with the NysB protein during polyketide assembly. To facilitate the biosynthesis of butyryl-CoA, the rimJ gene encoding a putative crotonyl-CoA reductase, which catalyzes the reduction of crotonyl-CoA to butyryl-CoA, was placed under the control of the nysRIVp promoter (30) and cloned together with the rimA-nysA hybrids into the pSOK804 integrative vector. The recombinant plasmids were then introduced into the S. noursei NDA59 mutant, and the recombinant strains were tested for the production of polyene macrolides. The recombinant plasmids expressing hybrid A along with rimJ were able to restore nystatin biosynthesis in the NDA59 mutant almost to the wild-type level (data not shown), while no polyene macrolide production could be detected in the case of hybrids B, C, and D. No new nystatin analogues could be detected in the extracts of the mutants harboring hybrid A.
Hybrid RimA/NysA proteins carrying the AT domain of NysA are able to restore rimocidin and CE-108 biosynthesis in the rimA disruption mutant.
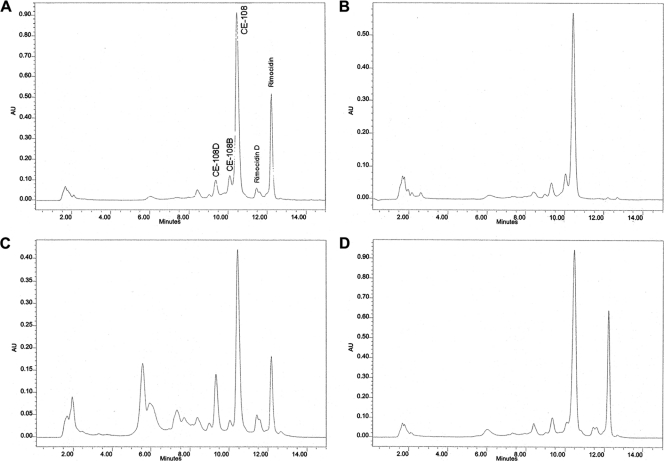
Hybrids C and D were tested for restoration of the polyene macrolide production in the rimA-deficient mutant of S. diastaticus. Both hybrid proteins differ only in the AT domains, which originate from NysA and RimA in hybrids C and D, respectively (Fig. 3). The corresponding hybrid genes were placed under the control of the ermEP* promoter in several steps as described in Table S1 in the supplemental material. The resulting plasmids (pLEC104C and pLEC106D for the hybrids C and D, respectively) were used to transform protoplasts of a new rimA disruptant (S. diastaticus variant 108::PM1/pLEC101). Several candidates were selected and confirmed by direct plasmid extraction (for the presence of recombinant plasmid) and by Southern blotting for the correct integration of PM1/pLEC101 phage into the chromosome. The HPLC analysis of the fermentation broth of the recombinant strains showed restoration of CE-108D, CE-108, rimocidin D, and rimocidin production in both cases (Fig. 4C and D), as evident from the appearance of peaks eluting, respectively, at ca. 9.5 min, 10.5 min, 11.5 min, and 12.3 min. The identities of these metabolites have previously been confirmed by LC-mass spectrometry (LC-MS) analyses of the extract from the wild-type S. diastaticus variant 108 (E. M. Seco et al., unpublished data). Some additional peaks appeared on the chromatogram within the ca. 2- to 8-min elution time upon complementation with hybrid C. However, we could not correlate these peaks with polyene UV spectra and do not know at present what they represent. In any case, successful complementation of the S. diastaticus rimA mutant with hybrids C and D strongly suggests that the AT0 domain of NysA under heterologous conditions is able to recognize and recruit both malonyl-CoA and butyryl-CoA. Moreover, hybrids C and D could functionally interact with the RimB PKS, which was surprising considering the presence of an intact NysA C terminus on both hybrids and the lack of such interaction between NysA and RimB (see above).
Fig. 4.
HPLC analysis of culture extracts of the S. diastaticus rimA mutant complemented with rimA (A), pimS0 (B), nysA-rimA hybrid C (C), and nysA-rimA hybrid D (D).
The loading module of pimaricin PKS is not able to prime RimB with a butyryl starter.
PimS0 is a loading PKS module responsible for the priming of pimaricin biosynthesis in S. natalensis (1). The loading modules RimA and PimS0 share a high degree of homology (70%) and have identical architectural organization with the catalytic domains CoL, ACP, KSS, AT and a second ACP similarly arranged (Fig. 2). Despite this similarity, PimS0 seems to be able to prime the pimaricin PKS only with acetyl-CoA in S. natalensis. However, this could also be due to the strict specificity of the accepting extender module of pimaricin PKS for the acetate primer. Considering the apparent relaxed specificity of RimB toward priming units, we used the S. diastaticus variant 108 rimA mutant as a host for analyzing the substrate specificity of AT domain in PimS0. For this purpose, the 6,229-bp XmnI-EcoRI fragment from the pimaricin biosynthetic cluster (containing the 3′ end of pimG, pimF, and pimS0 and 255 bp downstream the stop codon of pimS0) was excised from the Cos37 cosmid (1) and cloned under the control of the constitutive promoter ermE*P in several steps as described in Table S1 in the supplemental material. The resulting plasmid, pLEC108, was introduced by transformation into S. diastaticus variant 108::PM1/pLEC101 protoplasts. As a control, plasmid pSM783, carrying the rimA gene under the control of ermE*P, was also introduced into the rimA mutant. Plasmid extraction and Southern blotting confirmed that the rimA-disrupted mutant carries, as expected, the corresponding plasmids pLEC108, pSM783, and pSM780, giving rise to S. diastaticus variant 108::PM1/pLEC101-pLEC108, S. diastaticus variant 108::PM1/pLEC101-pSM783, and S. diastaticus variant 108::PM1/pLEC101-pSM780, respectively. HPLC analysis of the fermentation broth of the corresponding recombinant strains confirmed that the rimA mutant, carrying pimS0 under the control of ermE*P, produces CE-108 but not rimocidin (Fig. 4B), as evident from the absence of rimocidin D- and rimocidin-characteristic peaks eluting at ca. 11.5 min and 12.3 min, respectively. The same mutant transformed with the rimA gene produced both CE-108 and rimocidin (Fig. 4A). These results clearly indicate that the AT domain of PimS0 has a strict substrate specificity for either malonyl-CoA or acetyl-CoA and is not able to prime the rimocidin pathway with butyryl-CoA.
Site-specific mutagenesis of certain AT0 amino acid residues differentially affects the ability of recombinant NysA to initiate nystatin biosynthesis.
Several reports on the site-specific mutagenesis of the AT domains, along with the structural data for the Streptomyces coelicolor malonyl-CoA:ACP transacylase (14, 16) and the recently published crystal structure of erythromycin PKS fragment (32), suggest that only a limited number of amino acids play a critical role in determining specificity of these domains. The study by Del Vecchio et al. (9) demonstrated that changing only 2 amino acids (namely, those corresponding to His185 and Phe187) in the AT of the erythromycin PKS can drastically influence the choice of the substrate by this domain. Guided by these observations, we designed a strategy for stepwise mutagenesis of the AT0 domain in NysA aimed at changing its specificity from malonyl-CoA to methylmalonyl-CoA. We have focused on six amino acid residues marked in the alignment of the two AT domain regions supposedly important for substrate selectivity (see Fig. S1 in the supplemental material). The DNA fragment encoding these regions of AT0 was excised from the nysA gene, subjected to site-specific mutagenesis, and cloned back into the original plasmid expressing NysA (see Materials and Methods). The recombinant plasmids expressing the NysA mutants (see Table S1 in the supplemental material) were introduced into S. noursei NDA59, and the resulting strains were analyzed for polyene macrolide production in order to monitor the effect that particular substitutions had on the biosynthesis of nystatin and, eventually, new polyene macrolides.
The single and combined site-specific mutations in AT0 and their effects on nystatin biosynthesis are presented in Table 1. Although none of the recombinant strains were shown to produce novel nystatin analogues, most of the mutants were able to successfully initiate nystatin biosynthesis, albeit at different levels. Substitutions of several amino acids in region A of AT0 (see Fig. S1 in the supplemental material), in particular T56V, G57D, W58V, and V89Q, apparently had a mildly negative effect on the ability of NysA to initiate nystatin biosynthesis. No new polyene macrolides besides those already described for wild-type S. noursei (8) could be detected in the extracts of the recombinant strains expressing the above-mentioned mutants.
Table 1.
Effect of certain amino acid substitutions in the AT0 domain of NysA on the ability of the recombinant protein to restore nystatin biosynthesis in the S. noursei nysA mutanta
| AT0 mutation |
KS mutation |
Nystatin yield (% of WTb) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| T56V | G57D | W58V | V89Q | H185Y | F187S | M117I | KSC | KSQ | |
| x | x | 46 ± 2 | |||||||
| x | x | x | 46 ± 4 | ||||||
| x | x | x | x | 51 ± 5 | |||||
| x | 82 ± 6 | ||||||||
| x | x | 55 ± 5 | |||||||
| x | 72 ± 5 | ||||||||
| x | x | 62 ± 5 | |||||||
| x | x | 22 ± 2 | |||||||
| x | x | x | 2 ± 1 | ||||||
| x | x | x | x | x | 37 ± 4 | ||||
| x | x | x | x | x | 10 ± 2 | ||||
| x | x | x | x | x | x | 3 ± 1 | |||
| x | x | x | x | x | x | 11 ± 2 | |||
| x | x | 41 ± 4 | |||||||
| x | x | 72 ± 4 | |||||||
| x | x | 72 ± 5 | |||||||
| x | x | 0 | |||||||
| x | x | x | 0 | ||||||
| x | x | x | 17 ± 1 | ||||||
| x | x | x | 93 ± 6 | ||||||
“x” indicates the presence of the mutation in the NysA domain. Note that the data represent an average from 3 parallel experiments. The NDA59 mutant complemented with the wild-type nysA gene was used as a reference.
WT, wild type.
Next, the His185 and Phe187 residues in region B of the AT0 domain (see Fig. S1 in the supplemental material), which were shown to be important for substrate specificity of the AT domain from erythromycin PKS (9), were changed to Tyr and Ser, respectively. Both single mutations and combinations thereof had a negative effect on nystatin biosynthesis by NDA59 expressing these NysA mutants, reducing the nystatin production level by ca. 25% and 80%, respectively (Table 1). Introduction of the V89Q mutation into the H185Y F187S double mutant drastically (by ca. 98%) reduced the nystatin production, while these mutations had only a moderate effect when introduced into either of the single mutants. Next, the H185Y and F187S mutations were introduced on the T56V G57D W58V V89Q background. All resultant mutants were shown to provide for nystatin biosynthesis at the levels ranging from 3% to 37% of that seen for the control. Detailed analysis of the extracts from all the above-mentioned recombinant strains failed to detect production of any new nystatin analogues.
Finally, we constructed the T56V G57D W58V V89Q M117I F187S NysA AT0 mutant based on the suggestion by Keatinge-Clay et al. (14), who implied that the Met and Phe residues (corresponding to Met117 and Phe187) provide a selectivity filter for the malonyl-CoA:ACP transacylase of S. coelicolor. As in all other cases, however, this NysA mutant failed to initiate synthesis of new nystatin analogues, while being able to restore the level of nystatin production to ca. 11% compared to the wild-type NysA (Table 1).
Combination of the AT0 and KS-like domain mutations reveals interplay between certain residues in these domains in initiation of nystatin biosynthesis.
Next, we examined the possibility that there might exist an interplay between certain residues in AT0 and KS-like domains of NysA in terms of the ability of this protein to initiate nystatin biosynthesis. We then combined several AT0 mutations with the mutations in the KS-like domain of NysA constructed previously (6). The original KS-like domain of NysA harbors a Ser residue instead of Cys in its active site, and it was suggested by others that the amino acid residue in this position might be involved in decarboxylation during condensation with the first extender unit. As previously shown, however, changing this Ser residue in the NysA KS-like domain to either Glu or Cys does not significantly affect the ability of this protein to initiate nystatin biosynthesis. The KSQ and KSC mutations were first introduced into the H185Y and F187S single AT0 mutants, and the recombinant strains were tested for nystatin production. The data obtained clearly demonstrated that the KSC mutation completely abrogates the initiation of nystatin biosynthesis by the H185Y AT0 mutant, while the KSQ mutation has no effect on this background (Table 1). The situation was different on the F187S mutant background, where the KSQ mutation had no effect, while the KSC mutation had a moderately negative effect. The KS mutations were then introduced into the H185Y F187S double mutant. The KSC H185Y F187S mutant was unable to initiate nystatin biosynthesis, while the KSQ H185Y F187S mutant restored the nystatin production in NDA59 to ca. 17% (Table 1). Taken together, these results reveal a complex interplay between the KS-like and AT domains in the nystatin PKS loading module during the initiation of polyketide assembly (see Discussion).
DISCUSSION
Multiple attempts at changing the starter unit specificity of the nystatin PKS loading module NysA using domain swaps, heterologous expression of loading module with wider substrate specificities, construction of hybrids, and site-specific mutagenesis of the AT domain in this protein have failed to produce novel nystatin analogues. Since cross-complementation of the S. noursei nysA mutant with RimA (known to have substrate flexibility) yielded only nystatin, we suggest that the failure to produce new nystatin analogues must be due to the inability of the first extender module of NysB PKS to accept propionate or butyrate as starter units, rather than to the absence of flexibility of the AT0 domain of NysA in recognizing other carboxylic acids. This suggestion is strongly supported by the fact that hybrids C and D (both carrying the AT0 domain of NysA) can restore both rimocidin and CE-108 production in the S. diastaticus variant 108 rimA mutant. Considering these results along with the fact that S. noursei is capable of producing nystatin and its analogues in substantial quantities (8), the key factor for generating new nystatin derivatives most probably lies within the KS1 domain of NysB. The latter domain, which for some reason could not be deduced from the amino acid sequence of this domain, probably has a strong preference for processing an acetate unit as a biosynthetic primer. However, since our attempts on replacement of the KS1 domain in NysB with another KS domain from the nystatin PKS have so far been unsuccessful, we could not confirm this hypothesis experimentally. From the results obtained, we can also suggest that the AT0 domain of the NysA, unlike that of PimS0, shares with RimA similar flexibility for substrate recognition, at least for butyryl-CoA and malonyl-CoA (and/or acetyl-CoA). In addition, RimA was recently shown to accept methylmalonyl-CoA, priming the rimocidin PKS with a propionate starter (Seco et al., unpublished). A possible alternative strategy for engineering NysB toward greater flexibility with regard to the priming could be the replacement of KS1 with a heterologous KS domain known to accept a large number of different acyl-CoAs. From this point of view, the KS domain from the first module of avermectin PKS could be a good candidate, as it was shown to accept and extend over 40 starter units (19). If successful, such replacement can also be combined with the replacement of AT0 domain in NysA with the AT domain from the avermectin PKS loading module, providing an opportunity for the production of a wide range of nystatin analogues with alternative C-37 side chains.
Interestingly, while yielding no new nystatin analogues, one of the hybrids of the rimocidin/CE-108 PKS loading modules RimA and NysA was able to efficiently initiate nystatin biosynthesis. This ability was shown only for the hybrid containing the original NysA KSS-AT0 domain assembly. Despite the fact that KS-like domains in NysA and RimA are quite similar (62% identity), and the KS-like domain of RimA also contains Ser in the active site, the presence of the NysA KS, AT, and the KS-AT linker seems to be crucial for the functionality of the hybrid.
From the data on erythromycin PKS (9), it could have been expected that the H185 F187S double mutant would utilize methylmalonyl-CoA as a starter, facilitating the production of methylnystatin. The facts that methylnystatin was not detected in the extract of NDA59 expressing the latter mutation and that the nystatin yield was substantially reduced could be explained by a shift of specificity toward methylmalonyl-CoA combined with a reduced ability of NysB to be primed with propionate starter.
Results obtained after combination of the AT0 mutations with the alterations of the active site residue of the NysA KS-like domain provided clear evidence of cooperation of these domains in the process of initiation of nystatin biosynthesis. Previously, based on the results obtained upon site-specific mutagenesis of the KS-like domain of NysA (6), we demonstrated that the residue in the active site of the KS-like domain in NysA does not affect the functionality of this protein with the wild-type AT0. The latter implied that the active site residue in the KS-like domain of NysA, in contrast to the KSQ domains in other PKS loading modules (4), is not crucial for decarboxylase activity required for utilization of malonyl-CoA as a starter. The fact that the KSQ H185Y mutant is nonfunctional, in contrast to the H185Y, KSC H185Y, KSC F187S, and KSQ F187S mutants, strongly suggests an interplay between the active site residue of KS-like domain of NysA and His185 in AT0. From these data, one would expect the KSQ H185Y F187S mutant to be nonfunctional as well in terms of initiation of nystatin biosynthesis. Unexpectedly, this mutant provided for reduced but significant nystatin biosynthesis, while the KSC H185Y F187S mutant completely failed to do so. These results indicate that the “active site” residue in the KS-like domain of NysA cooperates with both His185 and Phe187 in AT0 during recruitment of a starter unit and that the composition of this “triad” is crucial for the successful initiation of nystatin biosynthesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Research Council of Norway and grant BIO2008-03683 from the Spanish MICINN.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Aparicio J. F., Colina A. J., Ceballos E., Martín J. F. 1999. The biosynthetic gene cluster for the 26-membered ring polyene macrolide pimaricin. A new polyketide synthase organization encoded by two subclusters separated by functionalization genes. J. Biol. Chem. 274:10133–10139 [DOI] [PubMed] [Google Scholar]
- 2. Aparicio J. F., Caffrey P., Gil J. A., Zotchev S. B. 2003. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 61:179–188 [DOI] [PubMed] [Google Scholar]
- 3. Reference deleted.
- 4. Bisang C., et al. 1999. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401:502–505 [DOI] [PubMed] [Google Scholar]
- 5. Brautaset T., et al. 2000. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7:395–403 [DOI] [PubMed] [Google Scholar]
- 6. Brautaset T., Borgos S. E., Sletta H., Ellingsen T. E., Zotchev S. B. 2003. Site-specific mutagenesis and domain substitutions in the loading module of the nystatin polyketide synthase, and their effects on nystatin biosynthesis in Streptomyces noursei. J. Biol. Chem. 278:14913–14919 [DOI] [PubMed] [Google Scholar]
- 7. Brautaset T., et al. 2008. Improved antifungal polyene macrolides via engineering of the nystatin biosynthetic genes in Streptomyces noursei. Chem. Biol. 15:1198–1206 [DOI] [PubMed] [Google Scholar]
- 8. Bruheim P., et al. 2004. Chemical diversity of polyene macrolides produced by Streptomyces noursei ATCC 11455 and recombinant strain ERD44 with genetically altered polyketide synthase NysC. Antimicrob. Agents Chemother. 48:4120–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Del Vecchio F., et al. 2003. Active-site residue, domain and module swaps in modular polyketide synthases. J. Ind. Microbiol. Biotechnol. 30:489–494 [DOI] [PubMed] [Google Scholar]
- 10. Haydock S. F., et al. 1995. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 374:246–248 [DOI] [PubMed] [Google Scholar]
- 11. Reference deleted.
- 12. Horton R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93–99 [DOI] [PubMed] [Google Scholar]
- 13. Keating T. A., Walsh C. T. 1999. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3:598–606 [DOI] [PubMed] [Google Scholar]
- 14. Keatinge-Clay A. T., et al. 2003. Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure 11:147–154 [DOI] [PubMed] [Google Scholar]
- 15. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. 1999. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 16. Koppisch A. T., Khosla C. 2003. Structure-based mutagenesis of the malonyl-CoA:acyl carrier protein transacylase from Streptomyces coelicolor. Biochemistry 42:11057–11064 [DOI] [PubMed] [Google Scholar]
- 17. Liu H., Reynolds K. A. 1999. Role of crotonyl coenzyme A reductase in determining the ratio of polyketides monensin A and monensin B produced by Streptomyces cinnamonensis. J. Bacteriol. 181:6806–6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long P. F., et al. 2002. Engineering specificity of starter unit selection by the erythromycin-producing polyketide synthase. Mol. Microbiol. 43:1215–1225 [DOI] [PubMed] [Google Scholar]
- 19. Marsden A. F., et al. 1998. Engineering broader specificity into an antibiotic-producing polyketide synthase. Science 279:199–202 [DOI] [PubMed] [Google Scholar]
- 20. McDaniel R., Welch M., Hutchinson C. R. 2005. Genetic approaches to polyketide antibiotics. 1. Chem. Rev. 105:543–558 [DOI] [PubMed] [Google Scholar]
- 21. Pérez-Zúñiga F. J., et al. 2004. CE-108, a new macrolide tetraene antibiotic. J. Antibiot. 57:197–204 [DOI] [PubMed] [Google Scholar]
- 22. Reeves C. D., et al. 2001. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry 40:15464–15470 [DOI] [PubMed] [Google Scholar]
- 23. Rodríguez S., Santamaría R. I., Fernández-Abalos J. M., Díaz M. 2005. Identification of the sequences involved in the glucose-repressed transcription of the Streptomyces halstedii JM8 xysA promoter. Gene 351:1–9 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Schwecke T., et al. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. U. S. A. 92:7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seco E. M., Perez-Zuniga F. J., Rolon M. S., Malpartida F. 2004. Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem. Biol. 11:357–366 [DOI] [PubMed] [Google Scholar]
- 27. Seco E. M., Fotso S., Laatsch H., Malpartida F. 2005. A tailoring activity is responsible for generating polyene amide derivatives in Streptomyces diastaticus var. 108. Chem. Biol. 12:1093–1101 [DOI] [PubMed] [Google Scholar]
- 28. Seco E. M., Cuesta T., Fotso S., Laatsch H., Malpartida F. 2005. Two polyene amides produced by genetically modified Streptomyces diastaticus var. 108. Chem. Biol. 12:535–543 [DOI] [PubMed] [Google Scholar]
- 29. Sekurova O., Sletta H., Ellingsen T. E., Valla S., Zotchev S. B. 1999. Molecular cloning and analysis of a pleiotropic regulatory gene locus from the nystatin producer Streptomyces noursei ATCC 11455. FEMS Microbiol. Lett. 177:297–304 [DOI] [PubMed] [Google Scholar]
- 30. Sekurova O. N., et al. 2004. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 186:1345–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sletta H., et al. 2005. Nystatin biosynthesis and transport: nysH and nysG genes encoding a putative ABC transporter system in Streptomyces noursei ATCC 11455 are required for efficient conversion of 10-deoxynystatin to nystatin. Antimicrob. Agents Chemother. 49:4576–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang Y., Kim C. Y., Mathews I. I., Cane D. E., Khosla C. 2006. The 2.7-A crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. U. S. A. 103:11124–11129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reference deleted.
- 34. Weissman K. J. 2004. Polyketide biosynthesis: understanding and exploiting modularity. Philos. Transact. A Math. Phys. Eng. Sci. 362:2671–2690 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.