Abstract
European Commission regulation 2073/2005 on the microbiological criteria for food requires that Escherichia coli is monitored as an indicator of hygienic conditions. Since verocytotoxigenic E. coli (VTEC) strains often cause food-borne infections by the consumption of raw food, the Biological Hazards (BIOHAZ) panel of the European Food Safety Authority (EFSA) recommended their monitoring in food as well. In particular, VTEC strains belonging to serogroups such as O26, O103, O111, O145, and O157 are known causative agents of several human outbreaks. Eight real-time PCR methods for the detection of E. coli toxin genes and their variants (stx1, stx2), the intimin gene (eae), and five serogroup-specific genes have been proposed by the European Reference Laboratory for VTEC (EURL-VTEC) as a technical specification to the European Normalization Committee (CEN TC275/WG6). Here we applied a “modular approach” to the in-house validation of these PCR methods. The modular approach subdivides an analytical process into separate parts called “modules,” which are independently validated based on method performance criteria for a limited set of critical parameters. For the VTEC real-time PCR module, the following parameters are being assessed: specificity, dynamic range, PCR efficiency, and limit of detection (LOD). This study describes the modular approach for the validation of PCR methods to be used in food microbiology, using single-target plasmids as positive controls and showing their applicability with food matrices.
INTRODUCTION
Microbial analysis of foodstuffs is an integrated part of the management of microbial safety in the food chain. Standardized detection methods exist, which are based on the growth and isolation of the microorganism of interest and are in general referred to as “classical” culture methods. Based on the general expertise and supported by an international consensus in the field of food microbiology, the current detection methods are harmonized and commonly recognized as the standard detection methods (17). These methods are considered precise, practical, and relatively inexpensive in terms of laboratory consumables and reagents but are rather time-consuming; 3 to 7 days may be required before isolation and characterization, e.g., serotyping of the pathogen, are obtained. Currently, standardized methods exist for the detection of Escherichia coli O157 but not for the top five verocytotoxigenic E. coli (VTEC) serogroups; therefore, the scientific opinion of the European Food Safety Authority (EFSA) panel on Biological Hazards (BIOHAZ) strongly recommended the development and validation of methods for the detection of the top five VTEC serogroups: O26, O103, O111, O145, and O157 (6, 7).
During the last decade, several food-borne outbreaks have been reported, e.g., the recent emergency of the E. coli O104:H4 outbreak, and a strong demand for “rapid methods” has risen. The term “rapid” refers especially to the need for a short delay between an emergency alarm due to a potential pathogen outbreak and the outcome and decision. Irrespective of the alarm situation, the detection and identification of pathogenic microorganisms should be obtained through fully validated methods, which should be applied under accredited conditions to guarantee the correct risk assessment.
A trend toward molecular methods, namely PCR-based methods for the rapid detection of food-borne pathogens in food and feed microbiology, has been observed in recent years. Several real-time PCR methods have been developed, aiming at the rapid detection of pathogenic bacteria in different matrices. E. coli O157:H7, its virulence traits, and its detection have been the subject of several studies in recent years because of its importance and implications to public health (1, 19, 20, 21, 22). In addition, Fratamico et al. (11) developed a multiplex real-time PCR assay detecting several E. coli virulence traits in foods.
All the studies mentioned above highlight two important issues: first, the need for rapid methods to be implemented in food microbiology, and second, the need for validated methods. The ISO 16140:2003 standard, “Microbiology of food and animal feeding stuffs—protocols for the validation of alternative methods,” offers the possibility to validate the so-called “alternative methods” for official controls in the area of food microbiology (15). The alternative protocols could be accepted, as long as they are in line with internationally recognized methods, e.g., from the European Committee for Standardization (CEN), or are agreed upon by official national bodies. They represent, by definition, methods of analysis that detect or estimate the same analyte as the one measured by the corresponding reference method for a given category of products. The basic premise in such a method validation approach remains nevertheless that a comparative analysis between the outcome using the alternative method and the results obtained by the classical reference method should be demonstrated. An alternative approach to the above-mentioned so-called “global approach” is the separate validation of distinct consecutive steps in the analytical process. This approach has been designated the so-called “modular approach” (14). For a comprehensive description of the modular approach, please refer to the glossary provided in File S1 in the supplemental material.
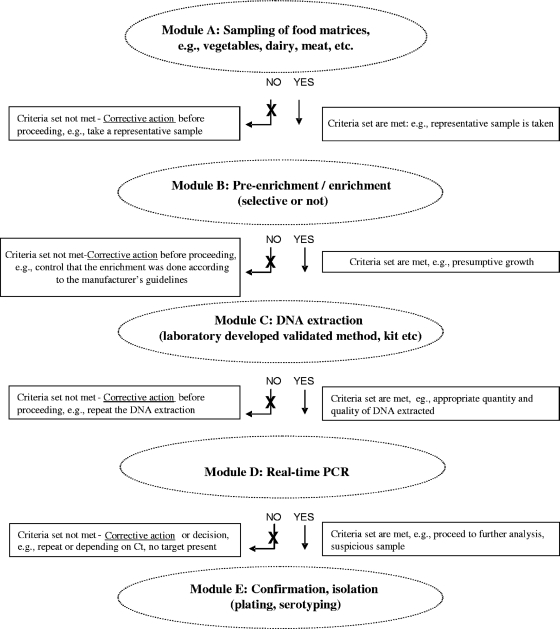
Here we wish to introduce the principle of a “modular approach” in microbial PCR method validation using some of the performance criteria of genetically modified organism (GMO) detection methods in the field of microbiology (4, 8, 18). A horizontal qualitative method for the detection of VTEC in foodstuffs submitted to CEN as the technical specification CEN/TC275/WG6 was chosen. This stepwise method consists of an enrichment step preventing the growth of Gram-positive bacteria, a DNA extraction, the real-time PCR analysis for the detection of the toxin and intimin genes (stx1, stx2, and eae genes), a serogroup determination by real-time PCR (only in case the stx/eae PCR is positive), the growth and isolation of suspected colonies, and confirmation of the pathogenicity traits by screening the colony itself. All eight real-time PCRs (stx1, stx2, eae, O26, O103, O111, O145, and O157) represent as such the real-time PCR “module” in the VTEC detection method (Fig. 1; see also File S1). These VTEC real-time PCRs were in-house tested for their specificity, dynamic range, PCR efficiency, and sensitivity (limit of detection [LOD]). This validation was performed on genomic DNA, while single-target plasmids were used as positive controls for the respective PCRs. The applicability of this VTEC PCR detection system on food matrices is also shown. The opportunities and limitations of the application of the modular approach in PCR method validation when applied to the detection of pathogenic microorganisms are discussed.
Fig. 1.
Schematic representation of the modular approach in food microbiology. The analytical process is subdivided into five steps, which are considered independent modules (here designated modules A, B, C, D, and E). In the modular approach, any method applied in a particular module should meet a minimal performance level for a number of critical method performance parameters. For each module, specific parameters can be defined/identified, allowing independent assessment of method performance for each of the different modules.
MATERIALS AND METHODS
Bacterial strains.
Reference E. coli strains of human or bovine origin, belonging to the five serogroups of interest and possessing different stx gene subtypes, were included in the study. Several strains containing none, one, two, or all three targets were analyzed. E. coli strains and respective targets are presented in Table 1. Specificity of the method was determined by testing the method against the following 16 closely related species: Hafnia alvei IZS 13 (Istituto Zooprofilattico Sperimentale), Enterobacter sakazakii CIP 103183 (Institut Pasteur), Listeria monocytogenes ATCC 19115, Salmonella enterica subsp. enterica ATCC 6994, Salmonella enterica serovar Senftenberg ATCC 43845, Salmonella enterica serovar Hadar (isolated by IZS), Salmonella enterica serovar Enteritidis IZS 581, Salmonella enterica serovar Cerro IZS 1138, Shigella boydii BAA-1247, Shigella dysenteriae ATCC 13313, Klebsiella pneumoniae ATCC 10031, Citrobacter freundii ATCC 43864, Yersinia enterocolitica ATCC 9610, Proteus mirabilis ATCC 7002, Campylobacter jejuni ATCC 49943, and Staphylococcus aureus ATCC 25923.
Table 1.
E. coli strains used to determine method specificitya
| Strain | VTEC PCR module specificity |
|||||||
|---|---|---|---|---|---|---|---|---|
| stx1 | stx2 | eae | O103 | O111 | O145 | O157 | O26 | |
| C210-03 (O157,stx1b,stx2c, eae) | + | + | + | − | − | − | + | − |
| ED 620 (O157, eae) | − | − | + | − | − | − | + | − |
| ED 621 (O157, eae) | − | − | + | − | − | − | + | − |
| C1178-04 (O145,stx1a,eae) | + | − | + | − | − | + | − | − |
| ED 645 (O145, stx2a, eae) | − | + | + | − | − | + | − | − |
| ED 657 (O145, stx2a, eae) | − | + | + | − | − | + | − | − |
| C125-06 (O103,stx2,eae) | − | + | + | + | − | − | − | − |
| ED 287 (O103, stx1a, eae) | + | − | + | + | − | − | − | − |
| ED 259 (O103, stx1a, eae) | + | − | + | + | − | − | − | − |
| MM13-02 (O111,eae) | − | − | + | − | + | − | − | − |
| ED 476 (O111, stx1a, stx2a, eae) | + | + | + | − | + | − | − | − |
| C1188-02 (O26,stx1a,stx2a,eae) | + | + | + | − | − | − | − | + |
| ED 643 (O26, stx1a, eae) | + | − | + | − | − | − | − | + |
| ED 654 (O26, stx2a, eae) | − | + | + | − | − | − | − | + |
Results derived from CT means ± SD from three repetitions. Reference strains are shown in boldface. The presence of the target gene(s) and serogroup for each strain is indicated in parentheses.
In silico bioinformatics analysis of primer, probes and amplicon DNA sequences.
Bioinformatics analysis of all DNA sequence information was performed by applying the NCBI software package (http://blast.ncbi.nlm.nih.gov/Blast.cgi). All relevant DNA sequence data were retrieved from public databases (NCBI). An in silico specificity analysis for each primer, probe, and amplicon DNA sequence by BLASTN analysis was performed by probing each nucleotide sequence against the databases and applying the default selection criteria.
Development of single-target VTEC reference plasmids.
The DNA sequences for the respective targets of the stx, the eae, and the serogroup genes were kindly provided by the European Reference Laboratory (EURL) for E. coli. The respective PCR target sequences covering stx1a, stx2a, stx2b, stx2c, stx2d, and the major eae gene variants, as well as the serogroup-specific genes (O157 [rfbE], O111 [wbdI], O26 [wzx], O145 [ihp1], and O103 [wzx]), were synthesized and inserted in the pUC19 vector (GeneArt, Regensburg, Germany). The correctness of the insert was verified by restriction enzyme analysis and by dideoxy sequencing. A total of nine single-target plasmids were constructed as follows: pCRL-ECstx1a, pCRL-ECstx2a,2c,2d, pCRL-ECstx2b, pCRL-ECeae, pCRL-ECwzx-O26, pCRL-ECihpl-O145, pCRL-ECwzx-O103, pCRL-ECwbdl-O111, and pCRL-ECrfb-O157.
Bacterial growth conditions, DNA extraction, and DNA quantification.
Pure overnight cultures of the bacterial strains to be used in the specificity testing were grown in tryptone soy broth (TSB) at 37°C. E. coli strains were grown in LB broth. Total DNA was extracted from overnight cultures using the DNA blood and tissue extraction kit (Qiagen, Milan, Italy). The DNA concentration was determined by fluorimetric means (PicoGreen; Invitrogen, Italy). All DNA extracts and the synthesized plasmids were diluted to a stock concentration of 20 ng/μl in molecular biology-grade water and stored at −20°C.
Serial decimal dilutions of the plasmid DNA were made in molecular biology-grade water. Each dilution was homogenized for 1 h before a subsequent aliquot was prepared. The estimated bacterial genome copy number at each dilution was calculated according to the following formula, number of copies = [ng amount × (6.023 × 1023)]/[number of base pairs × (1 × 109) × 660], where 660 is the average mass of a base pair in Daltons and 6.023 × 1023 is Avogadro's number (in molecules/mole).
Primers, probes, and real-time PCR conditions.
The sequences from the primers and probes for the respective targets were taken from the study by Nielsen and Andersen (19) and studies by Perelle et al. (21, 22), respectively. Primers and probes were purchased from Microsynth (Microsynth AG, Balgach, Switzerland). All probes were 6-carboxyfluorescein–6-carboxytetramethylrhodamine (FAM-TAMRA) labeled (Table 2).
Table 2.
List of the primers and probes used in this study and sizes of the expected amplicons
| Target (reference) | Sequence (5′–3′)a | Amplicon size (bp) | GenBank accession no. |
|---|---|---|---|
| stx1 (21) | Fw: TTT GTY ACT GTS ACA GCW GAA GCY TTA CG | 132 | M16625 |
| Rev: CCC CAG TTC ARW GTR AGR TCM ACR TC | |||
| Probe: CTG GAT GAT CTC AGT GGG CGT TCT TAT GTA A | |||
| stx2 (21) | Fw: TTT GTY ACT GTS ACAGCW GAA GCY TTA CG | 128 | X07865 |
| Rev: CCC CAG TTC ARW GTR AGR TCM ACR TC | |||
| Probe: TCG TCA GGC ACT GTC TGA AAC TGC TCC | |||
| eae (19) | Fw: CAT TGA TCA GGA TTT TTC TGG TGA TA | 102 | Z11541 |
| Rev: CTC ATG CGG AAA TAG CCG TTA | |||
| Probe: ATAGTC TCG CCA GTA TTC GCC ACC AAT ACC | |||
| rfbE (O157) (21) | Fw: TTT CAC ACT TAT TGG ATG GTC TCA A | 88 | AF163329 |
| Rev: CGA TGA GTT TAT CTG CAA GGT GAT | |||
| Probe: AGG ACC GCA GAG GAA AGA GAG GAA TTA AGG | |||
| wbdI (O111) (21) | Fw: CGA GGC AAC ACA TTA TAT AGT GCT TT | 146 | AF078736 |
| Rev: TTT TTG AAT AGT TAT GAA CAT CTT GTT TAG C | |||
| Probe-TTG AAT CTC CCA GAT GAT CAA CAT CGT GAA | |||
| wzx (O26) (21) | Fw: CGC GAC GGC AGA GAA AAT T | 135 | AF529080 |
| Rev: AGC AGG CTT TTA TAT TCT CCA ACT TT | |||
| Probe: CCC CGT TAA ATC AAT ACT ATT TCA CGA GGT TGA | |||
| ihp1 (O145) (21) | Fw: CGA TAA TAT TTA CCC CAC CAG TAC AG | 132 | AF531429 |
| Rev: GCC GCC GCA ATG CTT | |||
| Probe: CCG CCA TTC AGA ATG CAC ACA ATA TCG | |||
| wzx (O103) (22) | Fw: CAA GGT GAT TAC GAA AAT GCA TGT | 98 | AY532664 |
| Rev: GAA AAA AGC ACC CCC GTA CTT AT | |||
| Probe: CAT AGC CTG TTG TTT TAT |
In the sequences, Y is (C/T), S is (C/G), W is (A/T), R is (A/G), and M is (A/C). Fw, forward primer; Rev, reverse primer.
All PCR amplifications were performed in a final volume of 25 μl of reaction mixtures containing 1× of TaqMan universal PCR master mix (2×) (Applied Biosystems, Milan, Italy), 1 μM each primer, 200 nM probe, and the appropriate quantity of DNA template. All reactions were performed on an Applied Biosystems 7500 Fast real-time PCR system. The amplification conditions used were the following: 50°C for 2 min, 95°C for 10 min, and then 45 consecutive cycles of first 15 s at 95°C and then 1 min either at 55°C (wzx gene for serogroup O103 detection) or at 60°C (all other targets).
Determination of the specificity (inclusivity/exclusivity) of the respective VTEC PCR assays.
Primer pair specificity for each target was assessed by testing the amplification of plasmid and genomic DNA of target-containing and target-lacking strains. Four criteria were set to define what is considered a “specific signal” generated in TaqMan qPCR analysis, as follows: (i) an (exponential) amplification above the threshold level obtained with template DNA comprising the target sequence(s) (“inclusivity”), (ii) a lack of amplification with the negative controls, the so-called no-template controls (NTC), and the genomic DNA from strains reported to lack that particular target (Table 1) (“exclusivity”), and (iii) a single band on agarose gels after PCR amplification using target-containing template DNA, with (iv) a molecular weight corresponding to the predicted size for each PCR amplicon. In each analysis, 0.2 ng of DNA template was applied.
Statistical analysis, dynamic range, and LOD.
All statistical analyses were carried out using ProUCL 4.0 (http://www.epa.gov/esd/tsc/software.htm) and STATISTICA 9.1 (StatSoft Inc., Tulsa, OK). Unless stated otherwise, the criterion for significance was a P value of <0.05 for all comparisons.
The dynamic range of each of the PCR methods was determined over a 5-log concentration range (1 to 10,000 copies). Each dilution was assayed 6-fold, and a least-squares regression analysis was performed. In this study, the sensitivity of the PCR assays was expressed as the limit of detection (LOD) of bacterial genome copies. LOD was formally defined as the concentration which permits detection of the analyte at least 95% of the time.
In order to determine the LOD, the optimal number (N) of reactions that need to be carried out has to be estimated; here, we followed the procedure described in CRLVL04/08VP (3). As a starting point, we assume that at the limit of detection, X = 1 reaction out of N reactions is negative. The number of reactions was then determined by estimating a 95% confidence interval for a binomial distribution, whose upper bound for the probability of one negative result remains below α of 0.05. There are a number of ways to calculate confidence intervals for binomial proportions (2). Here, we used the normal approximation of the binomial distribution to estimate the confidence interval. Using p^ = X/N (where p^ is the sample proportion of successes), the upper confidence limit Lu, which is the only one of interest here, is given by
where z1 − α/2 is the 1 − α/2 percentile of the normal distribution, i.e., z = 1.96 for α = 0.05. A value of N has to be found so that Lu ≤ 0.05; a numerical solution for the above equation arrives at an N value of 60. Thus, a dilution series ranging from 20 genome copies down to theoretical 0.1 copies was analyzed by 60-fold. The LOD was set at the copy number range in which ≤59/60 positives were detected.
Several of the PCR data sets contained values designated nondetermined, meaning that no PCR amplification above the threshold was detected upon 45 amplification cycles. These values were treated as so-called “nondetects,” permitting us to include them in the computation of the LOD, summary statistics, and hypothesis tests (13). Means and standard deviations (SD) of threshold cycle CT values were thus computed in ProUCL 4.0 using maximum likelihood estimation, assuming that the data follow a normal distribution. This approach assumes that the distribution of CT values extends beyond the arbitrarily chosen cutoff point.
If multiple data sets were compared, the Gehan test (12) was employed. In this case, the global P value of 0.05 was adjusted for the number (n) of tests performed, i.e., the significance threshold was set at P′ = P/n.
Applicability of the method in food: artificial inoculation of food matrices, DNA extraction, and quantification.
Overnight cultures of each strain belonging to different serogroups were inoculated in LB broth and incubated at 37°C. Twenty-five grams or ml of three different matrices, namely, minced meat, ready-to eat salad, and pasteurized skimmed milk, were artificially inoculated with 2 to 10 CFU g−1 (or ml) of each serogroup, as determined after serial decimal dilutions and plating on selective agar (tryptone bile X-glucuronide [TBX]; Oxoid). Samples were subsequently mixed with 225 ml of modified TSB (mTSB) containing 1.5 g liter−1 bile salts N°3 and the appropriate supplement (16 mg liter−1 novobiocin or 12 mg liter−1 acriflavine), as suggested by CEN TC275/WG6, and homogenized for 5 min. One sample from each matrix was not inoculated but treated similarly to the inoculated ones, homogenized, and enriched; these samples were used as controls. Samples were incubated at 37°C for 18 to 24 h before 1 ml was taken twice from each sample, and DNA extractions were performed using the Pure Complete DNA and RNA purification kit (Epicentre, Madison, WI). All extractions were performed in duplicate, and the DNA extracted was quantified with PicoGreen (Invitrogen, Italy).
A loopful of each sample was streaked on TBX to confirm the presence or absence of the pathogen of interest. Absence of the pathogen before and after the enrichment in the control samples was assessed by TBX plating. The total bacterial load of each matrix was also estimated on plate count agar (Oxoid).
Real-time PCRs of the food matrices.
Matrix influences on the PCR efficiencies of all eight VTEC detection methods were investigated in two ways. The influence of the matrix itself was assessed prior to artificially inoculation using a commercially available kit (TaqMan exogenous internal positive control kit; Applied Biosystems). The kit was used for this purpose, according to the supplier's recommendations. Instead, for the inoculated samples, the presence of any PCR inhibitors was assayed by dilution analysis of the DNA extracted from 1 ml of the enriched cultures over at least a 3-log range and estimating the PCR efficiency by two-parameter regression analysis (CT value versus dilution factor). All analyses were performed in triplicate.
RESULTS
Determination of the specificity of VTEC PCR methods.
In silico bioinformatics analysis demonstrated that all VTEC PCR methods recognized only sequences corresponding to the respective toxin genes (data not shown). For the queries on the toxins, at least 100 different BLAST hits corresponding to the respective targets originating from different E. coli strains were obtained. For the serogroup genes, all 100% matching positive BLAST hits corresponded to the respective genes, and no other species apart from E. coli showed significant identities with the sequences in question. In the latter cases, however, fewer hits were obtained (data not shown).
The specificity of each real-time PCR module was experimentally confirmed by testing 16 closely related species and nine E. coli strains apart from the reference ones. No amplification was observed when DNA of non-E. coli strains was used for the reactions. For the E. coli strains, a total number of 336 analyses were performed, and no false-positive or false-negative results were obtained. The samples containing 0.2 ng/μl yielded CT values ranging between 22.13 and 34.11, depending on the method applied. The results presented are expressed as the means of three repetitions ± SD. Cross-reactivity of the serogroup modules was also investigated; only strains belonging to a specific serogroup annealed with the respective primer pairs, whereas no amplification was observed when the DNA was tested with the other serogroup methods.
Determination of the dynamic range by dilution series analysis.
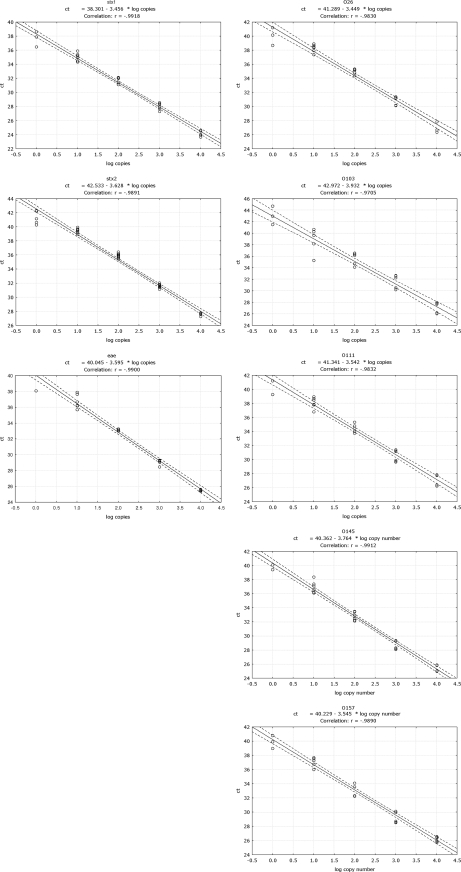
The term dynamic range refers to the range of concentrations over which a method performs in a linear manner with acceptable levels of accuracy and precision. The dynamic range of each target was determined based on six repetitions of DNA serial dilutions (Fig. 2).
Fig. 2.
Dynamic range of the real-time PCR methods for the detection of the stx1, stx2, and eae genes and the O26, O103, O111, O145, and O157 serogroups. Curves were obtained from six individual replicates for each concentration (expressed as the number of copies).
All methods performed in a satisfactory way over a range of 1 to 104 copies per reaction. Six out of six positive signals were obtained down to 10 copies per reaction; depending on the method, a positive signal was even obtained down to a concentration of an estimated single copy. Thus, the VTEC PCR methods performed well over a wide concentration range of target DNA (4 logs), and down to the 10 copies/reaction, a positive signal could always be obtained.
PCR efficiency of the pathogenic E. coli detection methods.
Based on the dynamic range of each reaction, the efficiency (E) for every single PCR method was calculated according to the following formula: E = [10(−1/slope)] − 1 (Fig. 2). Depending on the method, the PCR efficiency ranged between 80% and 95%. The best-performing method was the one of the serogroup O26, which had an efficiency of 95%; the least good method was the one targeting serogroup O103, whose efficiency was 80%. With the exception of the O103 and O145 PCR methods, whose efficiencies were below 90%, the remaining six methods had efficiencies above that value.
Interestingly, the efficiencies of the stx1 and stx2 methods were very similar (slopes equal to −3.534 and −3.628, respectively). In both reactions, the same pair of degenerated primers was applied, and specificity was thus provided only by the probe. In the cases in which both stx1 and stx2 genes were present in the same strain, e.g., serogroup O26, positive signals were obtained for both genes, implying no primer-probe selectivity for one target over the other and that no interference with the efficiency of the respective reactions occurred.
Determination of the LOD.
Based on the results of the dynamic range, in which the target was not always detected below 10 copies/reaction, an interval between 0.1 and 20 copies was chosen to determine the LOD of each PCR module. Sixty reactions, each containing a theoretical copy number of 15, 10, 5, and 0.1 target copies, were performed for each of the targets. In some cases (e.g., serogroups), no clear decision could be made regarding the LOD determination in that range, and therefore, a similar analysis at 20 copies was performed. Therefore, a total number of at least 240 reactions were performed for each method, which was statistically analyzed as described in Materials and Methods (Table 3).
Table 3.
LOD determinations for all VTEC PCR methodsa
| Copy no./method | Mean CT value ± SD (% positive) |
|||||||
|---|---|---|---|---|---|---|---|---|
| stx1 | stx2 | eae | O103 | O111 | O145 | O157 | O26 | |
| 20 | NAb | NA | NA | 38.64 ± 0.70 (100) | 35.11 ± 0.61 (100) | 35.7 ± 0.49 (100) | 36.5 ± 1.14 (95) | 36.2 ± 0.61 (100) |
| 15 | 34 ± 0.90 (100) | 34.1 ± 0.78 (100) | 34.8 ± 1.49 (100) | 36.8 ± 0.53 (97) | 38.4 ± 0.98 (100) | 37.5 ± 1.10 (100) | 38.6 ± 1.01 (95) | 38.7 ± 0.57 (98) |
| 10 | 36.5 ± 1.22 (95) | 36.4 ± 1.34 (98) | 35.9 ± 1.04 (100) | 39.86 ± 1.34 (98) | 37.9 ± 0.97 (93) | 36.1 ± 0.95 (98) | 36.4 ± 1.09 (98) | 37.7 ± 1.22 (97) |
| 5 | 37.5 ± 1.29 (87) | 38.3 ± 1.61 (93) | 38.0 ± 0.85 (98) | 38.9 ± 0.81 (98) | 39.7 ± 1.07 (97) | 39.0 ± 1.01 (95) | 40.1 ± 1.58 (82) | 40.2 ± 1.28 (97) |
| 0.1 | 42.3 ± 1.47 (5) | 41.4 ± 1.07 (12) | 41.1 ± 1.94 (8) | Undetermined | 41.6 (1.7) | 39.9 ± 0.56 (6.7) | Undetermined | 41.5 ± 0.08 (5) |
Based on 60 repetitions with genomic target DNA as the template. The copy number per reaction and the maximum likelihood estimates of the mean CT value ± SD are shown. The percentages of positives are presented in parentheses.
NA, not applicable.
The respective data sets for the different levels of the stx1, stx2, and eae targets were significantly different among each other (P = 0 for all pairs of comparisons). The LOD of each method was set at 1 dilution above the level in which only 59 out of 60 reactions gave a positive result. Thus, the LOD ranges between 10 and 15 copies for the stx1 and stx2 reaction and between 5 and 10 for that of the eae. Regarding the serogroup O26 method, all results were significantly different among each other (P = 0) except for the data set of 10 and 15 copies, which were not significantly different (P of 0.877 at a significance threshold of P′ of 0.0083). The LOD of this method was set between 5 and 15 copies. The copy numbers of the O111 method also presented significantly different CT values among all data sets (P = 0), except for the 5 and 10 copies (P of 0.016 at a significance threshold of P′ of 0.0083). The LOD of this method was set at 5 to 15 copies. All data sets for the serogroup O145 and O157 methods were significantly different; 10 copies versus 15 copies was considered significantly different at P of 0.0071 for O145 and at P of 0.0001 for O157 (both at a significance threshold of P′ of 0.0083). Moreover, 15 versus 20 copies was considered significantly different at P of 0.0002 for serogroup O157. The respective LODs were 10 to 15 copies for O145 and 10 to 20 copies for O157. Finally, O103 CT values were statistically different, except at levels 5 and 10, where no significant difference was observed (P of 0.0434 at a significance threshold of P′ of 0.0167). The LOD was thus set at 5 to 15 copies.
Applicability of the method on food matrices.
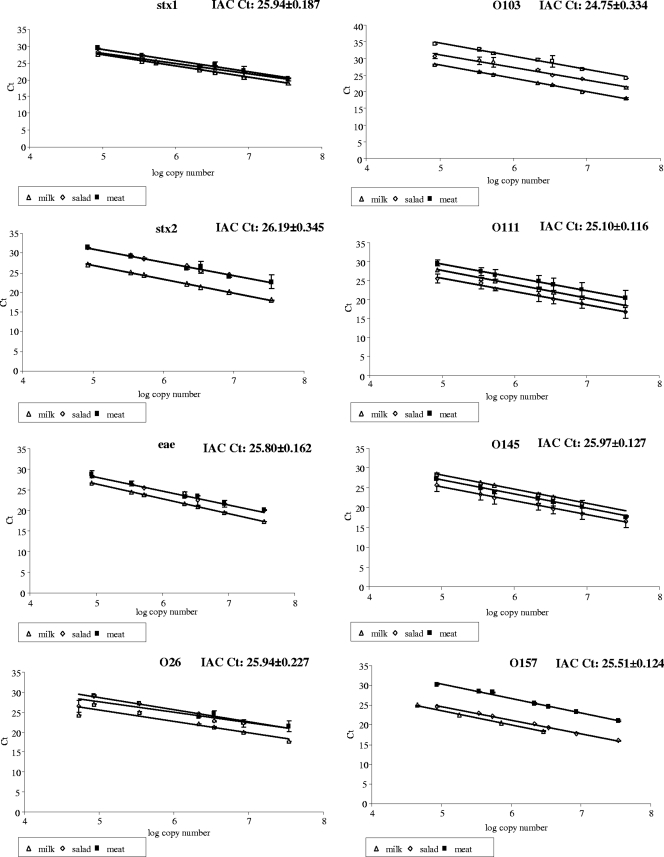
The initial total bacterial counts, as determined on perchloric acid (PCA), were 1.2 × 103 CFU g−1 in the case of meat and 6.6 × 102 CFU g−1 for salad, and none was detected in the pasteurized skimmed milk. DNA extracted from all matrices, inoculated and noninoculated ones, was quantified, and subsequently, 20 ng was used to determine the presence of inhibitors in the extract. No inhibition was present, since a similar CT value with the internal amplification control was always obtained among matrices and pure cultures (Fig. 3). Therefore, any negative results obtained with the VTEC PCR methods indicated the absence of the corresponding target in the sample. Regarding the inoculated samples, serial dilutions were prepared, starting from 20 ng of total DNA extracted down to 0.05 ng per reaction when possible, in the case in which less DNA was extracted (O157 strain inoculated in milk); 1:4 serial dilutions of the DNA were prepared and analyzed by real-time PCR (Fig. 3). All PCRs performed well, as demonstrated by the linearity of the curve and the efficiencies obtained, again indicating no interference of the tested matrices with the PCR analyses.
Fig. 3.
Dilution curves of the real-time PCR methods for the detection of the stx1, stx2, and eae genes and the O26, O103, O111, O145, and O157 serogroups in milk, salad, and meat. Curves were obtained from three individual replicates for each concentration (expressed as the number of copies).
All PCR results were in accordance with the ones obtained by plating on selective TBX plates. A positive PCR outcome matched the presence of only the correct pathogen that was inoculated, while no PCR amplification was detected, and characteristic E. coli colonies were not detected in any of the control samples.
DISCUSSION
The demand for standardized rapid analytical methods such as PCR-based methods for the detection of microbial organisms in food requires the establishment of an appropriate validation approach. Even though several PCR-based methods have been developed for food pathogen detection, to date, no official ISO method for using qualitative or real-time PCR with bacterial pathogens exists. A specific ISO standard is under development to set up the general requirements and definitions of real-time PCR when used for food-borne pathogens (ISO/FDIS 22119) (16).
In this study, a set of eight horizontal PCR methods submitted to CEN for the detection and characterization of five pathogenic E. coli serogroups was assessed. The technical specification focuses first on the demonstration of the presence of pathogenic bacteria, scored by detection of the combined presence of the toxin(s) and the intimin genes and second, in case of any positive outcome, on the further identification by PCR serotyping of the five serogroups. The selection of the virulence targets was based on the fact that Shiga toxins and intimin constitute two of the major virulence attributes of typical VTEC strains (1, 20). To our knowledge, no other CEN horizontal method for pathogen detection based on real-time PCR has been validated to date, nor have any method performance criteria been defined.
Here, we aimed at introducing a new concept in the validation of food microbiology methods, the modular approach. Modularity in method validation may greatly increase the flexibility of the analysis; moreover, it may represent a major cost reduction in process validation. The basic inference in this approach is that similar methods (here, real-time PCR) applied to one particular module are equally acceptable as long as a number of general predefined performance criteria for the module are met.
This study focused on the validation of the analytical PCR module per se using the VTEC methods as an example and assessed whether some of the PCR method performance criteria set for GMO detection could also serve in the case of microbial PCR detection methods. Four parameters were chosen as performance criteria for a real-time PCR method for microbial detection: the specificity, the dynamic range, the PCR efficiency, and the LOD. The specificity is a sine qua non criterion whenever a method aims at identifying the presence of particular targets in a sample. The dynamic range, the PCR efficiency, and the LOD represent characteristics of the sensitivity of the method. The dynamic range illustrates the linear correlation between the detection and the number of targets in the sample over a broad range of target concentrations. The PCR efficiency reflects how well a particular method amplifies a target compared to an ideal situation (efficiency at 100%). This numeric is particularly important when methods are applied in a combinatory detection approach in which the decisions depend on the presence of multiple targets. Finally, the LOD is a standard statistic of a method which determines the smallest amount of target that can be detected with a given certainty.
Both the in silico bioinformatics and the experimental analyses show that all methods are highly specific for their respective targets, covering a broad range of known VTEC strains. All methods perform very well over a wide range of target concentrations, maintaining acceptable PCR efficiencies (>80%). The limit of detection is also shown to be very low (between 5 and 20 copies) for all methods. Together, the results of these analyses demonstrate that the real-time PCR methods for detecting VTEC meet the strict criteria set for PCR methods in other fields (e.g., GMO detection [23]).
Pathogen detection procedures very often include an enrichment step, backing up the PCR results and allowing the proliferation and subsequent assignment of any positive target to its viability. In the case of the CEN horizontal method, during the enrichment, antibiotics which suppress the growth of the Gram-positive bacteria are used, therefore diminishing the background microflora. In this way, selective growth of the Gram-negative ones, such as E. coli, is obtained. Apart from the common practice of scoring viability of VTEC by plating, another inference to be taken into consideration is that both the intimin gene and at least one toxin gene have to be simultaneously present in the same cell. Therefore, positive signals for both genes do not necessarily indicate the presence of a pathogen, since the targets might be present in separate cells. Indeed, a VTEC strain potentially or highly pathogenic to humans should possess both the stx1 or/and stx2 and the intimin coding gene (CEN TC275/WG6). It is therefore highly recommended to always proceed in parallel with the isolation of the colonies and to assess for the presence of the pathogenicity traits in the same cell.
Even though these methods were initially developed for clinical sample analyses, our results demonstrated that they perform equally in food matrices. Indeed, upon artificial inoculation of minced meat, ready-to-eat salad, and skimmed milk, no PCR inhibition could be measured, and all expected targets were correctly detected (Fig. 3). These results show that when combined with a suitable extraction method, e.g., a validated test kit, no interference with the VTEC PCR module is observed. Also, background microflora present in these matrices did not interfere with the detection of the respective VTEC targets, even when the latter were inoculated at low levels, as was the case in our analyses. Finally, two recent interlaboratory trials organized by the EURL-E. coli fully support the findings of our study. In both trials, the results obtained by all the laboratories involved were in agreement with the true values of the samples (9, 10).
Together, the outcomes of these different studies support our initial assumption that a combination of independently validated methods from different modules provides a good basis for establishing an effective overall analytical process. Such a concept of “modularity” demands a minimal performance assessment of the methods of the respective modules. Our results indicate that for the PCR module, specificity, dynamic range, PCR efficiency, and LOD could be valuable PCR method performance criteria for microbial methods. An additional critical parameter for the performance of any PCR is the suitability of the DNA template used in the PCR analysis. Thus, the purity of the extracted DNA represents the most important criterion linked to the extraction module whenever PCR is used. The performance of the extraction module, however, can be assessed and validated independently from the PCR (and any other steps in the procedure) (Fig. 1). Since PCR is known to be prone to inhibition, any DNA extract should be verified through PCR inhibition monitoring, e.g., by including an internal amplification control or by dilution analysis of the extracted template DNA, as described in our study (24). It is always advised to perform a PCR inhibition analysis whenever unexpected negative results are obtained (for the possibility of false negatives).
A modular approach allows for establishing decision support systems that are independent of a matrix. Such an approach may be an advantage in all microbiological areas, including the food sector and the medical and clinical worlds, and also for enforcement control laboratories. In many cases, within a single experiment or within a single laboratory, a broad range of matrices are handled, and these could all be subject to multiple pathogen contaminations. To date, as stipulated by the ISO standard 16140:2003, each matrix/pathogen combination would require a separate validation applying the so-called global approach, meaning that each time either the matrix or the pathogen of interest changes, another validation has to be performed.
Modularity can thus be valuable to the validation of PCR screening methods for the detection of food-borne pathogens along the food chain. The validation within a modular approach, compared to the global one used so far, has the advantages that it allows the operator to determine at which individual stage of the procedure that biases are likely to occur, to decide whether they interfere with the final outcome, and if necessary, to take corrective actions within the module and reduce the overall cost of quality assurance over the process.
In conclusion, this study demonstrates the successful applicability of common performance criteria for PCR methods used in both GMO and food pathogen detection. Eight real-time PCR methods detecting and serotyping VTEC were assessed and performed appropriately. These results indicate that the so-called modular approach in method validation can be considered a valuable, cost-effective, and flexible asset for in-house validation. Our performance data for the VTEC PCR module strongly support its use as an analytical screening module for efficient assessment of the presence of VTEC in food.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M. N. Losio of the Istituto Zooprofilattico Sperimentale di Lombardia ed Emilia Romagna for kindly providing the DNA used in the study. We thank Luca Cocolin, Giuseppe Arcangeli, and Cristian Magnabosco for kindly allowing D.-M. Kagkli to use their facilities during some experimental procedures.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Beutin L., et al. 2007. Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by combination of serotyping and molecular typing of Shiga toxin genes. Appl. Environ. Microbiol. 73:4769–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown L. D., Cai T. T., DasGupta A. 2001. Interval estimation for a binomial proportion. Stat. Sci. 16:101–133 [Google Scholar]
- 3. Community Reference Laboratory for G. M. Food Feed (CRL-GMFF) 2009. Event-specific method for the detection of dried-killed bacterial biomass PT 73 (TM) derived from E. coli GM strain AG3139 using real-time PCR. CRLVL04/08VP. CRL-GMFF, Ispra, Italy [Google Scholar]
- 4. European Commission 2003. Regulation (EC) no. 1829/2003 of the European Parliament and of the Council on genetically modified food and feed. Off. J. Eur. Union L268:1–23 [Google Scholar]
- 5. European Commission 2003. Regulation (EC) no. 1830/2003 of the European Parliament and of the Council concerning the traceability and labeling of genetically modified organisms and the traceability of food and feed products produced from genetically modified organisms and amending Directive 2001/18/EC. Off. J. Eur. Union L268:24–28 [Google Scholar]
- 6. European Food Safety Authority 2007. Scientific opinion of the Panel on Biological Hazards on a request from EFSA on monitoring of verotoxigenic Escherichia coli (VTEC) and identification of human pathogenic VTEC types. EFSA J. 579:1–61 [Google Scholar]
- 7. European Food Safety Authority 2009. Technical specifications for the monitoring and reporting of verotoxigenic Escherichia coli (VTEC) on animals and food (VTEC surveys on animals and food) on request of EFSA. EFSA J. 7:1366–1409 [Google Scholar]
- 8. European Network of GMO Laboratories 2003. Definition of minimal performance requirements for analytical methods of GMO testing. http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_Analytical_methods.pdf
- 9. European Reference Laboratory for E. coli 2010. Report of the 4th inter-laboratory study on the detection of VTEC non-O157 in food samples. http://www.iss.it/binary/vtec/cont/Report_PT4_2010.pdf [Google Scholar]
- 10. European Reference Laboratory for E. coli 2010. Report of the 5th inter-laboratory study on verocytotoxin-producing E. coli (VTEC) identification and typing. http://www.iss.it/binary/vtec/cont/Report_PT5_2010.pdf [Google Scholar]
- 11. Fratamico P. M., DebRoy C., Miyamoto T., Liu Y. 2009. PCR detection of enterohemorrhagic Escherichia coli O145 in food by targeting genes in the E. coli O145 O-antigen gene cluster and the Shiga toxin 1 and Shiga toxin 2 genes. Foodborne Pathog. Dis. 6:605–611 [DOI] [PubMed] [Google Scholar]
- 12. Gehan E. A. 1965. Generalized two-sample Wilcoxon test for doubly censored data. Biometrika 52:650–653 [PubMed] [Google Scholar]
- 13. Helsel D. R. 2005. Nondetects and data analysis: statistics for censored environmental data. John Wiley and Sons, New York, NY [Google Scholar]
- 14. Holst-Jensen A., Berdal K. G. 2004. The modular analytical procedure and validation approach, and the units of measurement for genetically modified materials in foods and feeds. J. AOAC Int. 87:927–936 [PubMed] [Google Scholar]
- 15. International Organization for Standardization 2003. ISO 16140: microbiology of food and animal feeding stuffs—protocol for the validation of alternative methods. International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 16. International Organization for Standardization 2009. ISO/FDIS 22119: microbiology of food and animal feeding stuffs. Real-time polymerase chain reaction (PCR) for the detection of food-borne pathogens—general requirements and definitions. International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 17. Jasson V., Jacxsens L., Luning P., Rajkovic A., Uyttendaele M. 2010. Alternative microbial methods: an overview and selection criteria. Food Microbiol. 27:710–730 [DOI] [PubMed] [Google Scholar]
- 18. Joint FAO/WHO Food Standards Programme 2010. Codex Alimentarius Commission report of the thirty-first session of the codex committee on methods of analysis and sampling. ALINORM 10/33/23 (appendices II and III). Joint FAO/WHO Food Standards Programme, Rome, Italy: http://www.codexalimentarius.net [Google Scholar]
- 19. Nielsen E. M., Andersen M. T. 2003. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J. Clin. Microbiol. 41:2884–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paton J. C., Paton A. W. 1998. Pathogenesis and diagnosis of Shiga-toxin producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perelle S., Dilasser F., Grout J., Fach P. 2004. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Mol. Cell. Probes 18:185–192 [DOI] [PubMed] [Google Scholar]
- 22. Perelle S., Dilasser F., Grout J., Fach P. 2005. Detection of Escherichia coli serogroup O103 by real-time polymerase chain reaction. J. Appl. Microbiol. 98:1162–1168 [DOI] [PubMed] [Google Scholar]
- 23. Van den Eede G. L. 2010. Compendium of reference methods for GMO analyses. Publications Office of the European Union, Luxembourg, Luxembourg: http://publications.jrc.ec.europa.eu/repository/handle/111111111/15068 [Google Scholar]
- 24. Wilson I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.