Abstract
A new protocol employing nested PCR-restriction fragment length polymorphism (RFLP) based on the flaB gene and two restriction enzymes was worked out. This protocol allows the identification of all Borrelia species transmitted by Ixodes ricinus in Europe, including Borrelia miyamotoi and 3 genetic variants of B. garinii. A dendrogram of flaB sequence similarity was in accordance with RFLP variants.
TEXT
The Borrelia genus encompasses nearly 40 species of tick-transmitted pathogens, characterized by different host interactions and vector specificities. Those spirochetes are divided into two main groups: relapsing fever (RF) borreliae, transmitted by “soft ticks” (Argasidae), and the Borrelia burgdorferi sensu lato complex, also known as the Lyme disease (LD) borreliae, propagated by “hard ticks” (Ixodidae) (10). The host and vector specificities of the RF complex members facilitate their identification, with the exception of B. miyamotoi, which appears on three continents and which is transmitted by different tick species on each (3, 14, 15).
The spirochetes of the LD complex present host specificity (6), but they do not show strict vector requirements. In Europe, the main vector of all eight B. burgdorferi sensu lato species and of B. miyamotoi is Ixodes ricinus, whose hosts are at least 300 species of forest vertebrates (4, 5). It is therefore essential to precisely identify the Borrelia species spread on this continent by the same tick, since it determines correct risk assessment of different forms of borreliosis induced by different bacteria from the Lyme disease-inducing species.
Multiple markers are used for a DNA-based distinction of the Borrelia species, i.e., rss, encoding 16S rRNA, ospA, encoding surface protein A, and flaB, encoding flagellin, as well as noncoding genomic sequences rrfA-rrlB and rrs-rrlA for intergenic regions (1, 2, 8, 12, 13, 16). Some protocols for differentiation of Borrelia species involve the application of species-specific probes, but only five European species transmitted by I. ricinus have been identified using probes so far (9). Furthermore, the restriction analyses developed thus far permit the distinction of no more than 7 species of the B. burgdorferi sensu lato complex using 5 restriction enzymes, apart from B. bissettii and B. miyamotoi (8).
The aim of this study is the presentation of a possible application of the established PCR-restriction fragment length polymorphism (RFLP) protocol using two restriction enzymes for the identification of all Borrelia species transmitted by I. ricinus in biological material collected in Europe based on the flaB gene.
The examined material consisted of 52 Borrelia DNA isolates that had been obtained in previous studies from I. ricinus ticks collected from vegetation and removed from birds in west Poland and that were divided into 6 species of Borrelia (7, 17–20, 22).
The nested PCR method with genus-specific primer sets 132f/905r and 220f/823r used to detect the flaB gene fragment of Borrelia has been described earlier (21). DNA isolated from reference strains of 6 Borrelia species (Fig. 1 and 2) obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ; Germany) was used as positive controls. The PCR products were separated on 1.5% agarose gel (Prona, Spain) with addition of ethidium bromide (Sigma-Aldrich, Germany) at 5 V/cm for 1 h. A Nova 100-bp DNA ladder (Novazym, Poland) was applied for evaluation of the size of the obtained product. The results of the PCR were viewed under UV light and were archived in computer storage using BioCapt software (Vilber Lourmat, France).
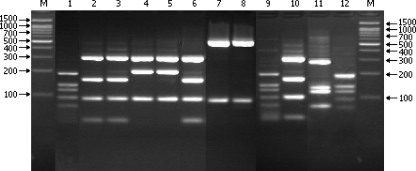
Fig. 1.
HpyF3I restriction patterns of the amplified flaB DNA (604 or 598 bp) from Borrelia strains. Lanes: M, marker DNA (Nova 100-bp DNA ladder); 1, 9, and 12, B. valaisiana; 2, 3, 6, and 10, B. afzelii; 4 and 5, B. lusitaniae; 7 and 8, B. miyamotoi; 11, B. bissettii. Positive controls: B. afzelii VS461 (lane 10), B. bissettii DN127 (lane 11), and B. valaisiana VS116 (lane 12).
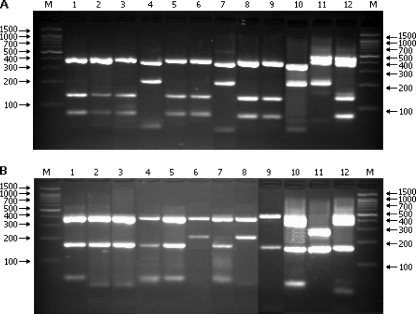
Fig. 2.
HpyF3I (A) and Ecl136II (B) restriction patterns of the amplified flaB DNA (604 or 598 bp) from Borrelia strains. Lanes: M, marker DNA (Nova 100-bp DNA ladder); 1, 5, and 7, B. garinii type Vb; 2, 3, and 12, B. garinii type Va; 4, 7, and 10, B. burgdorferi sensu stricto; 6 and 8, B. garinii type Vd; 9, B. bavariensis; 11, B. spielmanii. Positive controls: B. burgdorferi sensu stricto IRS (lane 10), B. spielmanii PC-Eq17 (lane 11), and B. garinii 20047 (lane 12).
The PCR-RFLP protocol with a single restriction enzyme, HpyF3I (Fermentas, Lituania), recognizing the CTNAG sequence inside a flaB gene fragment had been used previously for identification of Borrelia species (20, 21). This study constitutes an extension to that protocol with the use of an additional restriction enzyme, Ecl136II (Fermentas), which recognizes the GAGCTC sequence, allowing the identification of all Borrelia species vectored by I. ricinus. The procedure, based on analysis of unique sequences of the flaB gene from the European Borrelia species and strains obtained from GenBank (Table 1 ), was elaborated using DNAMAN 5.2.9 software (Biosoft, Canada), which performed in silico restriction analysis of the DNA sequences. Predicted restriction patterns are shown in Table 2. Digestion with HpyF3I generates 9 RFLP patterns, 6 of which are unique for 5 species: 2 for B. afzelii and 1 each for B. valaisiana, B. bissettii, B. lusitaniae, and B. miyamotoi. Therefore, no further analysis of the obtained results is necessary. The remaining 3 patterns require additional processing with the Ecl136II enzyme, which permits the distinction of B. burgdorferi sensu stricto from the unclassified strain Borrelia sp. SV1, of B. spielmanii from the south European strains of B. lusitaniae, and of B. garinii from B. bavariensis. The latter enzyme also allows strain differentiation within the heterogeneous species of B. garinii. The digestion products of both enzymes were analyzed on 3% agarose gel (Prona) and archived as described above.
Table 1.
Borrelia strains used in this study for working out the PCR-RFLP protocol based on the flaB gene for differentiation of Borrelia species and strains transmitted by I. ricinus
| Borrelia species | Strain | Source | Countrya (reference) | RFLP patternb with: |
Accession no. | |
|---|---|---|---|---|---|---|
| HpyF3I | Ecl136II | |||||
| B. burgdorferi sensu stricto | DG-1 | Canis lupus familiaris (blood) | Poland (21) | I | Ia | DQ016625 |
| D69-04 | I. ricinus | Poland (19) | I | Ia | DQ016620 | |
| ZS7 | I. ricinus | Germany | I | Ia | NC_011728 | |
| GL56-07 | I. ricinus | Poland† | I | Ia | HM345910 | |
| T90-5-02 | I. ricinus fed on Turdus merula | Poland† | I | Ia | HM345911 | |
| Borrelia sp. SV1 | SV1 | I. ricinus | Finland | I | Ib | NZ_ABJZ02000005 |
| B. afzelii | 9W10-04 | I. ricinus fed on Capreolus capreolus | Poland (19) | II | FJ874924 | |
| VS461 | I. ricinus | Switzerland | II | D63365 | ||
| ZL109-07 | I. ricinus | Poland† | II | HM345907 | ||
| ST19-05 | I. ricinus | Poland† | II | HM345908 | ||
| OS17-07 | I. ricinus | Poland† | II | HM345909 | ||
| P-Gau | Human | Germany | III | X63413 | ||
| B. bissettii | DN127 | Ixodes pacificus | United States | IV | D82857 | |
| B. garinii | 20047 | I. ricinus | France | V | Va | D82846 |
| PB35-99 | I. ricinus | Poland† | V | Va | HM345897 | |
| DB60-01 | I. ricinus | Poland† | V | Va | HM345899 | |
| ZL148-07 | I. ricinus | Poland† | V | Va | HM345900 | |
| D7-04 | I. ricinus | Poland (19) | V | Vb | DQ016622 | |
| D106-04 | I. ricinus | Poland (19) | V | Vb | DQ016621 | |
| T32-5-05 | I. ricinus fed on C. capreolus | Poland (19) | V | Vb | DQ650336 | |
| DB1F7-04 | I. ricinus | Poland (19) | V | Vb | DQ650331 | |
| DK29 | Human | Denmark | V | Vb | X69608 | |
| Far04 | Fratercula arctica | Faroe Islands | V | Vb | NZ_ABPZ02000016 | |
| HE | V | Vb | X69609 | |||
| K48 | I. ricinus | Slovakia | V | Vb | X69610 | |
| KL10 | V | Vb | L42881 | |||
| PBr | Human | Germany | V | Vb | NZ_ABJV02000003 | |
| DB74-01 | I. ricinus | Poland† | V | Vb | HM345898 | |
| T44-4-02 | I. ricinus fed on Turdus philomelos | Poland† | V | Vb | HM345904 | |
| ST12-05 | I. ricinus | Poland† | V | Vb | HM345905 | |
| RP54-05 | I. ricinus | Poland† | V | Vb | HM345906 | |
| T53-9-02 | I. ricinus fed on T. merula | Poland† | V | Vd* | HM345901 | |
| T40-10-02 | I. ricinus fed on T. philomelos | Poland† | V | Vd* | HM345902 | |
| T41-2-02 | I. ricinus fed on T. philomelos | Poland† | V | Vd* | HM345903 | |
| B. bavariensis sp. nov. | PBi | Human | Germany | V | Vc | NC_006156 |
| TRO | Human | Slovenia | V | Vc | X69614 | |
| DB18N6-04 | I. ricinus | Poland† | V | Vc | DQ650333 | |
| B. valaisiana | BA9F9-05 | I. ricinus | Poland (19) | VI | DQ650330 | |
| D58-04 | I. ricinus | Poland (19) | VI | DQ016624 | ||
| VS116 | I. ricinus | Switzerland | VI | D82854 | ||
| T107-7-02 | I. ricinus fed on T. merula | Poland† | VI | HM345913 | ||
| PR45-05 | I. ricinus | Poland† | VI | HM345912 | ||
| B. lusitaniae | D23-04 | I. ricinus | Poland (19) | VII | DQ016623 | |
| DB8-09-04 | I. ricinus | Poland† | VII | HM345916 | ||
| SP6-09 | I. ricinus | Poland† | VII | HM345914 | ||
| SP38-09 | I. ricinus | Poland† | VII | HM345915 | ||
| PotiB2 | I. ricinus | Portugal | VIII | VIIIa | D82856 | |
| B. spielmanii | A14S | Human | Netherlands | VIII | VIIIb | NZ_ABKB02000003 |
| B. miyamotoi | WL10-6-04 | I. ricinus fed on C. capreolus | Poland (19) | IX | DQ650338 | |
| 123T05-2 | I. ricinus fed on Cervus elaphus | Poland (19) | IX | FJ823229 | ||
| D110-07 | I. ricinus | Poland (20) | IX | FJ518804 | ||
| LB-M56 | I. ricinus | France | IX | AF529084 | ||
| OS109-07 | I. ricinus | Poland† | IX | HM345917 | ||
| PB111-09 | I. ricinus | Poland† | IX | HM345918 | ||
| SP48-09 | I. ricinus | Poland† | IX | HM345919 | ||
| ZL27-07 | I. ricinus | Poland† | IX | HM345920 | ||
†, this study.
*, new restriction pattern obtained in this study.
Table 2.
DNA fragments generated by digestion of the flaB gene amplified with 220f and 823r primers using HpyF3I and Ecl136II restriction enzymes
| Borrelia species | HpyF3I |
Ecl136II |
||
|---|---|---|---|---|
| Restriction fragment sizes (bp) | RFLP patterna | Restriction fragment sizes (bp) | RFLP pattern | |
| B. burgdorferi sensu stricto | 359, 207, 38 | I | 387, 166, 51 | Ia |
| Borrelia sp. SV1 | 359, 207, 38 | I | 387, 217 | Ib |
| B. afzelii PKo type | 305, 165, 92, 42 | II* | ||
| B. afzelii PGau type | 397, 165, 42 | III* | ||
| B. bissettii | 280, 135, 117, 72 | IV* | ||
| B. garinii 20047 type | 388, 135, 72, 9 | V | 387, 166, 36, 15 | Va |
| B. garinii PBr type | 388, 135, 72, 9 | V | 387, 166, 51 | Vb |
| B. bavariensis | 388, 135, 72, 9 | V | 438, 166 | Vc |
| B. valaisiana | 188, 135, 117, 92, 72 | VI* | ||
| B. lusitaniae Polish type | 305, 207, 92 | VII* | ||
| B. lusitaniae PotiB2 type | 397, 207 | VIII | 387, 166, 51 | VIIIa |
| B. spielmanii | 397, 207 | VIII | 279, 166, 159 | VIIIb |
| B. miyamotoi | 512, 86 | IX* | ||
*, unique restriction pattern, further differentiation is not required.
Application of the presented protocol led to the confirmation of the presence of 6 Borrelia species in Poland, i.e., B. burgdorferi sensu stricto (Fig. 2, lanes 4 and 7) (pattern Ia), B. afzelii (Fig. 1, lanes 2, 3, and 6) (pattern II), B. garinii (Fig. 2, lanes 1 to 3, 5, 6, 8, and 9) (pattern V), B. valaisiana (Fig. 1, lanes 1 and 9) (pattern VI), B. lusitaniae (Fig. 2, lanes 4 and 5) (pattern VII), and B. miyamotoi (Fig. 1, lanes 7 and 8) (pattern IX). Digestion of the samples displaying restriction pattern V (B. garinii/B. bavariensis) with the second enzyme indicated also the presence of B. bavariensis in Poland (Fig. 2, lane 9). Sequence variability of the flaB gene among the Polish isolates of B. garinii was higher than the variability estimated from the sequences available in GenBank (patterns Va and Vb). An additional restriction pattern (Vd) (Fig. 2, lanes 6 and 8), composed of fragments of 397 and 217 bp and different from the pattern predicted for B. garinii and B. bavariensis, was individuated. Pattern Vd was found only in tick samples removed from birds, whereas pattern Va was characteristic of strains obtained from host-seeking ticks and pattern Vb was generated in strains obtained from distinct sources (human and animal samples and ticks collected from vegetation and removed from mammals and birds) (Table 1). Further examinations are required to confirm the host-dependent genetic diversity within B. garinii species observed in this study. Within B. lusitaniae isolates, different restriction patterns reflect the geographical versatility of south and central European strains.
Partial sequencing of the flaB gene with primers 220f and 823r was performed for 25 positive samples and gave restriction patterns characteristic of different Borrelia species and strains. DNA sequencing was performed by dye termination cycle sequencing. Each strand was analyzed by using ABI fluorescence automated sequencers.
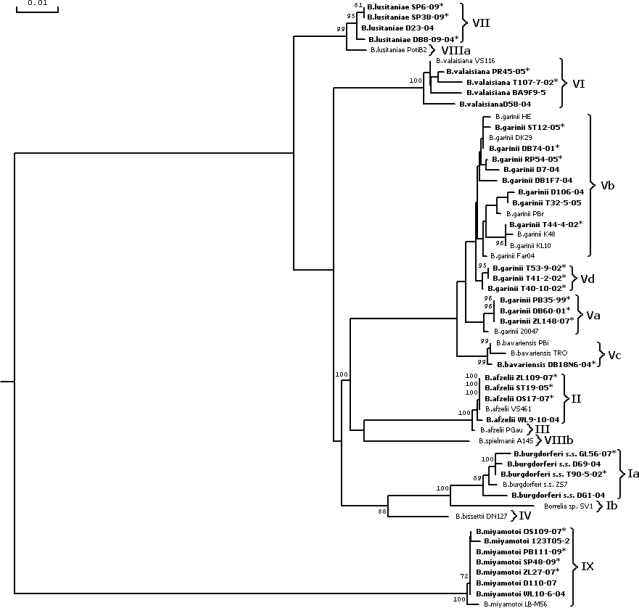
In order to compare the degrees of similarity of the examined samples within the restriction pattern groups, the above-mentioned sequences were analyzed and presented in a dendrogram indicating the RFLP patterns. The dendrogram fully confirmed the correctness of the Borrelia species identification carried out using the PCR-RFLP analysis by grouping the isolates in accordance with their taxonomic classifications (Fig. 3). The analysis revealed the similarity of the strain-generated Vd pattern to B. garinii, as these strains made a compact cluster, distinct from B. bavariensis (Fig. 3). This is evidence of the conserved character of the mutations in flaB in each species from the Borrelia genus. The grouping was also coherent among the isolates identified as B. garinii, as all three groups constituted separate clusters.
Fig. 3.
Dendrogram of genetic relationships among the flaB DNA sequences of the European Borrelia strains based on PCR-RFLP analysis. The tree was constructed by a maximum-likelihood method. The values in the tree represent bootstrap results. Polish strains are in boldface. *, sequences used in this study; Ia to IX, restriction patterns.
In conclusion, the application of the discussed protocol permits 14 easily resolvable genetic variants to be obtained, based on the analysis of the flaB gene fragment produced with the 220f and 823r primers from the DNA of 9 Borrelia species transmitted by I. ricinus: 1 for B. burgdorferi sensu stricto, B. valaisiana, B. bissettii, B. spielmanii, B. bavariensis, B. miyamotoi, and strain Borrelia sp. SV1; 2 for B. afzelii and B. lusitaniae; and 3 for B. garinii. Thus, the method not only is a precise tool for distinguishing between the Borrelia species but also indicates their variability, especially in case of the most heterogeneous strains of B. garinii. Furthermore, the method is fast and simple and does not require the use of specialized equipment, and the results are clear and easy to interpret.
Nucleotide sequence accession numbers.
flaB gene sequences obtained in this study were deposited in the GenBank/EMBL/DDBJ databases under accession numbers DQ650333 and HM345897 to HM345920 (Table 1).
Footnotes
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Derdáková M., Beati L., Pet'ko B., Stanko M., Fish D. 2003. Genetic variability within Borrelia burgdorferi sensu lato genospecies established by PCR-single-strand conformation polymorphism analysis of the rrfA-rrlB intergenic spacer in Ixodes ricinus ticks from the Czech Republic. Appl. Environ. Microbiol. 69:509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fukunaga M., Okada K., Nakao M., Konishi T., Sato Y. 1996. Phylogenetic analysis of Borrelia species based on flagellin gene sequences and its application for molecular typing of Lyme disease borreliae. Int. J. Syst. Bacteriol. 46:898–905 [DOI] [PubMed] [Google Scholar]
- 3. Fukunaga M., et al. 1995. Genetic and phenotypic analysis of Borrelia miyamotoi sp. nov., isolated from the ixodid tick Ixodes persulcatus, the vector for Lyme disease in Japan. Int. J. Syst. Bacteriol. 45:804–810 [DOI] [PubMed] [Google Scholar]
- 4. Gern L., Rouvinez E., Toutoungi L. N., Godfroid E. 1997. Transmission cycles of Borrelia burgdorferi sensu lato involving Ixodes ricinus and/or I. hexagonus ticks and the European hedgehog, Erinaceus europaeus, in suburban and urban areas in Switzerland. Folia Parasitol. (Praha) 44:309–314 [PubMed] [Google Scholar]
- 5. Gylfe A., et al. 1999. Isolation of Lyme disease Borrelia from puffins (Fratercula arctica) and seabird ticks (Ixodes uriae) on the Faeroe Islands. J. Clin. Microbiol. 37:890–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kurtenbach K., et al. 2002. Host association of Borrelia burgdorferi sensu lato—the key role of host complement. Trends Microbiol. 10:74–79 [DOI] [PubMed] [Google Scholar]
- 7. Michalik J., Wodecka B., Skoracki M., Sikora B., Stańczak J. 2008. Prevalence of avian-associated Borrelia burgdorferi s.l. genospecies in Ixodes ricinus ticks collected from blackbirds (Turdus merula) and song thrushes (T. philomelos). Int. J. Mol. Microbiol. 298(Suppl. 1):129–138 [Google Scholar]
- 8. Michel H., et al. 2004. An ospA-PCR/restriction fragment length polymorphism-based method for sensitive detection and reliable differentiation of all European Borrelia burgdorferi sensu lato species and OspA types. Med. Microbiol. Immunol. 193:219–226 [DOI] [PubMed] [Google Scholar]
- 9. Morán Cadenas F., et al. 2007. Identification of host bloodmeal source and Borrelia burgdorferi sensu lato in field-collected Ixodes ricinus ticks in Chaumont (Switzerland). J. Med. Entomol. 44:1109–1117 [DOI] [PubMed] [Google Scholar]
- 10. Parola P., Raoult D. 2001. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin. Infect. Dis. 32:897–928 [DOI] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12. Picken R. N. 1992. PCR primers and probes derived from flagellin gene sequences for specific detection of the agents of Lyme disease and North American relapsing fever. J. Clin. Microbiol. 30:99–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ranka R., Bormane A., Salmina K., Baumanis V. 2004. Identification of three clinically relevant Borrelia burgdorferi sensu lato genospecies by PCR-restriction fragment length polymorphism analysis of 16S–23S ribosomal DNA spacer amplicons. J. Clin. Microbiol. 42:1444–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rich S. M., Armstrong P. M., Smith R. D., Telford S. R. 2001. Lone star tick-infecting borreliae are most closely related to the agent of bovine borreliosis. J. Clin. Microbiol. 39:494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scoles G. A., Papero M., Beati L., Fish D. 2001. A relapsing fever group spirochete transmitted by Ixodes scapularis ticks. Vector Borne Zoonotic Dis. 1:21–34 [DOI] [PubMed] [Google Scholar]
- 16. Sparagano O. A., et al. 1999. Molecular detection of pathogen DNA in ticks (Acari: Ixodidae): a review. Exp. Appl. Acarol. 23:929–960 [DOI] [PubMed] [Google Scholar]
- 17. Wodecka B. 2003. Detection of DNA of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks in north-western Poland. Ann. Agric. Environ. Med. 10:171–178 [PubMed] [Google Scholar]
- 18. Wodecka B. 2008. Reservoir host identification in Borrelia-infected Ixodes ricinus ticks, p. 149–157.In Buczek A., Blaszak C. (ed.), Arthropods. Influence on host. Akapit, Lublin, Poland. [Google Scholar]
- 19. Wodecka B. 2007. Significance of red deer (Cervus elaphus) in the ecology of Borrelia burgdorferi sensu lato. Wiad. Parazytol. 53:231–237 [PubMed] [Google Scholar]
- 20. Wodecka B., Leońska A., Skotarczak B. 2010. A comparative analysis of molecular markers for the detection and identification of Borrelia spirochetes in Ixodes ricinus. J. Med. Microbiol. 59:309–314 [DOI] [PubMed] [Google Scholar]
- 21. Wodecka B., Rymaszewska A., Sawczuk M., Skotarczak B. 2009. Detectability of tick-borne agents DNA in the blood of dogs undergoing treatment for borreliosis. Ann. Agric. Environ. Microbiol. 16:9–14 [PubMed] [Google Scholar]
- 22. Wodecka B., Skotarczak B. 2005. First isolation of Borrelia lusitaniae DNA from Ixodes ricinus ticks in Poland. Scand. J. Infect. Dis. 37:27–34 [DOI] [PubMed] [Google Scholar]