Abstract
Pipelines transporting brackish subsurface water, used in the production of bitumen by steam-assisted gravity drainage, are subject to frequent corrosion failures despite the addition of the oxygen scavenger sodium bisulfite (SBS). Pyrosequencing of 16S rRNA genes was used to determine the microbial community composition for planktonic samples of transported water and for sessile samples of pipe-associated solids (PAS) scraped from pipeline cutouts representing corrosion failures. These were obtained from upstream (PAS-616P) and downstream (PAS-821TP and PAS-821LP, collected under rapid-flow and stagnant conditions, respectively) of the SBS injection point. Most transported water samples had a large fraction (1.8% to 97% of pyrosequencing reads) of Pseudomonas not found in sessile pipe samples. The sessile population of PAS-616P had methanogens (Methanobacteriaceae) as the main (56%) community component, whereas Deltaproteobacteria of the genera Desulfomicrobium and Desulfocapsa were not detected. In contrast, PAS-821TP and PAS-821LP had lower fractions (41% and 0.6%) of Methanobacteriaceae archaea but increased fractions of sulfate-reducing Desulfomicrobium (18% and 48%) and of bisulfite-disproportionating Desulfocapsa (35% and 22%) bacteria. Hence, SBS injection strongly changed the sessile microbial community populations. X-ray diffraction analysis of pipeline scale indicated that iron carbonate was present both upstream and downstream, whereas iron sulfide and sulfur were found only downstream of the SBS injection point, suggesting a contribution of the bisulfite-disproportionating and sulfate-reducing bacteria in the scale to iron corrosion. Incubation of iron coupons with pipeline waters indicated iron corrosion coupled to the formation of methane. Hence, both methanogenic and sulfidogenic microbial communities contributed to corrosion of pipelines transporting these brackish waters.
INTRODUCTION
Although the microbial communities in oil and gas fields have been analyzed extensively (3, 5, 12, 17, 21), those inhabiting the walls of pipelines transporting oil, water, or gas are only beginning to be characterized (10, 15, 18, 20, 25, 26, 31). Yet understanding these communities is highly relevant, as they contribute to microbially influenced corrosion (MIC), which, along with other forms of corrosion, can lead to pipeline failure. As a consequence, considerable effort is made to protect pipelines by injection of oxygen scavengers, corrosion inhibitors, and/or biocides.
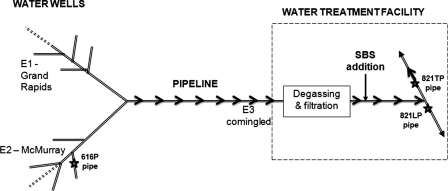
Corrosion accelerates when the anodic dissolution of metallic iron (Fe0 → Fe2+ + 2e) is effectively coupled with the cathodic reduction of an available electron acceptor, e.g., O2 under aerobic conditions (2H+ +1/2O2 + 2e → H2O) or protons under anaerobic conditions (2H+ + 2e → 2[H] and 2[H] → H2, where [H] represents atomic hydrogen). Because of the high reduction potential of O2, aerobic conditions are generally more corrosive than anaerobic conditions. Degassing and the addition of the oxygen scavenger sodium bisulfite (SBS) are used in the brackish-water-gathering system (investigated here) to prevent oxygen-mediated corrosion (Fig. 1).
Fig. 1.
Schematic of the brackish-water-gathering system. Water was collected through multiple pipelines, transporting 103 m3/day each from the Grand Rapids formation (E1) and the McMurray formation (E2). The comingled water (E3; 104 m3/day) was transported to a water treatment facility, where sodium bisulfite (SBS) was added to scavenge oxygen. Samples of pipe-associated solids (PAS) were obtained from sections of failed pipe collected either upstream (616P) or downstream (821TP of 821LP) from the SBS addition point. The main direction of flow is indicated by the arrows; the water in 821LP was often stagnant.
Under anaerobic conditions, the activity of sulfate-reducing bacteria (SRB) may accelerate corrosion by the use of cathodic H2 to reduce sulfate to sulfide, which precipitates 1/4 of the ferrous iron as FeS. In bicarbonate-rich brackish waters, as encountered here (Table 1), the remainder of the Fe2+ formed may precipitate as FeCO3, giving overall equations 1 and 2 as follows:
| (1) |
| (2) |
Table 1.
Concentrations (ppm) of selected analytes in brackish-water wellsa
| Analyte | Concn in well water (ppm) |
|
|---|---|---|
| Grand Rapids | McMurray | |
| Bicarbonate | 390–560 | 1,500–1,600 |
| Chloride | 2,500–3,600 | 5,200–5,300 |
| Sulfate | 0–1 | 2 |
| Sulfide | 0 | 0 |
| Nitrate + nitrite | 0 | 0 |
| Sodium | 1,700–2,300 | 3,500–3,600 |
| Calcium | 12–24 | 30–40 |
| Total iron | 0.16–0.61 | 2.6–2.8 |
| DOC | 0–10 | 0–10 |
| TOC | 0–10 | 0–10 |
Data represent the ranges observed for 10 Grand Rapids and two McMurray wells and were provided by the pipeline operator. DOC, dissolved organic carbon; TOC, total organic carbon.
In the absence of sulfate, methanogens may contribute to anaerobic corrosion of iron by using cathodic H2 for the reduction of CO2 to methane as indicated by equations 1 and 3:
| (3) |
However, because corrosion is a surface phenomenon, its mechanism is exceedingly complex and some have questioned whether microorganisms contribute in this way (6). The most detrimental forms of corrosion are localized. Given the ability of microorganisms to attach to surfaces and grow in favorable niches, it is understandable that this may lead to alteration of surface properties and ensuing pitting corrosion.
Here we focus on pipelines transporting brackish water from either the Grand Rapids or the McMurray formation near Fort McMurray, Alberta, Canada (Fig. 1), for use in the production of bitumen by steam-assisted gravity drainage (SAGD). All of the transported water is converted into steam, which is injected in the subsurface. Frequent corrosion failures in the cold part of this system require replacement of the affected pipe sections. This offers a unique opportunity to determine the microbial community composition at defined pipeline locations. Pipe wall community compositions are typically determined through analysis of pigging solids, obtained by scraping an entire pipeline with a spherical metal pig (10, 26), or by placing metal coupons in a pipeline that are removed after a certain period of time (15, 18, 20). Neither method represents sections of pipe that correspond to actual corrosion failures. Analysis of localized sections of failed pipe, as presented here, can indicate changes in the wall-attached microbial community as a result of treatment (e.g., injection of the oxygen scavenger bisulfite), allowing evaluation of whether the treatment has the desired effect of lowering corrosion risk.
MATERIALS AND METHODS
Sample description and handling.
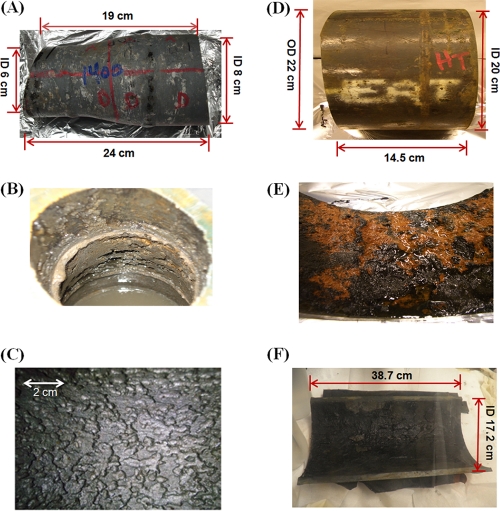
In addition to planktonic samples (E1, E2, and E3), three samples of failed pipe were obtained after being cut out of the leaking line from the locations indicated in Fig. 1. The samples were shipped in pipe-associated water (PAW) in an effort to preserve the microbial community that may have contributed to causing the leak. Samples were obtained both upstream and downstream of the SBS injection point, allowing us to focus on the effects of this treatment on the pipeline microbial community. The water and pipeline cutout samples analyzed in this study are listed in Table 2. The pipeline cutouts are shown in Fig. 2A, D, and F. Relevant dimensions, including inner diameter (ID) and length (L), are indicated in Fig. 2, as well as in Table 2, where the derived internal volume and pipe surface area are also given. Pipe section 616P was collected upstream of the SBS injection point, while pipe sections 821LP and 821TP were collected downstream, 821TP from a line with continuous flow and 821LP from a line that was often stagnant (Fig. 1). The subsurface temperature was 7 to 10°C, and the temperature of water in the pipeline was on average 13°C. Samples were shipped at ambient temperature (20°C) and were received within 1 to 2 days of collection. The pipe sections were transported in a bucket filled to the brim with PAW and closed with an air-tight lid. Upon arrival, parts of water samples E1, E2, and E3 were immediately filtered through 0.2-μm-pore-size sterile Millipore filters, with the remainder being stored in a Coy anaerobic hood (90% N2, 10% CO2). The filters with collected solids, including biomass, were stored frozen at −80°C. Pipe sections were removed from the bucket with PAW and transferred into the anaerobic hood. PAW was processed, as were E1, E2, and E3. Pipe-associated solids (PAS) were removed by scrubbing the internal pipe surface with a sterile spatula in the anaerobic hood and resuspending in PAW that had been subjected to filtration with a 0.2-μm-pore-size filter. Part of the PAS suspension was again filtered through a 0.2-μm-pore-size Millipore filter, and the filter was stored at −80°C, pending DNA extraction. The remainder of the PAS suspension was stored anaerobically at room temperature for further analyses.
Table 2.
Samples processed during the studya
| Date received | Description | Sample code | Amplicon code | MID | n | Groupb | No. of OTUs | SHIN | SIM | No. of taxac |
|---|---|---|---|---|---|---|---|---|---|---|
| 10/28/2009 | Pipe-associated solids | PAS-821LPd | 941 | TACGCTGTCT | 6,251 | a | 174 | 2.16 | 0.29 | 80 |
| PAS-821LPd | 940 | TACAGATCGT | 5,908 | a | 252 | 2.34 | 0.26 | 62 | ||
| PAS-821LPd | 929 | ATATCGCGAG | 4,850 | a | 313 | 2.35 | 0.28 | 53 | ||
| PAS-821LPd | 930 | CGTGTCTCTA | 5,522 | a | 268 | 2.59 | 0.2 | 70 | ||
| 10/28/2009 | Pipe-associated water | PAW-821LP | 939 | TACACGTGAT | 5,318 | b | 212 | 2.8 | 0.15 | 91 |
| PAW-821LP | 928 | ATCAGACACG | 4,653 | b | 294 | 3.19 | 0.11 | 96 | ||
| 8/13/2009 | Pipe-associated solids | PAS-616Pe | 516 | AGACGCACTC | 2,329 | c | 136 | 2.19 | 0.23 | 58 |
| PAS-616Pe | 1351 | TAGTATCAGC | 9,785 | c | 325 | 1.91 | 0.38 | 79 | ||
| 1/21/2010 | Pipe-associated solids | PAS-821TPf | 1718 | ATACGACGTA | 8,678 | d | 152 | 1.51 | 0.32 | 65 |
| 9/24/2009 | McMurray water | E2-2 | 938 | TACACACACT | 3,796 | e | 47 | 0.34 | 0.9 | 26 |
| E2-2 | 926 | ACGCTCGACA | 4,829 | e | 94 | 0.65 | 0.8 | 34 | ||
| 9/24/2009 | Comingled water | E3-2 | 927 | AGACGCACTC | 4,372 | e | 122 | 0.7 | 0.82 | 60 |
| 1/27/2010 | Grand Rapids water | E1-3 | 1714 | TGATACGTCT | 11,755 | e | 136 | 0.88 | 0.66 | 69 |
| 9/24/2009 | Grand Rapids water | E1-2 | 937 | CGACGTGACT | 5,456 | e | 156 | 1.69 | 0.42 | 91 |
| E1-2 | 925 | ACGAGTGCGT | 4,200 | e | 194 | 1.62 | 0.52 | 79 | ||
| 1/21/2010 | Pipe-associated water | PAW-821TP | 1717 | CGAGAGATAC | 9,488 | f | 354 | 2.82 | 0.16 | 169 |
| 1/27/2010 | Comingled water | E3-3 | 1716 | CATAGTAGTG | 10,450 | f | 267 | 2.45 | 0.22 | 104 |
| 1/27/2010 | McMurray water | E2-3 | 1715 | TACTGAGCTA | 9,396 | f | 187 | 1.61 | 0.29 | 99 |
| 8/13/2009 | Pipe-associated water | PAW-616P | 515 | ACGCTCGACA | 2,352 | g | 210 | 3.62 | 0.06 | 133 |
| PAW-616P | 514 | ACGAGTGCGT | 2,065 | g | 352 | 4.16 | 0.05 | 151 | ||
| 9/7/2009 | Comingled water | E3-1 | 365 | TGATACGTCT | 3,211 | h | 250 | 3.57 | 0.06 | 99 |
| 9/7/2009 | McMurray water | E2-1 | 364 | TCTCTATGCG | 6,055 | i | 522 | 4.15 | 0.04 | 179 |
Multiple table entries for a given sample were for amplicons obtained through different DNA isolation and/or PCR procedures as explained in the text. The multiplex identification sequence (MID) and the number of good 16S pyrotags obtained for each amplicon library (n) are indicated. The number of OTUs observed, the Shannon index (SHIN) and the Simpson index (SIM), and the number of derived taxa, describing microbial community diversity, are also indicated. The data are ordered according to the dendrogram in Fig. 3. Dates are given as month/day/year.
Dendrogram subgroup (see Fig. 3).
Data represent numbers of entries with at least one good read (see Table S1 in the supplemental material).
Pipe section 821LP had an inner diameter (ID) of 20 cm, a length (L) of 14.5 cm (Fig. 2C), an internal surface area (A) of 910 cm2, and an internal volume (V) of 4,553 cm3.
Pipe section 616P had an ID of 6 to 8 cm, an L of 21.5 cm (Fig. 2A), an A of 473 cm2, and a V of 827 cm3.
Pipe section 821TP had an ID of 17.2 cm, an L of 38.7 cm (Fig. 2E), an A of 810 cm2, and a V of 23763.
Fig. 2.
Pipe sections analyzed in this study. (A) Pipe section 616P (6 and 8 cm are the inner diameters [ID]). (B) Corrosion and scale on the inside pipe surface of 616P. (C) Closeup image of the pipe surface from a pipe section near 616P after removal of scale and cleaning. (D) Pipe section 821LP (22 cm is the outer diameter [OD]). (E) Corrosion and scale on the inside pipe surface of 821LP. (F) Corrosion and scale on the inside pipe surface of 821TP.
DNA extraction from the water and pipe samples.
Genomic DNA was isolated using a procedure modified from that of Marmur (19). Biomass-containing filtered solids were thawed, resuspended in 280 μl of 0.15 M NaCl and 0.1 M EDTA (pH 8), and treated with lysozyme. Samples were lysed by treatment with 25% sodium dodecyl sulfate (SDS) and three freeze-thaw cycles (at −70°C and 68°C), except those with amplicon codes 925, 926, 927, and 1351 (Table 2). The latter were lysed using a bead-beating procedure in a FastPrep Instrument for 60 s at a speed setting of 6.0 in 2-ml lysing matrix E tubes (MP Biomedicals). Following further processing, all samples were treated with DNase-free RNase and proteinase K (Roche Diagnostics GmbH, Germany) and by phenol extraction, precipitation with two volumes of 2.5 M sodium acetate in 95% ethanol, washing with 70% ethanol, and drying. The DNA was then dissolved in 10 mM Tris-Cl (pH 8.5) (buffer EB from a Qiagen QIAquick kit; Qiagen).
Community analysis by pyrosequencing.
DNA samples were generally amplified by two-step PCR amplification. The first PCR was performed with 16S primers 926Fw (AAACTYAAAKGAATTGRCGG) and 1392R (ACGGGCGGTGTGTRC). The PCR mixture (50 μl) contained 2 μl of 20-pmol/μl each forward and reverse primer, 25 μl of 2× PCR master mix containing 0.05 U/μl Taq DNA polymerase, reaction buffer, 4 mM MgCl2, 0.4 mM (each) deoxynucleoside triphosphates (dNTP; Fermentas), 21 μl of nuclease-free water, and 2 μl of DNA template (10 to 100 ng). PCR was performed for 3 min at 95°C, followed by 25 cycles of 30 s at 95°C, 45 s at 55°C, and 1.5 min at 72°C, and then 10 min at 72°C. The PCR product was checked on an 0.7% agarose gel and purified with a QIAquick PCR purification kit (Qiagen), and its concentration was determined using a Qubit fluorometer (Invitrogen) and a Quant-iT double-stranded DNA (dsDNA) HS assay kit (Invitrogen). The second PCR (10 cycles) was performed with 100 ng of PCR product and the FLX titanium amplicon primers 454T_RA_X and 454T_FwB, which have the sequences of 16S primers 926Fw and 1392R as their 3′ ends. Primer 454T_RA_X has a 25-nucleotide A-adaptor sequence (CGTATCGCCTCCCTCGCGCCATCAG) and a 10-nucleotide multiplex identifier barcode sequence (“MID” in Table 2), whereas primer 454T_FwB has a 25-nucleotide B-adaptor sequence (CTATGCGCCTTGCCAGCCCGCTCAG). The second-round PCR product was similarly checked and purified, and its DNA concentration was similarly determined. Amplicons for DNAs with sequence codes 937, 939, 940, and 941 (Table 2) were obtained by a single 35-cycle amplification with bar-coded primers. 16S PCR amplicons (typically 25 μl of 5 ng/μl) were sent for pyrosequencing (16S profiling) to the Genome Quebec and McGill University Innovation Centre, Montreal, Quebec, Canada. Pyrosequencing was performed with a Genome Sequencer (GS) FLX instrument, using a GS FLX Titanium Series XLR70 kit (Roche Diagnostics Corporation).
Analysis of pyrosequencing data.
Analysis was conducted with Phoenix 2, a 16S rRNA data analysis pipeline, developed in-house (see Fig. S1 in the supplemental material). Raw pyrosequencing data were subjected to stringent systematic checks to remove low-quality reads and minimize sequencing errors, which could have been introduced during the pyrosequencing process (13). Eliminated sequences included those that (i) did not perfectly match the adaptor and primer sequences, (ii) had ambiguous bases, (iii) had an average quality score below 27, (iv) contained homopolymer lengths greater than 8, or (v) were shorter than 200 bp after primer removal. The remaining high-quality sequences were compared to those in the nonredundant small-subunit (SSU) SILVA102 reference data set (22) using the Tera-BLAST algorithm and a 16-board TimeLogic Decypher system (Active Motif, Inc.). Sequences exhibiting an alignment of less than 90% of the trimmed read to the best BLAST match with more than 90% sequence identity were identified as potential chimeras and were excluded from further analysis. Sequences that passed quality control and chimeric sequence removal were clustered into Operational Taxonomic Units (OTUs) at a 3% distance by using the average linkage algorithm (24). After grouping sequences into OTUs, alpha diversity indices, including the total number of OTUs, the Simpson's diversity index, and the Shannon diversity index, were calculated for each sample. A taxonomic consensus of all representative sequences from each OTU was derived from the recurring species within 5% of the best bit score from a BLAST search of the SILVA database.
Amplicon libaries (Table 2) were clustered into a Newick-formatted tree by the unweighted-pair group method using average linkages algorithm (UPGMA), with the distance between communities calculated using the thetaYC coefficient (30) as a measurement of similarity between the structures of two communities in the Mothur software package (23). The Newick format of the sample relation tree was visualized using Dendroscope (14). Differences and similarities among amplicon libraries were also explored with Non-Metric Multidimensional Scaling (NMDS) ordination analysis using the thetaYC distance matrix in Mothur and the majorization algorithm method of Borg and Groenen (4). Statically significant differences in community structures were determined using analysis of molecular variance (AMOVA) based on the thetaYC coefficient.
XRD analysis of PAS.
PAS samples were sent to DNX Inc., Calgary, Alberta, Canada, for analysis of crystalline components by X-ray diffraction (XRD). Powder diffraction analysis using reference intensity ratio determinations was performed with a Rigaku Miniflex instrument and iron K-alpha radiation. The diffraction patterns for wall or pit scale were matched to standard patterns for siderite (iron carbonate), iron carbonate hydroxide, goethite (iron oxide hydroxide), elemental sulfur, mackinawite (iron sulfide), and elemental iron. Findings are presented as fractions (percent) of these components in the scale.
Iron corrosion under methanogenic conditions.
Iron coupons (4 by 1 by 0.1 cm) were cut from ASTM A366 carbon steel with 0.015% (wt/wt) carbon (ASTM International Designation A 1008/A). These were cleaned according to a standard protocol (NACE RP0775-2005). Briefly, the coupons were polished with 400-grit sandpaper and then placed in a dibutyl-thiourea-HCl solution for 2 min. The coupons were then neutralized in a saturated bicarbonate solution for 2 min, rinsed with deionized water and then with acetone, and finally dried in a stream of air. Coupons were weighed three times, and the average weight was recorded as the starting weight. Duplicate coupons were placed in small plastic holders to prevent them from contacting the glass wall or each other during tests.
For the corrosion experiments, 120-ml serum bottles were filled with 70 ml of brackish water (E1-3, E2-3, or E3-3) or with a Milli-Q purified water control (MQ) under anaerobic conditions. Two coupons were added, and the bottles were closed with a butyl rubber stopper. The headspace was 90% (vol/vol) N2 and 10% CO2. Each sample was incubated in duplicate at 32°C while being shaken at 100 rpm. After incubation for a number of hours (T), coupons were cleaned and dried according to the NACE standard protocol. The corrosion rate (CR) was determined from the metal weight loss (ΔW) according to the equation CR = 87,600 × ΔW/(A × T × D) mm/yr, where A represents the coupon area (cm2) and D represents the density of steel (7.85 g/cm3). A typical coupon weighed 4 g, had area of 8.9 cm2, and had a weight loss of 0.019 g over 2,688 h, giving a CR = 0.009 mm/yr.
During the course of the corrosion experiments, methane production was detected by injection of 0.2 ml of culture headspace into an HP 5890 gas chromatograph equipped with a stainless steel Porapak R 80/100 column (0.049 cm by 5.49 m). Injector and detector temperatures were 150 and 200°C, respectively. For determination of dissolved ferrous iron levels, a 100-μl aqueous sample was taken without shaking and added to 500 μl of 0.5 M HCl and mixed with 3 ml of ferrozine solution (1g/liter in 50 mM HEPES). Following incubation for 10 min at room temperature, the optical density at 562 nm was determined.
Nucleotide sequence accession numbers.
The entire set of sequences from the raw reads is available at the Sequence Read Archive (SRA) at NCBI under accession number SRP005237.
RESULTS
XRD analysis of pipeline scale.
Brackish waters from the Grand Rapids and McMurray formations had significant concentrations of sodium ion, chloride, and bicarbonate but lacked sulfate, sulfide, nitrate, nitrite, and organic carbon (Table 1). Concentrations in McMurray waters were generally higher than in Grand Rapids waters.
The surface of pipeline cutouts was covered by scale (Fig. 2B, E, and F). Many deep pits were observed once this scale was removed (Fig. 2C). In the case of pipe that was close to the subsurface water intake point (Fig. 1 [616P]), erosion corrosion through impingement of suspended sand, cavitation by bubbles of gaseous CO2 (formed when the water depressurizes at the surface), and CO2 corrosion were possible mechanisms for pit formation. Electrochemical carbonic acid or hydrogen sulfide corrosion (Fe0 + H2CO3 → FeCO3 + H2; Fe0 + H2S → FeS + H2), potentially stimulated by adhering microorganisms (see equations 2 and 3), must also have been occurring to explain the presence of ferrous iron-containing minerals [FeCO3, Fe2(OH)2CO3, and FeS] in the scale covering the corroded pipe (Table 3).
Table 3.
Composition of pipe wall scale upstream or downstream from the SBS injection point, as determined by XRD analysis
| Mineral or compound | Chemical formula | % upstream | % downstream |
|---|---|---|---|
| Siderite | FeCO3 | 85–95% | 55–65% |
| Iron carbonate hydroxide | Fe2(OH)2CO3 | 4–8% | 0% |
| Quartz (sand) | SiO2 | 4-8% | 0% |
| Sulfur | S8 | 0% | 20–30% |
| Mackinawite | FeS | 0% | 10–20% |
| Iron | Fe0 | Trace | 0% |
The pipeline scale upstream from the SBS addition point near well 616P (Fig. 1) had a high fraction of iron carbonates [FeCO3, 85 to 95%; Fe2(OH)2CO3, 4 to 8%], some sand (SiO2) (4 to 8%), and trace metallic iron. The presence of sand and metallic iron is consistent with an erosion corrosion process, i.e., sand abrasion of the iron surface. Sulfur (S8) and mackinawite (FeS) were not observed; however, these were major (20% to 30% and 10% to 20%, respectively) components of the scale downstream from the SBS addition point (Table 3), which had a smaller (55% to 65%) fraction of FeCO3. The S8 and FeS components clearly arose from the injected SBS, and microbial community analysis indicated that bacteria were likely involved in the formation of these scale components.
Dendrogram and NMDS ordination of microbial community compositions.
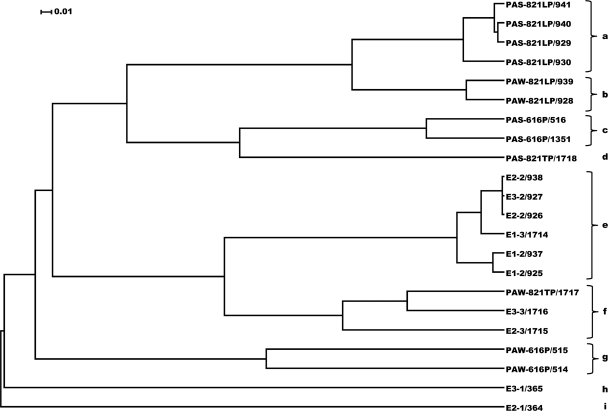
PCR amplification and sequencing of 14 samples gave 22 amplicon libraries (Table 2). The number of sequences in each library following processing of the Phoenix 2 pipeline is indicated in Table 2. Their relatedness was determined through the generation of the amplicon library tree (Fig. 3) as visualized with the UPGMA dendrogram program and NMDS ordination analysis (Fig. 4). Multiple amplicon libraries generated from the same sample always clustered together (Fig. 3), even with variations in DNA isolation methods (with or without bead beating) or PCR methods (two-step or single-step amplification), as indicated in Materials and Methods. The tree also indicated that amplicon libraries from sessile, pipe-associated solid samples PAS-616P, PAS-821LP, and PAS-821TP, as well as that from water sample PAW-821LP, clustered separately from those of all other planktonic water samples (Fig. 3). The latter were all from pipe sections experiencing rapid flow, except PAW-821LP, which was from a stagnant-water pipe section. Apparently stagnancy led to decreased differences between the planktonic and sessile communities, causing PAW-821LP to cluster together with PAS-821LP (Fig. 3).
Fig. 3.
Amplicon library relational tree. The tree was generated using the UPGMA algorithm with the distance between communities calculated using the thetaYC coefficient in the Mothur software package. The tree was visualized with Dendroscope. Dendrogram subgroups a to i are indicated. Amplicon libraries from sessile PAS samples (a, c, and d) formed trees that were separate from those for planktonic samples of fast-flowing water (e to i). Amplicon libraries for PAW-821LP (c), the single sample of stagnant water, formed a tree with those for the PAS samples. Bar, 0.01 substitutions per nucleotide position.
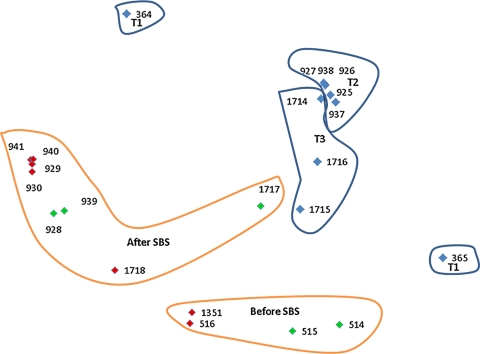
Fig. 4.
NMDS ordination of amplicon libraries calculated using the majorization algorithm based on the thetaYC coefficient matrix. Each point in the plot represents a single amplicon library, as indicated with its amplicon library code (Table 1). The distances between points represent the differences between the amplicon libraries. The red and green points represent amplicon libraries from sessile samples (PAS) and planktonic samples (PAW), respectively, which were shipped together. There was no clear separation between the sessile and planktonic amplicon libraries. However, there was a significant community structure shift between amplicon libraries from samples collected before and after the point of SBS addition. The blue points represent amplicon libraries from planktonic water samples E1, E2, and E3 (Fig. 1), which were received at times T1 (28 October 2009), T2 (24 September 2009), and T3 (27 January 2010). Amplicon libraries from samples collected at T1 are more distant from the amplicon libraries from samples collected at T2 and T3 than those are from each other.
The AMOVA of differences between amplicon libraries of planktonic and sessile (biofilm) samples was restricted to samples of PAS and PAW (pipe-associated solids and pipe-associated water), which arrived in the laboratory in the same bucket. Planktonic water samples E1, E2, and E3 were excluded from this analysis. Hence, we compared amplicon libraries 941, 940, 929, 930, 516, 1351, and 1718 from sessile PAS samples (Table 2) with libraries 939, 928, 1717, 515 and 514 from planktonic PAW samples. Alternatively, differences between amplicon libraries obtained from samples collected either upstream (516, 1351, 515, and 514) or downstream (941, 940, 929, 930, 939, 928, 1718, and 1717) of the point of bisulfite injection were analyzed. The observed separation of amplicon libraries before and after SBS addition (Fig. 4) was statistically significant, as determined by the AMOVA results (P < 0.001) (see Table S2 in the supplemental material). However, planktonic and biofilm communities exhibited no significant differences in community structures and membership at the given sample size (P = 0.116 [0.212]) (see Table S2 in the supplemental material). Planktonic community libraries from water samples E1, E2, and E3, collected at different times (T1 [libraries 364 and 365], T2 [938, 927, 926, 937, and 925], and T3 [1714, 1716, and 1715], as indicated in Table 1), were also compared (Fig. 4 [blue points]). Amplicon libraries from samples collected at T1 were more distant from the amplicon libraries from samples collected at T2 and T3 than those were from each other, and their spatial separation was statistically significant, as tested with AMOVA (see Table S2 in the supplemental material).
Microbial community composition of brackish waters.
The grouping of pyrotags into taxa by BLAST searches of the SILVA database is shown for all amplicons in Table S1 in the supplemental material. Sequences of multiple amplicon libraries from the same sample were combined into a single taxon distribution for each of the 14 samples (Table 4 ). Amplicons from subgroup e (Fig. 3) contained a very high fraction of taxa of the genus Pseudomonas (70% to 97%), giving this subgroup a low level of species richness and evenness, with a Shannon index (SHIN) of 0.34 to 1.69 and a Simpson index (SIM) of 0.42 to 0.90 (Table 2). Subgroup f contained 28 to 57% Pseudomonas spp. and 28 to 37% Methanobacteriaceae, increasing the evenness and richness of the community (Table 2 [SHIN = 1.61 to 2.82]). The taxon distributions of subgroups g, h, and i were distinct (Fig. 3) and among the most even (Table 2 [SHIN = 3.57 to 4.16]). All three subgroups contained 1.8% to 3.4% Pseudomonas, but only subgroup g had a high (8.5%) fraction of Methanobacteriaceae. This subgroup also had high fractions of the methanogenic genera Methanolobus (11.3%) and Methanocalculus (12.9%). Subgroup h contained high fractions of Methylophilaceae (30%) and Flavobacterium (10.8%), whereas subgroup i had high fractions of Anaerolinaceae (11.3%), Rhodocyclaceae (10.1%), and Streptococcus (13%).
Table 4.
Phylogenetic classification and distribution of pyrotags into taxa in combined amplicon librariesa
| Ranking | Taxonomical classification |
f (%) of taxonb for indicated sample code |
Total no. of reads (n, 130,721) | fall (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAS-821LP (amplicon codes 929, 930, 940, 941; dendrogram subgroup a; 22,533 reads) | PAW-821LP (amplicon codes 928, 939; dendrogram subgroup b; 9,971 reads) | PAS-616P (amplicon codes 516, 1351; dendrogram subgroup c; 12,114 reads) | PAS-821TP (amplicon code 1718; dendrogram subgroup d; 8,678 reads) | E2-2 (amplicon codes 926, 938; dendrogram subgroup e; 8,625 reads) | E3-2 (amplicon code 927; dendrogram subgroup e; 4,372 reads) | E1-3 (amplicon code 1714; dendrogram subgroup e; 11,755 reads) | E1-2 (amplicon codes 925, 937; dendrogram subgroup e; 9,656 reads) | PAW-821TP (amplicon code 1717; dendrogram subgroup f; 9,488 reads) | E3-3 (amplicon code 1716; dendrogram subgroup f; 10,450 reads) | E2-3 (amplicon code 1715; dendrogram subgroup f; 9,396 reads) | PAW-616P (amplicon codes 514, 515; dendrogram subgroup g; 4,417 reads) | E3-1 (amplicon code 365; dendrogram subgroup h; 3,211 reads) | E2-1 (amplicon code 364; dendrogram subgroup i; 6,055 reads) | ||||
| 1 | Genus Pseudomonas | 0.191 | 0.431 | 0.669 | 0.023 | 96.812 | 92.704 | 81.140 | 69.874 | 27.593 | 42.450 | 56.779 | 3.373 | 2.149 | 1.800 | 41,573 | 31.803 |
| 2 | Family Methanobacteriaceae | 0.644 | 13.569 | 56.241 | 40.551 | 0.325 | 0.069 | 13.569 | 0.238 | 28.215 | 21.033 | 36.739 | 8.467 | 0.031 | 0.000 | 22,181 | 16.968 |
| 3 | Genus Desulfomicrobium | 47.783 | 28.663 | 0.000 | 18.414 | 0.012 | 0.091 | 0.000 | 0.041 | 2.551 | 0.019 | 0.000 | 0.136 | 0.000 | 0.000 | 15,482 | 11.844 |
| 4 | Genus Desulfocapsa | 21.737 | 13.108 | 0.000 | 35.262 | 0.000 | 0.000 | 0.485 | 0.373 | 7.757 | 0.010 | 0.011 | 0.136 | 0.000 | 1.453 | 10,190 | 7.795 |
| 5 | Family Anaerolineaceae (uncultured) | 11.019 | 7.111 | 0.545 | 1.636 | 0.000 | 0.046 | 0.009 | 0.083 | 3.457 | 0.029 | 0.074 | 0.838 | 0.062 | 11.329 | 4,474 | 3.423 |
| 6 | Genus Desulfurivibrio | 3.533 | 13.810 | 0.008 | 0.346 | 0.000 | 0.183 | 0.706 | 7.457 | 0.685 | 0.048 | 0.000 | 0.023 | 0.000 | 0.231 | 3,100 | 2.371 |
| 7 | Family Coriobacteriaceae (marine group) | 3.386 | 2.928 | 13.398 | 0.012 | 0.197 | 0.023 | 0.247 | 0.476 | 0.632 | 0.029 | 0.149 | 1.200 | 0.000 | 0.264 | 2,918 | 2.232 |
| 8 | Family Rhodocyclaceae (uncultured) | 4.642 | 0.963 | 0.066 | 0.300 | 0.012 | 0.137 | 0.026 | 0.031 | 0.991 | 0.555 | 0.000 | 0.136 | 0.062 | 10.058 | 1,958 | 1.498 |
| 9 | Genus Desulfuromonas | 0.284 | 0.532 | 9.782 | 0.012 | 0.046 | 0.274 | 0.357 | 2.589 | 0.401 | 0.124 | 0.064 | 1.200 | 0.062 | 1.156 | 1,793 | 1.372 |
| 10 | Genus Rhodoferax | 0.000 | 0.020 | 0.000 | 0.000 | 0.000 | 0.206 | 0.009 | 5.468 | 3.868 | 6.641 | 0.000 | 0.068 | 1.028 | 1.453 | 1,725 | 1.320 |
| 11 | Family Methylophilaceae (uncultured) | 0.000 | 0.000 | 0.008 | 0.000 | 0.000 | 0.252 | 0.000 | 0.145 | 0.601 | 2.775 | 0.000 | 0.045 | 30.364 | 0.231 | 1,364 | 1.043 |
| 12 | Order Desulfuromonadales (Sva1033) | 0.164 | 0.632 | 7.586 | 0.000 | 0.104 | 0.160 | 0.434 | 1.367 | 0.569 | 0.163 | 0.021 | 0.815 | 0.000 | 0.000 | 1,327 | 1.015 |
| 13 | Family Comamonadaceae (uncultured) | 0.036 | 0.130 | 0.066 | 0.000 | 0.000 | 1.372 | 0.136 | 0.145 | 0.896 | 3.722 | 0.043 | 4.302 | 9.903 | 1.288 | 1,183 | 0.905 |
| 14 | Genus Thiobacillus | 0.036 | 0.060 | 0.239 | 0.000 | 0.000 | 0.412 | 0.017 | 2.185 | 5.902 | 0.794 | 0.011 | 0.974 | 0.062 | 3.369 | 1,167 | 0.893 |
| 15 | Genus Thauera | 0.000 | 0.020 | 3.219 | 0.000 | 0.012 | 0.274 | 0.000 | 0.041 | 0.105 | 0.038 | 0.021 | 4.981 | 0.062 | 3.898 | 883 | 0.675 |
| 16 | Genus Streptococcus | 0.004 | 0.020 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.031 | 0.021 | 0.000 | 0.011 | 0.702 | 0.000 | 13.113 | 834 | 0.638 |
| 17 | Genus Acetobacterium | 0.027 | 0.481 | 1.139 | 0.046 | 0.128 | 0.000 | 0.621 | 0.456 | 0.516 | 0.038 | 1.171 | 7.086 | 0.000 | 0.033 | 802 | 0.614 |
| 18 | Family Rhizobiaceae | 0.004 | 0.030 | 1.106 | 0.000 | 0.000 | 0.114 | 0.179 | 0.000 | 0.653 | 2.287 | 0.096 | 0.181 | 8.004 | 0.066 | 743 | 0.568 |
| 19 | Genus Methanolobus | 0.146 | 0.281 | 0.025 | 0.265 | 0.023 | 0.046 | 0.000 | 0.021 | 0.569 | 0.153 | 0.287 | 11.297 | 0.000 | 0.000 | 689 | 0.527 |
| 20 | Genus Hydrogenophaga | 0.004 | 0.010 | 0.033 | 0.000 | 0.000 | 0.526 | 0.009 | 0.031 | 0.812 | 2.995 | 0.000 | 0.091 | 6.571 | 0.595 | 674 | 0.516 |
| 21 | Genus Proteiniphilum | 0.377 | 3.901 | 0.297 | 0.012 | 0.012 | 0.000 | 0.000 | 0.145 | 0.011 | 0.000 | 0.011 | 2.830 | 0.000 | 0.017 | 654 | 0.500 |
| 22 | Genus Polaromonas | 0.000 | 0.060 | 0.000 | 0.000 | 0.000 | 0.023 | 0.000 | 4.381 | 0.042 | 0.000 | 0.000 | 0.113 | 0.000 | 3.534 | 653 | 0.500 |
| 23 | Genus Methanocalculus | 0.000 | 0.010 | 0.116 | 0.000 | 0.000 | 0.000 | 0.000 | 0.021 | 0.074 | 0.153 | 0.404 | 12.905 | 0.000 | 0.000 | 648 | 0.496 |
| 24 | Genus Leptolinea | 0.683 | 0.592 | 0.000 | 0.115 | 0.000 | 0.000 | 0.000 | 0.000 | 0.084 | 0.000 | 0.000 | 0.294 | 0.000 | 6.639 | 646 | 0.494 |
| 25 | Genus Flavobacterium | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.137 | 0.000 | 0.000 | 0.970 | 0.517 | 0.000 | 0.023 | 10.838 | 0.017 | 503 | 0.385 |
| 26 | Genus Methylomicrobium | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.046 | 0.000 | 0.000 | 1.781 | 2.794 | 0.000 | 0.000 | 0.623 | 0.000 | 483 | 0.369 |
| 27 | Family Methylococcaceae | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.206 | 0.000 | 0.000 | 0.105 | 2.335 | 0.000 | 0.023 | 6.571 | 0.000 | 475 | 0.363 |
| 28 | Genus Brachymonas | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.009 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 7.680 | 466 | 0.356 |
| 29 | Genus Fusibacter | 0.444 | 1.544 | 0.660 | 0.507 | 0.023 | 0.000 | 0.000 | 0.021 | 0.263 | 0.000 | 0.011 | 0.543 | 0.156 | 0.000 | 437 | 0.334 |
| 30 | Family Rikenellaceae | 0.244 | 0.933 | 0.124 | 0.565 | 0.093 | 0.000 | 0.000 | 0.010 | 0.263 | 0.000 | 0.106 | 2.106 | 0.031 | 0.000 | 350 | 0.268 |
| 31 | Genus Acholeplasma | 0.475 | 0.261 | 0.305 | 0.438 | 0.000 | 0.023 | 0.000 | 0.000 | 0.727 | 0.000 | 0.085 | 0.770 | 0.000 | 0.083 | 325 | 0.249 |
| 32 | Order Spirochaetales (PL-11B10) | 0.169 | 0.732 | 0.685 | 0.000 | 0.058 | 0.046 | 0.000 | 0.000 | 0.538 | 0.038 | 0.287 | 0.181 | 0.031 | 0.000 | 292 | 0.223 |
| 33 | Genus Aquabacterium | 0.027 | 0.090 | 0.116 | 0.000 | 0.000 | 0.000 | 0.043 | 0.155 | 0.053 | 0.000 | 0.043 | 5.026 | 0.000 | 0.000 | 280 | 0.214 |
| 34 | Genus Dechloromonas | 0.240 | 0.431 | 0.008 | 0.012 | 0.000 | 0.595 | 0.000 | 0.010 | 0.295 | 0.919 | 0.000 | 0.226 | 0.498 | 0.050 | 279 | 0.213 |
| 35 | Genus Methanosaeta | 0.000 | 0.211 | 0.000 | 0.012 | 0.000 | 0.000 | 0.009 | 0.041 | 0.137 | 0.000 | 0.043 | 0.249 | 0.000 | 3.683 | 278 | 0.213 |
All good reads n for multiple amplicon libraries from a single sample were combined, as indicated by the amplicon code(s). The fraction of reads, f (%), is indicated for each taxon. The sum of all reads is 130,721. The total number of reads (nall) and the sum fraction of reads, fall (%), for each taxon are also indicated and were used to rank the table. Results representing an fall (%) of <0.2 are not shown but can be found in Table S1 in the supplemental material.
Overall, the data indicated that the 10 planktonic samples of rapidly flowing water harbored diverse communities in which minimally 47 OTUs (26 taxa) and maximally 522 OTUs (179 taxa) were observed at the given sequence depth (Table 2).
Microbial community composition of pipe wall samples.
Pseudomonas is usually a dominant genus in amplicon libraries from planktonic water samples, but Pseudomonas bacteria were only a minor (0.02 to 0.67%) community component in amplicon libraries from sessile pipeline samples (Table 4 [PAS-821LP, PAS-616P, and PAS-821TP]). We have used the sample codes to refer to these libraries, as these can be directly related to their positions in the brackish-water-production operation (Fig. 1). The pipe wall upstream from the SBS addition point (Table 4 [PAS-616P]) harbored a very high fraction of Methanobacteriaceae (56%) as well as of Coriobacteriaceae (13%) and Desulfuromonas (9.8%). The physiological function of the latter is unknown, because no sulfur is present in the pipe scale at 616P (Table 3). Desulfuromonas acetoxidans couples oxidation of acetate to the reduction of sulfur to sulfide. The pipe wall downstream from the SBS addition point (Table 4 [PAS-821TP]) showed Methanobacteriaceae (41%) but also Desulfocapsa (35%) and Desulfomicrobium (18%) as major community components. Desulfocapsa derives energy for growth from the disproportionation of sulfur, thiosulfate, or bisulfite to sulfide and sulfate. For bisulfite, the reaction is as follows:
| (4) |
Desulfocapsa sulfoexigens is an autotroph that requires only CO2 as the carbon source (11). Some SRB, including Desulfovibrio sulfodismutans, can also catalyze reactions (4), but in keeping with their status as incomplete oxidizers, these require acetate and CO2 as the carbon source (1, 2). Desulfurivibrio, which was found in PAS-821TP at 0.3% of the reads (Table 4), can reduce sulfur and polysulfides, with H2 as the electron donor. The reactions reported to be catalyzed by these microorganisms do not lead to formation of elemental sulfur (S0). The finding of 20 to 30% sulfur in the scale near 821TP (Table 2) suggests that bisulfite may also be disproportionated to sulfur and sulfate:
| (5) |
This reaction was not proven by our study, and alternative explanations for the observed production of sulfur must be considered. The SRB Desulfomicrobium may use cathodic hydrogen to reduce sulfate to sulfide (reactions 1 and 2), forming FeS and FeCO3, of which 10% to 20% and 55% to 65% were present in the scale downstream of the SBS injection point (Table 2).
The community in PAS-821LP, which often experienced stagnant conditions, had a low fraction of Methanobacteriaceae (0.6%), while harboring high fractions of Desulfomicrobium (48%), and Desulfocapsa (22%), as well as some Desulfurivibrio (3.5%). Hence, this community was dominated by Deltaproteobacteria capable of reducing sulfate, sulfur, or polysulfide or of disproportionating sulfur and bisulfite. Overall, the results indicated a large community change as a result of the injection of SBS, with Deltaproteobacteria of the genera Desulfocapsa and Desulfomicrobium becoming major community components. Pipe sections subjected to rapid flow (821TP) appeared to maintain a high fraction of Methanobacteriaceae in the sessile population not seen in stagnant-water pipe sections (821LP).
Contribution of methanogens to iron corrosion.
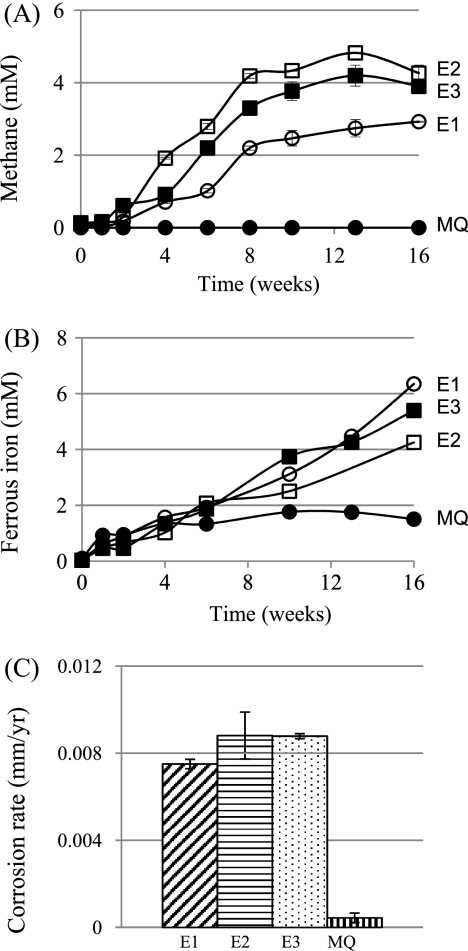
In view of the high fraction of methanogens in the brackish waters and on the pipeline walls (Table 4), it was of interest to check whether these contribute to corrosion. Incubation of metal coupons with brackish water samples E1-3, E2-3, and E3-3 resulted in methane evolution as shown in Fig. 5A. Production of 3 to 4 mM methane was observed in the headspace over a 16-week period. No methane production was seen in the MQ control (Fig. 5A) or in incubations of brackish waters without metal coupons (results not shown). The organic carbon content of brackish waters is only 0 to 10 μg/liter (Table 1); if that carbon were to be converted into methane, it would give rise to a maximum of 1 μM methane. Hence, the methane formed in the incubations in Fig. 5A resulted from using Fe0 as an electron donor for CO2 reduction.
Fig. 5.
(A and B) Headspace methane (A) and ferrous iron formation by brackish waters incubated with iron coupons (B). (C) Corrosion rate of iron coupons as determined by metal weight loss. Data represent average results from two separate incubations with two coupons each. Brackish water samples E1-3, E2-3, and E3-3 (Table 2) were used for the experiment.
The corrosion rates observed by weight loss determinations were low (Fig. 5B [0.008 to 0.009 mm/year]), although they were significantly higher than those observed for the MQ control. The average weight loss of the coupons used (Fig. 5C [E1, E2, and E3]) was 0.0188 g, which corresponds to a corroded iron concentration of 3.3 mM. This value is lower than the observed ferrous ion concentration of 4 to 6 mM (Fig. 4B), which in turn is lower that the maximal ferrous ion concentration of 6 to 10 mM expected based on the concentration of methane in the headspace (Fig. 4A [2.4 to 4 mM]). Precipitation of ferrous ions as iron carbonate and its incomplete removal during coupon cleaning, leading to underestimation of the actual weight loss, are possible causes of these discrepancies.
DISCUSSION
The results of the pyrotag sequencing strategy used here to determine community compositions in pipelines transporting brackish waters have indicated that (i) the pipelines harbor a significant fraction of Methanobacteriaceae archaea, which colonize the pipeline surface and contribute to corrosion, and that (ii) injection of SBS gives rise to a significant change of the wall-attached microbial community to one which has a high fraction of the bisulfite-disproportionating genus Desulfocapsa and the SRB Desulfomicrobium. Although NMDS ordination analysis indicated that, overall, the biofilm and planktonic communities were not significantly different (see Table S2 in the supplemental material), some genera, e.g., Pseudomonas, which was predominantly planktonic, showed a strongly biased distribution. Stevenson et al. (26) also reported that the sessile and planktonic communities in a flowing pipeline were different. Flow appeared to be important, as similar sessile and planktonic communities were observed under stagnant conditions (Fig. 3 [subgroups a and b, in contrast to subgroups c and g or subgroups d and f]).
We have shown that the Methanobacteriaceae archaea, present in a substantial fraction on the pipeline surface in pipe sections 616-P and 821-TP, likely contributed to the corrosion. The potential of methanogenic Archaea to use cathodic hydrogen to reduce CO2 to methane was first demonstrated by Daniels et al. (7), who showed that a number of pure cultures of hydrogenotrophic methanogens were able to form methane from Fe0 and CO2. The strains tested included Methanococcus thermolithotrophicus, Methanobacterium thermoautotrophicum, and Methanobacterium bryantii, Methanosarcina barkeri, and Methanospirillum hungatei. All were able to form methane from Fe0 in anaerobic tubes with an N2-CO2 headspace. Because these strains were not preselected, it would seem that the ability of methanogens to oxidize Fe0 is widespread. In contrast, Dinh et al. (9) used enrichment techniques to isolate strain IM1, which was able to produce up to 4 mM CH4 from iron granules in 20 days, a level of activity comparable to that found here (Fig. 5A). Strain IM1 belonged to either the Methanobrevibacter or the Methanobacterium genus. Likewise, Uchiyama et al. (27) isolated the corrosive methanogen Methanococcus maripaludis strain KA1 from an Fe0-corroding enrichment culture, using a sample obtained from the bottom of a crude oil storage tank. Interestingly, type strain Methanococcus maripaludis JJ1 was found not to be corrosive, indicating that this trait can be strain dependent.
The effect of the oxygen scavenger bisulfite on the microbial community of a high-temperature (50°C) wash tank in the Berkel oil field in the Netherlands was reported recently (28). The water, oil and, gas produced were essentially free of sulfide and sulfate. Ammonium bisulfite (ABS), injected at 15 ppm, represented the only input of inorganic sulfur into the system. A downstream wash tank had elevated levels of Thermodesulfovibrio yellowstonii, a thermophilic SRB not found in other field samples. Increased corrosion was observed downstream of the ABS injection point, which was ascribed to this SRB and caused the authors of that study to question the utility of ABS as an oxygen-scavenging, corrosion-preventing agent. The wash tank had decreased levels of Deltaproteobacteria, indicating that the bacteria were unlikely to contribute to sulfite disproportionation and/or sulfite or sulfate reduction.
In the brackish-water-transporting lines studied here, sodium bisulfite (SBS) was injected to maintain a residual concentration of 25 ppm downstream from the injection point. This led to a proliferation of Deltaproteobacteria disproportionating sulfite to sulfate and sulfide (equation 3) or to sulfate and sulfur (equation 4). The latter reaction has not been previously described for known isolates (2, 11, 16) but is thermodynamically feasible and appears likely in view of the high fraction of sulfur in the scale downstream from the SBS injection point (Table 3). If the injected SBS were to react exclusively with O2, as intended, only sulfate would be formed (H+ + HSO3− + 1/2 O2 → SO42− + H2O), which could then be used by SRB such as the Desulfomicrobium species found as a major community component in 821TP and 821LP. The high sulfur content in pipeline scale indicates the presence of excess SBS beyond what is required for O2 scavenging.
In conclusion, it appears that culture-independent methods to determine pipeline microbial community composition contribute to the understanding of which groups potentially participate in iron corrosion. These methods also indicate microbially mediated disproportionation of bisulfite, a process not previously described for this environment. The collected information is thus of both fundamental and practical interest.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant contracts as well as by an NSERC Industrial Research Chair Award to G.V.; the work was also supported by Baker Hughes Incorporated, Commercial Microbiology Limited (Intertek), the Computer Modeling Group Limited, ConocoPhillips Company, YPF SA, Aramco Services, Shell Canada Limited, Suncor Energy Developments Inc., and Yara International ASA, as well as by Alberta Innovates-Energy and Environment Solutions. This work was also supported by funding from Genome Canada, Genome Alberta, the government of Alberta, and Genome BC.
We thank Roberto Allende-Garcia for providing us with all the water and pipe samples and Rhonda Clark, other members of the Petroleum Microbiology Research Group, and Peter Dunfield for their support.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Bak F., Cypionka H. 1987. A novel type of energy metabolism involving fermentation of inorganic sulfur compounds. Nature 326:891–892 [DOI] [PubMed] [Google Scholar]
- 2. Bak F., Pfennig N. 1987. Chemolithotrophic growth of Desulfovibrio sulfodismutans sp. nov. by disproportionation of inorganic sulfur compounds. Arch. Microbiol. 147:184–189 [Google Scholar]
- 3. Bødtker G., Lysnes K., Torsvik T., Bjørnestad EØ., Sunde E. 2009. Microbial analysis of backflowed injection water from a nitrate-treated North Sea oil reservoir. J. Ind. Microbiol. Biotechnol. 36:439–450 [DOI] [PubMed] [Google Scholar]
- 4. Borg I., Groenen P. 2010. Modern multidimensional scaling: theory and applications. Springer, New York, NY [Google Scholar]
- 5. Cornish Shartau S. L., et al. 2010. Ammonium concentrations in produced waters from a mesothermic oil field subjected to nitrate injection decrease through formation of denitrifying biomass and anammox activity. Appl. Environ. Microbiol. 76:4977–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crolet J.-L. 2005. Microbial corrosion in the oil industry: a corrosionist's view, p. 143–169.In Ollivier B., Magot M. (ed.), Petroleum microbiology. ASM Press, Washington, DC. [Google Scholar]
- 7. Daniels L., Belay N., Rajagopal B. S., Weimer P. 1987. Bacterial methanogenesis and growth from CO2 with elemental iron as the sole source of electrons. Science 237:509–511 [DOI] [PubMed] [Google Scholar]
- 8. DeSantis T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dinh H. T., et al. 2004. Iron corrosion by novel anaerobic microorganisms. Nature 427:829–832 [DOI] [PubMed] [Google Scholar]
- 10. Duncan K. E., et al. 2009. Biocorrosive thermophilic microbial communities in Alaskan North Slope oil facilities. Environ. Sci. Technol. 43:7977–7984 [DOI] [PubMed] [Google Scholar]
- 11. Finster K., Liesack W., Thamdrup B. 1998. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment.. Appl. Environ. Microbiol. 64:119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gieg L. M., Davidova I. A., Duncan K. E., Suflita J. M. 2010. Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ. Microbiol. 12:3074–3086 [DOI] [PubMed] [Google Scholar]
- 13. Huse S., Huber J., Morrison H., Sogin M., Welch D. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huson D., et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jan-Roblero J., Romero J. M., Amaya M., Le Borgne S. 2004. Phylogenetic characterization of a corrosive consortium isolated from a sour gas pipeline. Appl. Microbiol. Biotechnol. 64:862–867 [DOI] [PubMed] [Google Scholar]
- 16. Janssen P. H., Shuhmann A., Bak F., Liesack W. 1996. Disproportionation of inorganic sulfur compounds by the sulfate-reducing bacterium Desulfocapsa thiozymogenes gen. nov. sp. nov. Arch. Microbiol. 166:184–192 [Google Scholar]
- 17. Kaster K. M., Bonaunet K., Berland H., Kjeilen-Eilertsen G., Brakstad O. G. 2009. Characterization of culture-independent and -dependent microbial communities in a high-temperature offshore chalk petroleum reservoir. Antonie Van Leeuwenhoek 96:423–439 [DOI] [PubMed] [Google Scholar]
- 18. López M. A., Zavala-Díaz de la Serna F. J., Jan-Roblero J., Romero J. M., Hernández-Rodríguez C. 2006. Phylogenetic analysis of a biofilm bacterial population in a water pipeline in the Gulf of Mexico. FEMS Microbiol. Ecol. 58:145–154 [DOI] [PubMed] [Google Scholar]
- 19. Marmur J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208–218 [Google Scholar]
- 20. Neria-González I., Wang E. T., Ramirez F., Romero J. M., Hernandez-Rodriguez C. 2006. Characterization of bacterial community associated to biofilms of corroded oil pipelines from the southeast of Mexico. Anaerobe 12:122–133 [DOI] [PubMed] [Google Scholar]
- 21. Pham V. D., et al. 2009. Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ. Microbiol. 11:176–187 [DOI] [PubMed] [Google Scholar]
- 22. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schloss P. D., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schloss P. D., Westcott S. L. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77:3219–3226 doi:10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwermer C. U., et al. 2008. Impact of nitrate on the structure and function of bacterial biofilm communities in pipelines used for injection of seawater into oil fields. Appl. Environ. Microbiol. 74:2841–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stevenson B. S., et al. 2011. Microbial communities in bulk fluids and biofilms of an oil facility have similar composition but different structure. Environ. Microbiol. 13:1078–1090 [DOI] [PubMed] [Google Scholar]
- 27. Uchiyama T., Ito K., Mori K., Tsurumaru H., Harayama S. 2010. Iron-corroding methanogen isolated from a crude-oil storage tank. Appl. Environ. Microbiol. 76:1783–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van der Kraan G. M., Bruining J., Lomans B. P., van Loosdrecht M. C., Muyzer G. 2010. Microbial diversity of an oil-water processing site and its associated oil field: the possible role of microorganisms as information carriers from oil-associated environments. FEMS Microbiol. Ecol. 71:428–443 [DOI] [PubMed] [Google Scholar]
- 29. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yue J. C., Clayton M. K. 2005. A similarity measure based on species proportions. Commun. Stat. Theor. Methods 34:2123–2131 [Google Scholar]
- 31. Zhu X. Y., Lubeck J., Kilbane J. J., II 2003. Characterization of microbial communities in gas industry pipelines. Appl. Environ. Microbiol. 69:5354–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.