Abstract
Display of heterologous antigens on the cell surface is considered a useful technique for vaccine delivery by recombinant lactobacilli. In this study, two recombinant Lactobacillus acidophilus derivatives displaying Salmonella flagellin (FliC) were constructed using different anchor motifs. In one instance, the FliC protein was fused to the C-terminal region of a cell envelope proteinase (PrtP) and was bound to the cell wall by electrostatic bonds. In the other case, the same antigen was conjugated to the anchor region of mucus binding protein (Mub) and was covalently associated with the cell wall by an LPXTG motif. These two recombinant L. acidophilus cell surface displays resulted in dissimilar maturation and cytokine production by human myeloid dendritic cells. The surface-associated antigen was highly sensitive to simulated gastric and small intestinal juices. By supplementation with bicarbonate buffer and soybean trypsin inhibitor, the cell surface antigen was protected from proteolytic enzymes during gastric challenge in vitro. The protective reagents also increased the viability of the L. acidophilus cells upon challenge with simulated digestive juices. These results demonstrate the importance of protecting cells and their surface-associated antigens during oral immunization.
INTRODUCTION
Vaccine delivery systems using lactic acid bacteria (LAB) are under development. Many Lactobacillus and Lactococcus strains are generally regarded as safe (GRAS) because they have been consumed for centuries in fermented foods, or as probiotics originating as commensals of the human intestinal tract. It is now well documented that LAB can provide immune-modulating/immune-stimulating activities and contribute to health maintenance (8, 38). Recent studies have revealed that several cell surface components of the probiotic bacteria are recognized by immune cells via pattern recognition receptors (PRRs) (25). In particular, lipoteichoic acid (LTA), peptidoglycan (PG), and muramyl dipeptide (MDP), the subcomponent of PG, are known as the major immune stimulators recognized by Toll-like receptor 2 (TLR2) and nucleotide-binding oligomerization domain 2 (NOD2) (18, 30, 53). The combination of safety and immunogenicity of LAB satisfy key requirements for vaccine delivery vehicles. Several studies have demonstrated the potential of Lactococcus lactis and Lactobacillus strains for immunization, as described in previous reviews (33, 46, 51).
The probiotic LAB strain Lactobacillus acidophilus NCFM is particularly promising as a vaccine delivery vector. Besides the favorable characteristics of LAB described above, L. acidophilus NCFM is proven to have both acid and bile tolerance (3, 40), which may allow association with the intestinal mucosa and facilitate an oral delivery strategy. Importantly, it has been shown that L. acidophilus interacts with dendritic cells (DCs) through DC-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN) (23). The fact that the whole-genome sequence is known is also a key advantage for designing oral vaccines to be delivered by probiotic LAB (1). Proof of principle has been demonstrated by Mohamadzadeh et al., who constructed a recombinant L. acidophilus strain producing the protective antigen of Bacillus anthracis and succeeded in inducing protective immunity in a murine model (34).
For the construction of recombinant LAB as vaccine candidates, there are three strategies for the subcellular distribution of antigens: cytoplasmic accumulation, secretion, and cell surface display. Intracellular expression is the simplest strategy and has several advantages. For instance, high levels of antigen can be expressed from a strong promoter and can be encapsulated within the bacterium (43, 52). Importantly for mucosal immunization, the cell envelope may protect intracellular protein antigens from digestive enzymes present in mucosal fluids. Secretion of antigens has infrequently been used for LAB-based vaccines, since antigen secretion does not allow accumulation of the proteins during cultivation prior to immunization. Nevertheless, there are studies demonstrating that antigen-secreting LAB can induce protective immunity (34, 35, 45). Surface display systems for antigen delivery have been used more commonly. Hypothetically, the merit of this strategy is that the exposed antigens could interact directly with immune cells and might be processed by antigen-presenting cells more efficiently than intracellular antigens that were encapsulated within the robust cell walls of the Gram-positive bacteria. In fact, two independent studies showed that surface displays provided better immunogenicity than intracellular accumulation (6, 37). However, there are contradictory results in the case of intragastric (i.g.) immunization (39, 43). When delivered orally, extracellular antigens are exposed to proteolytic enzymes, such as pepsin and trypsin, present in gastric and pancreatic fluids. These intensive digestions likely lower the immunopotency of cell surface antigens, although the sensitivity to those digestive juices is unclear. If cell surface antigens are highly sensitive to the digestive process, additional measures must be taken to mitigate degradation.
Surface display systems in LAB have been developed by mimicking natural surface components and proteins. In Gram-positive bacteria, there are roughly five types of surface proteins: membrane proteins, lipoproteins, covalently bound cell wall-associated proteins, noncovalently bound cell wall-associated proteins, and surface layer proteins (12, 27). Several bacterial proteins have been exploited to construct recombinant LAB expressing anchor-fused antigens on the cell surfaces, including the Bacillus subtilis membrane protein PgsA and the covalently bound cell wall-associated proteins Lactococcus lactis PrtP, Lactobacillus casei PrtP, and Streptococcus pyogenes M6 (14, 26, 37, 41). Noncovalently bound cell wall-associated proteins, including Lactobacillus delbrueckii subsp. bulgaricus PrtB, the Lactobacillus brevis surface layer protein SlpA, and an L. acidophilus surface layer protein, have also been used for these applications (2, 17, 22, 48). Although these proteins are found on the cell surface, their physical/biochemical properties are dissimilar. Therefore, even if the same antigens were displayed, the anchoring motifs might elicit different impacts on the net immunogenicity of LAB displaying heterologous antigens; however, this has not been proven empirically.
The main purpose of this study was to develop surface display systems in L. acidophilus and to investigate whether different anchoring motifs affect the immunogenicity of antigen displays in L. acidophilus. A C-terminal protein fragment of the surface proteinase (PrtP) and a binding domain from the mucus binding protein (Mub) from L. acidophilus NCFM were employed to develop two different cell surface antigen display systems. Although both anchor motifs bind to the cell wall, PrtP is anchored by electrostatic bonds, while Mub is anchored through a covalent bond. Salmonella enterica serovar Typhimurium flagellin (FliC) is not only a protective antigen for immunization but also the agonist of TLR5 (20, 49); thus, recombinant L. acidophilus strains displaying surface FliC were predicted to be highly immunogenic. In order to determine their immunological properties, the recombinant L. acidophilus strains producing FliC were cocultured with human myeloid DCs. The sensitivities of surface-exposed antigens to proteolysis by digestive juices were also determined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Lactobacillus acidophilus NCFM and derivative recombinant strains were grown statically in MRS broth (Difco Laboratories, Detroit, MI) alone or supplemented with 5 μg/ml of erythromycin (Em) at 37°C. MRS agar (1.5% agar) plates were incubated anaerobically. Escherichia coli DH5α and other recombinant strains were grown aerobically with shaking in LB medium (Difco) with or without 200 μg/ml of Em at 37°C. All bacterial strains used in this study are listed in Table 1.
Table 1.
Bacterial strains and plasmids
| Bacterial strain | Plasmid | Description of bacterial strain and/or plasmid | Reference or source |
|---|---|---|---|
| E. coli DH5α | Cloning host | 19 | |
| NCK2149 | pTRK1029 | pTRK882 backbone, FliC-Amub inserted | 15; this study |
| NCK2157 | pTRK1033 | pTRK1029 backbone; Sprt inserted | This study |
| NCK2159 | pTRK1034 | pTRK1033 backbone, Amub replaced with Aprt | This study |
| L. acidophilus NCFM | Human intestinal isolate; expression host | 1 | |
| NCK1895 | pTRK882 | Erythromycin resistance | 15 |
| NCK2158 | pTRK1033 | Cell surface display of FliC-Amub by covalent bond | This study |
| NCK2160 | pTRK1034 | Cell surface display of FliC-Aprt by electrostatic bond | This study |
Construction of plasmids.
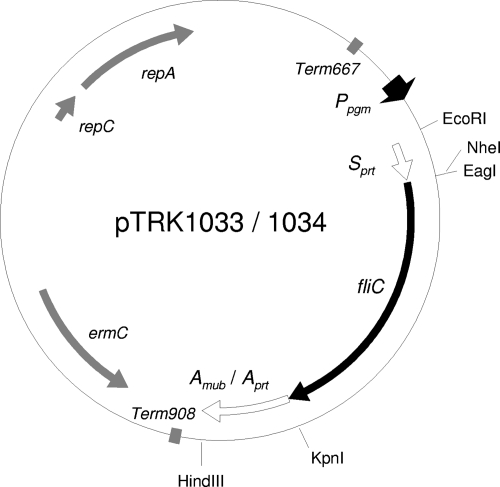
Regular and overlap PCRs were performed using the primers shown in Table 2. Gene fragments encoding Salmonella serovar Typhimurium FliC and the 160 C-terminal amino acid residues of L. acidophilus NCFM Mub (LBA1709) (Amub) were amplified with specific primer pairs AK_21–AK_13 for FliC and AK_11–AK_12 for Amub, as well as with pFliCP (L. L. Stoeker et al., submitted for publication) as the template DNA for FliC and L. acidophilus NCFM chromosomal DNA as the template DNA for Amub. These two PCR products were subsequently connected by overlap PCR using primers AK_21 and AK_12. The resulting chimeric DNA fragment (FliC-Amub) was digested and inserted into the EcoRI-HindIII site of pTRK882 (15). After the cloning of FliC-Amub (pTRK1029), a gene fragment encoding the ribosome binding site and the 70 N-terminal amino acid residues of L. acidophilus NCFM PrtP (Sprt) was obtained by PCR using the pair of primers AK_38 and AK_39. The Sprt DNA fragment was inserted into the EcoRI-EagI site of pTRK1029 to create pTRK1033. The Amub-encoding site of pTRK1033 was replaced with the DNA fragment encoding the 160 C-terminal amino acid residues of PrtP (LBA1512), which was generated by PCR using primers AK_36 and AK_37. The plasmid was digested with KpnI and HindIII and was ligated with the digested Aprt-encoding DNA fragment (pTRK1034). A map of pTRK1033 and pTRK1034 is shown in Fig. 1.
Table 2.
Sequences of PCR primers
| Primer | Sequence | Restriction site(s) |
|---|---|---|
| AK_21 | ACACGAATTCCGGCCGATGGCACAAGTCATTAATAC | EcoRI, EagI |
| AK_13 | GTTGGAGTAACTGTTGGCACGGTACCACGCAGTAAAGAGAGGAC | |
| AK_11 | CGTCCTCTCTTTACTGCGTGGTACCGTGCCAACAGTTACTCCAAC | |
| AK_12 | ACACCTCGAGAAGCTTCTGCAGTTATCTCCGTTTCCTATCAG | PstI, HindIII, XhoI |
| AK_38 | GCATGAATTCAGGAGTTAATAATATGAGAAATAAAAAAGTGGGG | EcoRI |
| AK_39 | GCATCGGCCGGCTAGCATCATCAACTTCATCTGCC | EagI, NheI |
| AK_36 | GCATGGTACCGTACCAAGAAAAGTTCAACCTC | KpnI |
| AK_37 | GCATAAGCTTTTATTTCAAAGCTGTATTAGCTAC | HindIII |
Fig. 1.
Gene map of pTRK1033 and pTRK1034.
Transformation of L. acidophilus NCFM.
The procedures for the preparation of competent cells and electroporation have been described previously (50). Briefly, bacterial cells at exponential phase were cultivated and were washed sufficiently with isosmotic buffer. Approximately 1 μg of plasmid was mixed with the competent cells, and the mixture was transferred to a 2-mm-gap electroporation cuvette. Cells received an electrical pulse (2.5 kV, 400 Ω, 25 μF) and recovered in MRS broth. Plasmid-loaded cells were subsequently grown on MRS plates supplemented with Em.
Western blotting.
Log-phase cultures of recombinant L. acidophilus were centrifuged to remove cells. The culture supernatant was concentrated 20 times by precipitation with trichloroacetic acid (TCA). Precipitated proteins were washed with acetone and were redissolved in 8 M urea in 50 mM Tris-Cl (pH 7.0). Cells were washed sufficiently and were suspended in lysis buffer consisting of 500 U/ml of mutanolysin (Sigma-Aldrich, Detroit, MI), 250 U/ml of Benzonase (Sigma-Aldrich), 1× protease inhibitors (Sigma-Aldrich), and 50 mM Tris-Cl (pH 7.0). After incubation at 60°C for 2 h, cells were homogenized by bead beating. To extract cell surface proteins from lactobacilli, cells were suspended in phosphate-buffered saline (PBS) or 8 M urea buffer. The cell suspensions were incubated at room temperature for 30 min, followed by centrifugation to collect a cell-free protein solution. Each protein sample was mixed with Laemmli buffer (Sigma-Aldrich) and was boiled for 10 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). A MagicMark XP Western protein standard (Invitrogen, Carlsbad, CA) and purified recombinant flagellin (Invivogen, San Diego, CA) were also applied. The protein blot was treated with anti-FliC monoclonal IgG (BioLegend, San Diego, CA) and horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Invitrogen). Chemiluminescence using ECL Plus (GE Healthcare Life Sciences, Piscataway, NJ) was visualized by a VersaDoc MP system (Bio-Rad, Hercules, CA).
Enzyme-linked immunosorbent assay (ELISA).
A MaxiSorp flat-bottom 96-well plate (Nalge Nunc International, Rochester, NY) was coated with 5 μg/ml of purified flagellin overnight. Bacterial cell suspensions (1 × 109 CFU/ml) prepared from an overnight culture were serially diluted (dilution factor, 2). One volume of anti-FliC solution (dilution, 1/25,000 in PBS supplemented with 1% bovine serum albumin [BSA] and 0.05% Tween 20) was added to each cell suspension, and the mixture was incubated at room temperature for 1 h. At the same time, the antigen-coated microplate was blocked with 1% BSA in PBS. The mixture of the bacteria and the antibody solution was centrifuged, and the cell-free supernatant was transferred to the blocked microplate. After 1 h of incubation, the microplate was washed and incubated with a secondary-antibody solution, including 1/50,000-diluted HRP-conjugated anti-mouse IgG, for 1 h. A tetramethylbenzidine (TMB) peroxidase substrate (Kirkegaard & Perry Laboratories [KPL], Gaithersburg, MD) was used for color development. The reaction was terminated with H2SO4, and optical absorbance at 450 nm was measured with a Sunrise microplate reader (Tecan US, Durham, NC).
TLR5 stimulating assay.
The reporter assay using HEK293 cells expressing TLR5 (Invivogen) has been described previously (Stoeker et al., submitted). Freshly prepared bacteria were added to the culture of TLR5-expressing cells and were incubated overnight. TLR5 activation was determined by NF-κB induction using the secreted embryonic alkaline phosphatase (SEAP) reporter plasmid pNiFty. SEAP activity in the coculture supernatants was detected with a luminometer after development with the SEAP reporter gene assay according to the manufacturer's instructions (Roche Applied Science, Indianapolis, IN).
Isolation of human myeloid DCs and coculture assay.
Untested human blood buffy coats were obtained from the American Red Cross (Durham, NC). Human myeloid DCs were isolated in accordance with the method described previously (Stoeker et al., submitted). In brief, lymphocytes were separated from whole blood using Ficoll-Plaque Plus (GE Healthcare) and centrifugation. CD1c+ cells were enriched with a magnetic-activated cell sorting (MACS) CD1c (BDCA-1)+ dendritic cell isolation kit (Miltenyi Biotec, Auburn, CA). The isolated DCs suspended in filtered human peripheral blood mononuclear cell (PBMC) medium (10% fetal bovine serum [FBS], 1.1% sodium pyruvate, 1% GlutaMAX, 100 U/ml penicillin, 100 μg/ml streptomycin) were plated in 96-well plates at a concentration of 2 × 105 cells/well. Fresh bacterial cells at stationary phase were washed with PBS, suspended in the same PBMC medium, and then inoculated into the DC cultures at a concentration of 2 × 106 CFU/well (DC/bacterium ratio, 1:10). After 24 h of incubation, DCs were centrifuged and collected for antibody staining, while supernatants were stored at −80°C for cytokine analysis.
Flow cytometric analysis.
An overnight culture of bacteria at stationary phase was collected and washed with PBS. The polyclonal anti-FliC rabbit antibody RY544 (dilution, 1/50) was added to the cell suspension (2 × 106 CFU in 20 μl of PBS supplemented with 1% BSA) and was incubated on ice for 30 min. The cells were washed and suspended in the same buffer, including 1/100-diluted phycoerythrin (PE)-conjugated anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). After a 30-min incubation, the stained bacterial cells were washed and suspended in PBS. For the staining of DCs, fluorescein isothiocyanate (FITC)-conjugated anti-human TLR5, PE/Cy7-conjugated anti-human CD80, PE-conjugated anti-human CD86, peridinin chlorophyll protein (PerCP)-conjugated anti-human CD83, and PerCP-conjugated anti-human CD40 (Biolegend) suspended in PBS were used. Flow cytometry was performed using a LSRII flow cytometer, and the results were analyzed with FACSDiva software (Becton Dickinson [BD], Franklin Lakes, NJ).
Multiple cytokine quantification.
Multiple cytokines in the culture supernatants of DCs were detected with a customized Milliplex MAP kit (Millipore) in accordance with the manufacturer's instructions. In brief, antibody-immobilized beads for the detection of interleukin 1β (IL-1β), IL-6, IL-8, IL-10, IL-12 (p70), tumor necrosis factor alpha (TNF-α), and alpha-2 interferon (IFN-α2) were incubated with 5-times-diluted culture supernatants of DCs. The beads were subsequently treated with detection antibodies as well as with PE-conjugated streptavidin and were analyzed with a Bio-Plex system (Bio-Rad).
Simulated-digestive-juice assays.
Bacterial cell suspensions of recombinant L. acidophilus were prepared from an overnight culture. The preparation of simulated gastric juice (SGJ) and simulated small intestinal juice (SSIJ) has been described previously (16). Briefly, SGJ consists of pepsin (Fisher Scientific, Pittsburgh, PA), NaCl, and HCl, while SSIJ includes oxgall (Difco), NaCl, and porcine pancreatin (MP Biomedicals, Solon, OH). Adjusted concentrations of bacterial cells (1 × 109 CFU/ml) either in water, in bicarbonate buffer (NaHCO3 solution), or in soybean trypsin inhibitor (SBTI) solution were mixed with 5 volumes of SGJ or SSIJ and were incubated at 37°C. For the counting of viable cells, cells treated with the digestive juice (SGJ or SSIJ) were properly diluted and plated onto MRS agar supplemented with erythromycin. In order to detect FliC displayed on the cell surface, cells treated with the digestive juices were collected by centrifugation and were suspended in methanol. The whole-cell suspensions in methanol were then mounted on a PVDF membrane (2 μl/spot). The dried membrane was incubated in PBS supplemented with 1% BSA and anti-FliC monoclonal IgG (diluted 1/5,000). After the membrane was washed with PBS, the blot was incubated with HRP-conjugated anti-mouse IgG in the same buffer. FliC-specific chemiluminescence signals were detected by the same system as described above.
Statistical analysis.
The Wilcoxon signed-rank test was applied to evaluate significant differences in assays using DCs. For multiple comparisons, the level of significance was adjusted by Bonferroni's correction. Judgments on rejecting a null hypothesis were suspended if the P value was higher than 0.017.
RESULTS
Production and surface association of FliC on L. acidophilus cells using two different anchoring motifs.
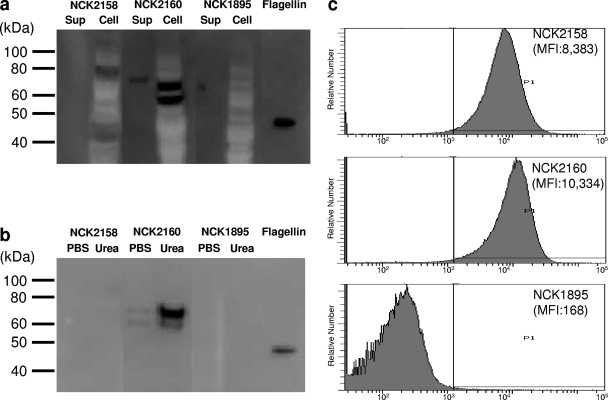
Plasmids with FliC fused to Sprt as well as either Amub or Aprt were successfully introduced into L. acidophilus. The production of FliC fusion proteins was confirmed by Western blotting (Fig. 2a). In NCK2158, producing FliC-Amub protein, a specific band was detected in the cell fraction but not in the culture supernatant. Meanwhile, in NCK2160, expressing FliC-Aprt, specific bands were observed in both the culture supernatant and the cell fraction. The expected molecular masses of FliC-Amub and FliC-Aprt are 70 kDa and 67 kDa, respectively, which were fairly reflected in the positions of the bands detected, although an additional, smaller band was observed in the NCK2160 cell fraction. Surface-associated FliC-Aprt on NCK2160 was efficiently extracted with 8 M urea buffer (Fig. 2b). These results indicate that FliC-Aprt was attached to the cell wall by noncovalent bonds. FliC-Amub was recovered with neither 8 M urea buffer nor PBS, as expected, because this protein should be covalently bound to the cell wall. Flow cytometric analysis confirmed that FliC-fused proteins were located on the cell surface (Fig. 2c). These data indicate that these anchoring motifs worked properly and efficiently, because almost all cells displayed FliC-fused proteins. The mean fluorescence intensity (MFI) of NCK2160 was approximately 20% higher than that of NCK2158. Thus, the density of the FliC-fused protein at the cell surface was likely higher in NCK2160 than in NCK2158.
Fig. 2.
Expression and cell surface display of FliC by recombinant L. acidophilus strains. (a) Concentrated culture supernatants (Sup) and cell extracts (Cell) were analyzed by Western blotting using an anti-FliC monoclonal antibody. Flagellin prepared from Salmonella serovar Typhimurium was also applied as a reference protein. Molecular sizes are shown on the left. (b) Cell surface proteins extracted with either PBS or 8 M urea were subjected to Western blotting for the detection of FliC. (c) FliC located on the cell surface was detected by flow cytometry. The fluorescence of PE represents surface-associated FliC. MFI, mean fluorescence intensity.
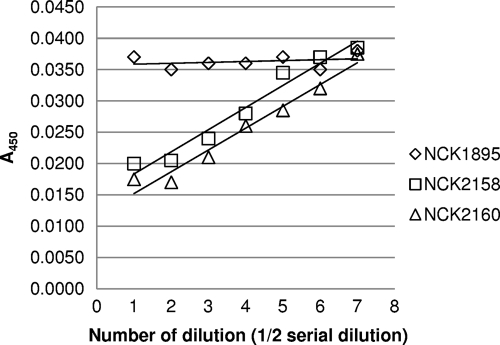
Relative amounts of FliC-fused proteins in NCK2158 and NCK2160.
In order to determine the relative amounts of FliC on the recombinant L. acidophilus strains, a FliC-specific monoclonal IgG was absorbed by FliC-displaying lactobacilli, and the remaining antibody was detected by ELISA. As shown in Fig. 3, the absorbance values were inversely proportional to the concentration of either NCK2158 or NCK2160 but were unchanged by NCK1895 regardless of its concentration. The antibody absorption capacity per cell was higher in NCK2160 than in NCK2158, with the difference estimated at approximately 20%. This number is close to the difference between the MFI values described above. Hence, the estimated relative amounts of surface-associated FliC were confirmed.
Fig. 3.
Relative amounts of surface-associated FliC. Recombinant L. acidophilus cell suspensions (dilution 0, 1 × 109 CFU/ml) were serially diluted (dilution 1, 5 × 108 CFU; dilution 2, 2.5 × 108 CFU; and so forth) and were incubated with anti-FliC monoclonal IgG (dilution, 1:50,000). The remaining anti-FliC antibody that was not absorbed by the bacteria was detected by ELISA. The data represent three separate experiments, and mean values for duplicate samples are shown. A450, absorbance at 450 nm.
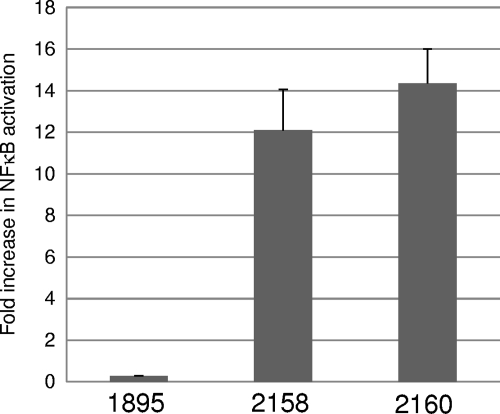
TLR5-stimulating activities of FliC-producing L. acidophilus.
A reporter gene assay using TLR5-expressing HEK293 cells was performed. The cells were incubated with the recombinant lactobacilli, and SEAP released in response to NF-κB activation was detected. As shown in Fig. 4, both NCK2158 and NCK2160 clearly activated NF-κB through stimulation of TLR5. The average TLR5-stimulating activity of NCK2160 was 19% higher than that of NCK2158, once again corresponding to the expected ratio of cell-associated FliC fusion proteins between the two recombinant lactobacilli. These results indicate that the surface-associated FliC was recognized by TLR5 and that the magnitude of the TLR5-stimulating activity of FliC-producing L. acidophilus depends on the quantity of surface-located FliC.
Fig. 4.
TLR5-stimulating activities of recombinant lactobacilli NCK1895, NCK2158, and NCK2160. The activity of SEAP released from HEK293 cells was measured after incubation with the recombinant bacteria. Values are means + standard errors from 3 experiments.
TLR5 expression and maturation of human myeloid DCs stimulated by recombinant lactobacilli.
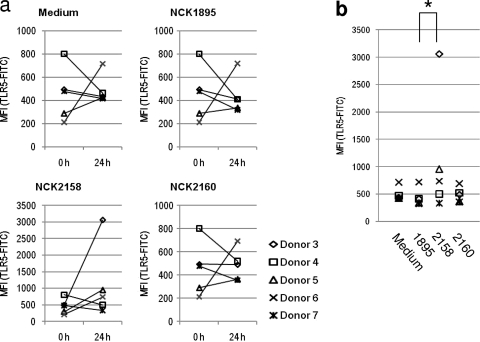
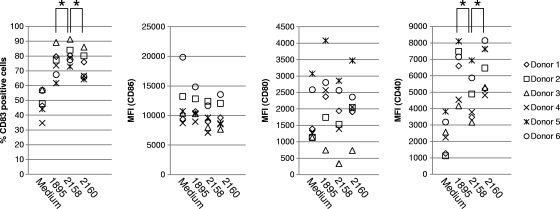
Freshly isolated myeloid DCs from human blood were stimulated with live L. acidophilus strains. Antibiotics were added to the culture medium in order to prevent bacterial growth. Under this condition, bacterial cells were alive at the early stages but completely dead at the end of the assay (data not shown). TLR5 expression by the DCs before and after incubation with the recombinant lactobacilli was determined by flow cytometry. Freshly isolated DCs (0 h) expressed low levels of TLR5 and upregulated or downregulated TLR5 expression after 24 h of incubation depending on the blood donor (Fig. 5a). The similarity of TLR5 expression patterns among NCK1895-stimulated, NCK2160-stimulated, and unstimulated (Medium) DCs demonstrates that these two L. acidophilus strains have minimal impact on TLR5 expression by the DCs. Meanwhile, NCK2158-stimulated DCs, especially the DCs from donors 3 and 5, clearly upregulated TLR5 expression after 24 h of incubation (Fig. 5a and b). Coculture with the recombinant L. acidophilus strains enhanced the maturation of the human myeloid DCs. By flow cytometry, the specific fluorescent intensities of CD40, CD80, CD83, and CD86 were analyzed. Among the four types of maturation markers tested in this study, CD83 and CD40 were clearly upregulated in response to the recombinant lactobacilli (Fig. 6). NCK2158 always induced CD83 expression most efficiently and CD40 least efficiently among the three L. acidophilus strains tested (P < 0.01). Meanwhile, no difference in DC maturation was observed between NCK2160 and NCK1895 stimulation. In other words, the pattern of DC maturation induced by NCK2160, but not by NCK2158, was similar to that induced by the control, NCK1895. This assay revealed significant immunological differences between covalently and noncovalently bound FliC on L. acidophilus cells.
Fig. 5.
TLR5 expression by human myeloid DCs stimulated with recombinant lactobacilli. Flow cytometry was used to analyze TLR5 expression on the DCs. (a) The isolated DCs were stained before (0 h) or after (24 h) incubation with or without stimulation. (b) TLR5 expression levels were compared after 24 h of incubation. MFI, median fluorescence intensity. *, P < 0.01.
Fig. 6.
Maturation of human myeloid DCs by stimulation with FliC-displaying L. acidophilus strains. Freshly isolated myeloid DCs from 6 different donors were incubated with or without recombinant lactobacilli for 24 h. The stimulated DCs were collected and stained with fluorescently labeled anti-CD83, anti-CD86, anti-CD80, and anti-CD40 antibodies. The fluorescence intensities of the stained DCs were determined by flow cytometry. MFI, median fluorescence intensity. *, P < 0.01.
Multiple cytokine release from stimulated DCs.
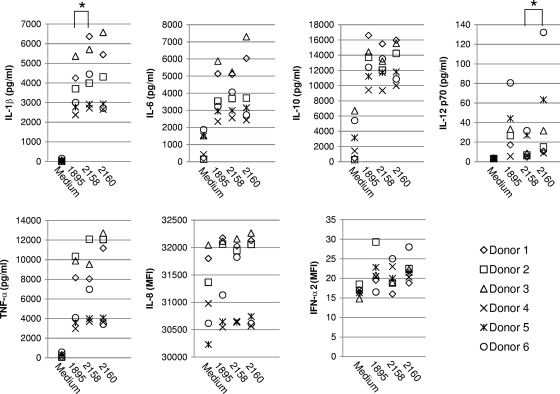
Stimulation of human myeloid DCs with recombinant lactobacilli induced the secretion of cytokines. Among seven kinds of cytokines, the secretion of IL-1β, IL-6, IL-10, IL-12 (p70), and TNF-α was clearly accelerated in response to any recombinant L. acidophilus strain (Fig. 7). The DCs stimulated with NCK2158 always secreted a greater amount of IL-1β than the DCs stimulated with NCK1895 (P < 0.01). Comparison of NCK2158 and NCK2160 stimulation showed that the amounts of IL-12 (p70) in the culture supernatants were significantly different. Regardless of whether the DCs were stimulated with bacteria, large amounts of IL-8 were detected in the culture supernatant. IFN-α2 was barely detected in the DC cultures.
Fig. 7.
Analysis of multiple cytokines secreted by lactobacillus-stimulated DCs. Freshly isolated human myeloid DCs (2 × 105 cells in 0.2 ml) were incubated with or without recombinant lactobacilli (2 × 106 CFU in 0.2 ml) for 24 h. Culture supernatants were collected and analyzed with a Milliplex MAP kit. The concentration (in picograms per milliliter) of each cytokine is shown along the y axis. The values for IL-8 and IFN-α2 are expressed as MFI because these numbers were out of the range of standard curves. *, P < 0.01.
Sensitivities of recombinant L. acidophilus strains and their surface-displayed FliC proteins to simulated digestive juice.
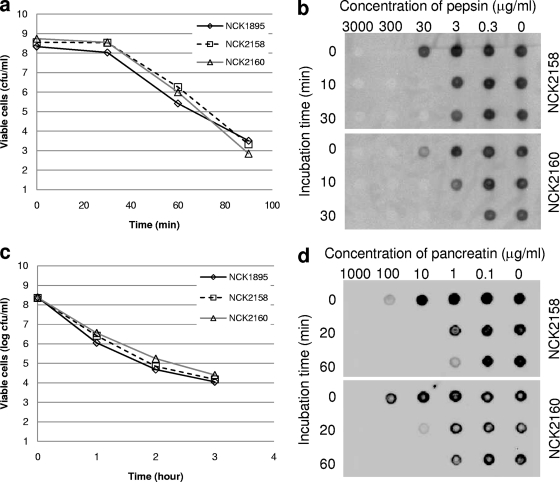
The recombinant lactobacilli were treated with either SGJ or SSIJ. The viability of the bacteria and the stability of the FliC fusion protein on the cell surfaces were monitored at different time points. All three L. acidophilus strains survived equally well at all time points, indicating that the expression and surface association of FliC did not affect the sensitivity of the bacteria to digestive processes (Fig. 8a and c). However, the surface protein itself was highly sensitive to the proteolytic effects of the digestive juices (Fig. 8b and d). Even after a 100× dilution of proteolytic enzymes, the FliC-fused proteins were digested quickly. The two differently bound surface-exposed FliC proteins exhibited similar sensitivities to SGJ and SSIJ. Hence, the different physical properties of cell wall-associated proteins had a minimal impact on their sensitivities to enzymatic proteolysis.
Fig. 8.
Sensitivities of recombinant bacteria and FliC to simulated digestive juice. (a and c) Recombinant L. acidophilus cells were incubated with simulated gastric juice (a) or simulated small intestinal juice (c). Viable cells (colony-forming cells) at several time points were counted. (b and d) Cell surface FliC was detected by dot blot analysis after treatment with either simulated gastric juice, including different concentrations of pepsin (b), or simulated small intestinal juice, including different concentrations of pancreatin (d).
Protective effects of bicarbonate and SBTI against the digestive process.
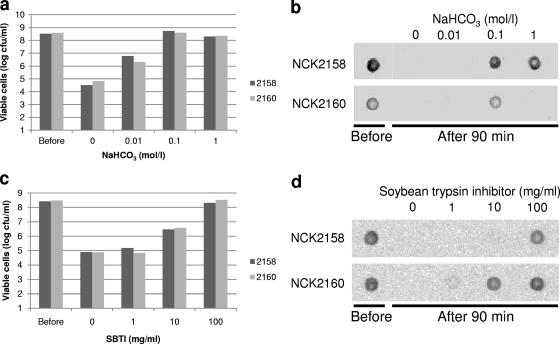
In theory, pepsin is inactive under neutral or basic pH conditions, and SBTI inhibits trypsin and chymotrypsin, which are major proteolytic enzymes in pancreatin. Thus, supplementation with bicarbonate buffer or SBTI prior to incubation with SGJ or SSIJ could protect surface-associated antigens from proteolytic degradation. As expected, digestion of the surface FliC was successfully inhibited by those protectants (Fig. 9b and d). Addition of 0.1 M NaHCO3 completely neutralized the acid in simulated gastric juice and provided a protective effect. Although a high concentration of bicarbonate could inactivate trypsin, the high-salt solution interfered with the electrostatic bonds of the cell surface protein anchored by Aprt. In fact, no specific signal was detected from NCK2160 cells pretreated with 1 M NaHCO3, but soluble FliC-Aprt was found in the cell-free supernatant (data not shown). In order to achieve full protection of surface FliC against SSIJ, high concentrations of SBTI were required. After a 90-min incubation, the FliC-specific signal of NCK2158 was observed only if 100 mg/ml of SBTI was added to the cell suspension, while the specific signal of NCK2160 was detected by adding less protectant. Thus, the antigen on NCK2160 in combination with SBTI seemed to be more resistant to proteolysis. The reagents used as the protectants collaterally contributed to the survivability of recombinant L. acidophilus strains (Fig. 9a and c). The survival rates of the lactobacilli increased in a concentration-dependent manner. With 0.1 M NaHCO3 and 100 mg/ml of SBTI, almost 100% of the cells remained viable.
Fig. 9.
Protective effects of bicarbonate and SBTI against simulated digestive juices. Recombinant lactobacilli were suspended in either bicarbonate buffer or SBTI solution prior to treatment with simulated digestive juice. Viable cells and surface FliC were detected after 90 min of incubation with either simulated gastric juice (a and b) or simulated small intestinal juice (c and d).
DISCUSSION
In the present study, two types of antigen display systems were developed in L. acidophilus using the cell wall anchor of either PrtP or Mub. Cell envelope proteinases are commonly found in LAB strains, but their structures, including the domains for wall attachment, are diverse (47). In particular, the proteinase of Lactococcus lactis binds to the cell wall covalently, while those of Lactobacillus delbrueckii and Lactobacillus helveticus associate with peptidoglycan by a noncovalent bond. According to the analysis by protein BLAST search, the C-terminal end of L. acidophilus PrtP is most related to the corresponding part of PrtH and is likely able to associate with the cell wall electrostatically. The present study showed that the FliC proteins fused to the 70 N-terminal amino acid residues and the 160 C-terminal amino acid residues of PrtP were efficiently displayed on the cell surface. This chimeric protein was removed by treatment with a chaotropic reagent, which confirmed the noncovalent bond of PrtP. The other surface protein employed in this study, Mub, has a mucus-binding domain, which is found in several LAB strains (7). This type of protein is considered to play an important role in adherence to gastrointestinal (GI) mucosa (44). L. acidophilus NCFM has nine Mub proteins, three of which possess an N-terminal signal peptide and a C-terminal anchor with a LPXTG sequence. LBA1709 was used in the present study. The LPXTG motif is cleaved by the specific serine protease sortase and eventually combines with peptidoglycan via covalent bond (27). As expected, the FliC protein fused to the C-terminal end of Mub was clearly displayed on the cell surface. This surface protein was not removed by a chaotropic reagent, but treatment with N-acetylmuramidase did successfully extract the protein. These results indicated that the FliC-Mub fusion protein was covalently associated with the L. acidophilus cell wall. Both surface-displayed FliC proteins in the recombinant L. acidophilus strains retained their TLR5-stimulating activities. FliC is known to have adjuvant effects due to its TLR5-stimulating activity (5, 31). Coadministration or protein fusion with FliC improves the efficiency of immunization (4, 11, 21). Therefore, the currently constructed FliC-producing L. acidophilus strains might be more robust vaccine delivery agents than the wild-type strain, due to the additional TLR agonist. For instance, a recombinant LAB expressing both FliC and a target antigen in the same cell might be more immunogenic than an isogenic strain producing the antigen only.
The two types of recombinant L. acidophilus derivatives producing FliC exhibited unequal impacts on DC maturation and cytokine production. In the present study, human myeloid DCs were stimulated with recombinant lactobacilli, and subsequently, four major maturation markers of DCs—CD40, CD80, CD83, and CD86—were upregulated. Among these molecules, the expression of CD40 and CD83 was clearly upregulated in response to any of the L. acidophilus strains. Similar effects of Lactobacillus strains on DC maturation have been reported previously (36). Interestingly, the L. acidophilus strain expressing covalently bound surface FliC induced a higher percentage of CD83+ cells and a lower percentage of CD40+ cells than the other two strains. One of the functions of CD83 on DCs is to activate T cells (42). CD40 is involved in cross talk between DCs and other lymphocytes (29). Hence, those differences in the upregulation of these molecules might affect downstream immune responses. Secretion of cytokines from DCs also plays a decisive role in adaptive immune responses. Among seven cytokines tested in the present study, the secretion of IL-1β, IL-6, IL-10, IL-12, and TNF-α from DCs was clearly upregulated by stimulation with L. acidophilus. The cytokine responses of human DCs stimulated with L. acidophilus NCFM have been reported (23). Although the DC/bacterium ratio (1:10) and incubation time (24 h) were the same, the cells employed in the previous study were monocyte-derived DCs. The monocyte-derived DCs tended to induce IL-10, IL-12, and TNF-α predominantly, while the CD1c+ myeloid DCs used in the current assay secreted large amounts of IL-1β, IL-6, IL-10, and TNF-α but much less IL-12. These different cytokine profiles may be interpreted in different ways. For example, cytokine induction in monocyte-derived DCs could represent a Th1-biased immune response or a mixed Th1 and Th2 response, but cytokine expression by CD1c+ myeloid DCs would be more consistent with a Th2 and/or Th17 response. This differential effect on DC subtypes should be taken into consideration when lactobacilli or other bacteria are assessed in vitro using DCs. Expression of FliC on the L. acidophilus cell surface induced additional secretion of IL-1β from human myeloid DCs. It is known that recognition of flagellin by TLR5 triggers the production of IL-1β precursor, which is subsequently processed by caspase-1 and secreted to the extracellular space (9, 32). As described above, the FliC on the recombinant lactobacilli was confirmed to activate NF-κB via TLR5. Thus, an increase in the level of total pro-IL-1β may be the reason for the additional IL-1β secretion. The only difference in cytokine responses between the recombinant lactobacilli displaying FliC with a covalent bond and those displaying FliC with a noncovalent bond was IL-12 production. Overall, display of FliC on the cell surface does not drastically affect interaction between L. acidophilus and human myeloid DCs, but the immunological properties of two different FliC-expressing L. acidophilus strains were dissimilar.
The current study also showed that the cell wall-associated antigens were highly sensitive to simulated digestive juices. This result suggested that such recombinant LAB might easily lose surface antigens if the strains were orally administered. As described above, LAB expressing surface antigens are highly immunogenic when administered parenterally but showed relatively lower immunogenicity when given orally (6, 37, 39, 43). The current study might help explain those results. If the hypothesis is correct, the surface antigens should be protected in the case of immunization via the oral route. In this study, supplementation with bicarbonate or SBTI enabled FliC on the bacterial cell surfaces to elude proteolysis in simulated digestive juices. Because pepsin works only at acidic pHs, neutralization with an alkalescent buffer completely inactivates the activity of the enzyme. Moreover, neutralized gastric fluid is not toxic to the bacterial cells, and thus, the viability of the cells increases as a collateral positive effect. In past studies, bicarbonate buffer has been used for oral immunization (10, 43). The current data support the effectiveness of this approach. SBTI forms a complex with the catalytic site of trypsin or chymotrypsin and inhibits their activities (13). This trypsin inhibitor has been used previously as a protectant for the oral administration of insulin (54). The surface FliC eluded pancreatic digestion in combination with SBTI, although a high concentration of the inhibitor was needed for full protection. SBTI also provided a protective effect against cell damage caused by simulated small intestinal juice in a concentration-dependent manner. This was not expected, because even if proteases were inactivated, bile salt should still be toxic to the cells. One possible reason for this protection from bile is that the high concentration of SBTI protein absorbs bile and interferes with the binding of bile to bacterial cells. Improvement of cell viability in the GI tract may result in increases in the antigen yield and in the persistence of cells in vivo. Thus, supplementation with these protective reagents could possibly improve the efficiency of oral immunization. Encapsulation would be another approach to the protection of antigens and delivery agents. For example, the use of whey protein-gum arabic microencapsulation protected bile salt hydrolase (Bsh) produced by Lactobacillus plantarum WCFS1 from digestive fluids in vitro (24). In addition, Lei et al. reported that enteric coating of recombinant Lactococcus lactis secreting influenza virus H5N1 hemagglutinin actually increased the efficiency of oral immunization (28). If bacterial cells are encapsulated, protective reagents against gastric juices are no longer required. Meanwhile, protective reagents against pancreatic enzymes may still be important, because surface-displayed antigens are probably exposed to proteolytic enzymes in the small intestine after the enteric coat is dissolved.
In conclusion, recombinant lactobacilli displaying antigens on the cell surface by use of different anchoring motifs are basically comparable but can have unequal immunological properties. This phenomenon should be taken into consideration when antigen delivery systems using lactic acid bacteria are developed. This study also showed high sensitivities of surface antigens to digestive enzymes and demonstrated that bicarbonate and SBTI protected both bacterial cells and surface antigens from simulated digestive juices. In the case of oral immunization, coadministration of such protective reagents would likely improve the efficiency of vaccination.
ACKNOWLEDGMENTS
We are grateful to Shizunobu Igimi (National Institute of Health Sciences, Tokyo, Japan) for kindly providing the polyclonal rabbit antibody against Salmonella serovar Typhimurium (RY544). We also thank Evelyn Durmaz, Sara B. Gunderson, and Yong Jun Goh for technical assistance and scientific discussions.
This study was supported by National Institutes of Health grants R01DE019069 and R21AI077409 and was partially supported by the North Carolina Dairy Foundation.
Footnotes
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Altermann E., et al. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 102:3906–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avall-Jääskeläinen S., Kylä-Nikkilä K., Kahala M., Miikkulainen-Lahti T., Palva A. 2002. Surface display of foreign epitopes on the Lactobacillus brevis S-layer. Appl. Environ. Microbiol. 68:5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Azcarate-Peril M. A., Altermann E., Hoover-Fitzula R. L., Cano R. J., Klaenhammer T. R. 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70:5315–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bargieri D. Y., et al. 2008. New malaria vaccine candidates based on the Plasmodium vivax merozoite surface protein-1 and the TLR-5 agonist Salmonella Typhimurium FliC flagellin. Vaccine 26:6132–6142 [DOI] [PubMed] [Google Scholar]
- 5. Bates J. T., Uematsu S., Akira S., Mizel S. B. 2009. Direct stimulation of tlr5+/+ CD11c+ cells is necessary for the adjuvant activity of flagellin. J. Immunol. 182:7539–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bermúdez-Humarán L. G., et al. 2004. An inducible surface presentation system improves cellular immunity against human papillomavirus type 16 E7 antigen in mice after nasal administration with recombinant lactococci. J. Med. Microbiol. 53:427–433 [DOI] [PubMed] [Google Scholar]
- 7. Boekhorst J., Helmer Q., Kleerebezem M., Siezen R. J. 2006. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152:273–280 [DOI] [PubMed] [Google Scholar]
- 8. Borchers A. T., Selmi C., Meyers F. J., Keen C. L., Gershwin M. E. 2009. Probiotics and immunity. J. Gastroenterol. 44:26–46 [DOI] [PubMed] [Google Scholar]
- 9. Ciacci-Woolwine F., McDermott P. F., Mizel S. B. 1999. Induction of cytokine synthesis by flagella from gram-negative bacteria may be dependent on the activation or differentiation state of human monocytes. Infect. Immun. 67:5176–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corthésy B., Boris S., Isler P., Grangette C., Mercenier A. 2005. Oral immunization of mice with lactic acid bacteria producing Helicobacter pylori urease B subunit partially protects against challenge with Helicobacter felis. J. Infect. Dis. 192:1441–1449 [DOI] [PubMed] [Google Scholar]
- 11. Delaney K. N., Phipps J. P., Johnson J. B., Mizel S. B. 2010. A recombinant flagellin-poxvirus fusion protein vaccine elicits complement-dependent protection against respiratory challenge with vaccinia virus in mice. Viral Immunol. 23:201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Desvaux M., Dumas E., Chafsey I., Hébraud M. 2006. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol. Lett. 256:1–15 [DOI] [PubMed] [Google Scholar]
- 13. De Vonis Bidlingmeyer U., Leary T. R., Laskowski M., Jr 1972. Identity of the tryptic and alpha-chymotryptic reactive sites on soybean trypsin inhibitor (Kunitz). Biochemistry 11:3303–3310 [DOI] [PubMed] [Google Scholar]
- 14. Dieye Y., Usai S., Clier F., Gruss A., Piard J. C. 2001. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 183:4157–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duong T., Miller M. J., Barrangou R., Azcarate-Peril M. A., Klaenhammer T. R. 1 September 2010. Construction of vectors for inducible and constitutive gene expression in Lactobacillus. Microb. Biotechnol. [Epub ahead of print.] doi:10.1111/j.1751–7915.2010.00200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frece J., et al. 2005. Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J. Appl. Microbiol. 98:285–292 [DOI] [PubMed] [Google Scholar]
- 17. Germond J. E., Delley M., Gilbert C., Atlan D. 2003. Determination of the domain of the Lactobacillus delbrueckii subsp. bulgaricus cell surface proteinase PrtB involved in attachment to the cell wall after heterologous expression of the prtB gene in Lactococcus lactis. Appl. Environ. Microbiol. 69:3377–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Girardin S. E., et al. 2003. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278:8869–8872 [DOI] [PubMed] [Google Scholar]
- 19. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 20. Hayashi F., et al. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099–1103 [DOI] [PubMed] [Google Scholar]
- 21. Honko A. N., Sriranganathan N., Lees C. J., Mizel S. B. 2006. Flagellin is an effective adjuvant for immunization against lethal respiratory challenge with Yersinia pestis. Infect. Immun. 74:1113–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim T. W., Igimi S., Kajikawa A., Kim H. Y. 2008. Display of heterologous proteins on the surface of Lactococcus lactis using the H and W domain of PrtB from Lactobacillus delbrueckii subsp. bulgaricus as an anchoring matrix. J. Appl. Microbiol. 104:1636–1643 [DOI] [PubMed] [Google Scholar]
- 23. Konstantinov S. R., et al. 2008. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U. S. A. 105:19474–19479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lambert J. M., Weinbreck F., Kleerebezem M. 2008. In vitro analysis of protection of the enzyme bile salt hydrolase against enteric conditions by whey protein-gum arabic microencapsulation. J. Agric. Food Chem. 56:8360–8364 [DOI] [PubMed] [Google Scholar]
- 25. Lebeer S., Vanderleyden J., De Keersmaecker S. C. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8:171–184 [DOI] [PubMed] [Google Scholar]
- 26. Lee J. S., et al. 2006. Mucosal immunization with surface-displayed severe acute respiratory syndrome coronavirus spike protein on Lactobacillus casei induces neutralizing antibodies in mice. J. Virol. 80:4079–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leenhouts K., Buist G., Kok J. 1999. Anchoring of proteins to lactic acid bacteria. Antonie Van Leeuwenhoek 76:367–376 [PubMed] [Google Scholar]
- 28. Lei H., Xu Y., Chen J., Wei X., Lam D. M. 2010. Immunoprotection against influenza H5N1 virus by oral administration of enteric-coated recombinant Lactococcus lactis mini-capsules. Virology 407:319–324 [DOI] [PubMed] [Google Scholar]
- 29. Ma D. Y., Clark E. A. 2009. The role of CD40 and CD154/CD40L in dendritic cells. Semin. Immunol. 21:265–272(Erratum, 22:190, 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuguchi T., et al. 2003. Lipoteichoic acids from Lactobacillus strains elicit strong tumor necrosis factor alpha-inducing activities in macrophages through Toll-like receptor 2. Clin. Diagn. Lab. Immunol. 10:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McDonald W. F., et al. 2007. A West Nile virus recombinant protein vaccine that coactivates innate and adaptive immunity. J. Infect. Dis. 195:1607–1617 [DOI] [PubMed] [Google Scholar]
- 32. Miao E. A., Andersen-Nissen E., Warren S. E., Aderem A. 2007. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 29:275–288 [DOI] [PubMed] [Google Scholar]
- 33. Mohamadzadeh M., Duong T., Hoover T., Klaenhammer T. R. 2008. Targeting mucosal dendritic cells with microbial antigens from probiotic lactic acid bacteria. Expert Rev. Vaccines 7:163–174 [DOI] [PubMed] [Google Scholar]
- 34. Mohamadzadeh M., Duong T., Sandwick S. J., Hoover T., Klaenhammer T. R. 2009. Dendritic cell targeting of Bacillus anthracis protective antigen expressed by Lactobacillus acidophilus protects mice from lethal challenge. Proc. Natl. Acad. Sci. U. S. A. 106:4331–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mohamadzadeh M., et al. 2010. Targeted expression of anthrax protective antigen by Lactobacillus gasseri as an anthrax vaccine. Future Microbiol. 5:1289–1296 [DOI] [PubMed] [Google Scholar]
- 36. Mohamadzadeh M., et al. 2005. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. U. S. A. 102:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Norton P. M., et al. 1996. Factors affecting the immunogenicity of tetanus toxin fragment C expressed in Lactococcus lactis. FEMS Immunol. Med. Microbiol. 14:167–177 [DOI] [PubMed] [Google Scholar]
- 38. Pagnini C., et al. 2010. Probiotics promote gut health through stimulation of epithelial innate immunity. Proc. Natl. Acad. Sci. U. S. A. 107:454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perez C. A., Eichwald C., Burrone O., Mendoza D. 2005. Rotavirus vp7 antigen produced by Lactococcus lactis induces neutralizing antibodies in mice. J. Appl. Microbiol. 99:1158–1164 [DOI] [PubMed] [Google Scholar]
- 40. Pfeiler E. A., Azcarate-Peril M. A., Klaenhammer T. R. 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189:4624–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pouwels P. H., Leer R. J., Boersma W. J. 1996. The potential of Lactobacillus as a carrier for oral immunization: development and preliminary characterization of vector systems for targeted delivery of antigens. J. Biotechnol. 44:183–192 [DOI] [PubMed] [Google Scholar]
- 42. Prechtel A. T., Steinkasserer A. 2007. CD83: an update on functions and prospects of the maturation marker of dendritic cells. Arch. Dermatol. Res. 299:59–69 [DOI] [PubMed] [Google Scholar]
- 43. Reveneau N., Geoffroy M. C., Locht C., Chagnaud P., Mercenier A. 2002. Comparison of the immune responses induced by local immunizations with recombinant Lactobacillus plantarum producing tetanus toxin fragment C in different cellular locations. Vaccine 20:1769–1777 [DOI] [PubMed] [Google Scholar]
- 44. Roos S., Jonsson H. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433–442 [DOI] [PubMed] [Google Scholar]
- 45. Scavone P., et al. 2007. Intranasal immunisation with recombinant Lactococcus lactis displaying either anchored or secreted forms of Proteus mirabilis MrpA fimbrial protein confers specific immune response and induces a significant reduction of kidney bacterial colonisation in mice. Microbes Infect. 9:821–828 [DOI] [PubMed] [Google Scholar]
- 46. Seegers J. F. 2002. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 20:508–515 [DOI] [PubMed] [Google Scholar]
- 47. Siezen R. J. 1999. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Van Leeuwenhoek 76:139–155 [PubMed] [Google Scholar]
- 48. Smit E., Jager D., Martinez B., Tielen F. J., Pouwels P. H. 2002. Structural and functional analysis of the S-layer protein crystallisation domain of Lactobacillus acidophilus ATCC 4356: evidence for protein-protein interaction of two subdomains. J. Mol. Biol. 324:953–964 [DOI] [PubMed] [Google Scholar]
- 49. Strindelius L., Filler M., Sjöholm I. 2004. Mucosal immunization with purified flagellin from Salmonella induces systemic and mucosal immune responses in C3H/HeJ mice. Vaccine 22:3797–3808 [DOI] [PubMed] [Google Scholar]
- 50. Walker D. C., Aoyama K., Klaenhammer T. R. 1996. Electrotransformation of Lactobacillus acidophilus group A1. FEMS Microbiol. Lett. 138:233–237 [DOI] [PubMed] [Google Scholar]
- 51. Wells J. M., Mercenier A. 2008. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 6:349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wells J. M., Wilson P. W., Norton P. M., Gasson M. J., Le Page R. W. 1993. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol. Microbiol. 8:1155–1162 [DOI] [PubMed] [Google Scholar]
- 53. Zeuthen L. H., Fink L. N., Frøkiaer H. 2008. Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology 124:489–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ziv E., et al. 1994. Oral administration of insulin in solid form to nondiabetic and diabetic dogs. J. Pharm. Sci. 83:792–794 [DOI] [PubMed] [Google Scholar]