Abstract
This study investigated the effects of dietary supplementation with a prebiotic mannan oligosaccharide (MOS) on broiler performance, bacterial community structure, and phylogenetic populations of cecal contents. Bird performance data were collected, and cecal samples were extracted from randomly caught poults from each treatment group every 7 days from hatching to the age of 42 days. Weight gain, feed consumption, and feed efficiency ratios did not differ significantly between groups. Automated ribosomal intergenic spacer analysis (ARISA) of the bacterial communities in birds receiving MOS-supplemented diets indicated that dietary supplementation with MOS at either of 2 levels significantly altered the bacterial community structure from that of the control group on all sample days. The phylogenetic identities of bacteria contained within the cecum were determined by constructing a 16S rRNA gene clone library. A total of 594 partial 16S rRNA gene sequences from the cecal contents were analyzed and compared for the three dietary treatments. The dominant bacteria of the cecum belonged to three phyla, Firmicutes, Bacteroidetes, and Proteobacteria; of these, Firmicutes were the most dominant in all treatment groups. Statistical analysis of the bacterial 16S rRNA gene clone libraries showed that the compositions of the clone libraries from broilers receiving MOS-supplemented diets were, in most cases, significantly different from that of the control group. It can be concluded that in this trial MOS supplementation significantly altered the cecal bacterial community structure.
INTRODUCTION
The health and nutritional status of poultry is largely interlinked with the gastrointestinal (GI) microflora, which directly or indirectly affects gut morphology, nutrition, the pathogenesis of intestinal disease, and immune responses (26). The gut microflora is considered to be relatively unstable and can easily be disturbed by various environmental factors, such as pathogenic challenge. Bacterial disease outbreaks impose significant constraints on poultry production, adversely impacting the poultry industry by reducing animal welfare and productivity through disease, poor digestion, and poor nutrient absorption. This, in turn, can lead to significant losses for the farmer and can increase the potential for the contamination of poultry products marketed for human consumption (37).
Antibiotic growth promoters have traditionally been used in poultry feed at subtherapeutic levels to prevent clinical and subclinical levels of disease, leading to improved growth rates and feed efficiencies (13). An increase in food safety concerns resulting from extensive antibiotic use has seen the poultry industry challenged in recent years as safer meat, free from antibiotics and disease, has become a requirement within the EU (16a). Consumer demand for antibiotic-free meat has also increased within the United States and other antibiotic-using countries as a result of concerns about the spread of antibiotic resistance, making it necessary for poultry producers to find suitable replacements for antibiotic growth promoters (45). Alternatives to antibiotic growth promoters include prebiotics, such as mannan oligosaccharides (MOS). Prebiotics are defined as “nondigestible food ingredients that increase the growth or activity of select members of the endogenous intestinal microbial community and the new community benefits the host” (32). Mannan oligosaccharides have been found to have beneficial effects on broilers. They have been shown to have the potential to stimulate beneficial bacteria while also having a negative effect on pathogenic bacteria (e.g., Salmonella spp. and Escherichia coli) in the broiler gut, which can have a positive effect on the health of the animals (4, 46). The effects of MOS supplementation on bird health and performance have also been studied comprehensively, and MOS have proven effective at improving weight gain and feed conversion efficiencies while also protecting against infection through pathogen binding (16, 21, 35, 46). However, little is known about the effect of MOS supplementation on the unculturable bacterial community of birds.
The objective of this study was to determine the effect of supplementation with MOS on the complex microbial ecology of the chicken cecum throughout the growth phase of broilers. Automated ribosomal intergenic spacer analysis (ARISA) was utilized because it generates substantial data on the microbial community that are less amenable to the conventional microbial analytical techniques used in the past to study microbial community structure. Because ARISA offers no phylogenetic information on changing bacterial communities, this technique was coupled with 16S rRNA gene clone library analysis in order to allow the phylogenetic identification of bacteria within the cecum and to determine what bacterial groups may be affected by dietary supplementation.
These molecular approaches have several limitations, including the possibility that DNA isolation, PCR amplification, and cloning might be biased in favor of certain bacteria and sequences. Furthermore, different species can have multiple rrn operon copy numbers, leading to overestimation of species richness and diversity (23, 47, 52). However, these techniques still provide an overview of the microbial community structure of a cecal sample and allow one to verify statistically the effect of dietary supplementation on the bacterial communities of the cecum. The understanding and description of intestinal microbial communities are very important for the development of these new feed additives and for the appropriate manipulation of diets to improve poultry performance, health, and welfare.
MATERIALS AND METHODS
Experimental design, sample collection, and preservation.
A total of 288 1-day-old male broiler chickens (Cobb strain; Cobb hatchery, Monticello, KY) were used in this study. Clean concrete-floor pens (3 ft by 5 ft) were used to house the birds. Animals were randomly split into three groups of eight pens, with 12 birds per pen (96 birds/group). The pens were divided into three treatment groups: group 1, fed a basal diet; group 2, fed a basal diet supplemented with 1 kg MOS per metric ton of basal diet (t−1); and group 3, fed a basal diet supplemented with 2 kg MOS t−1. Basal diets were prepared by a commercial feed mill and consisted primarily of corn and soybean meal, as outlined in Table 1; MOS was added on-site (Bio-Mos; supplied by Alltech Inc., Nicholasville, KY). The MOS doses chosen are typical commercial diet inclusion rates, and previous studies have proven these doses to be beneficial for broilers (9, 33, 35, 53). Starter feed was fed from day 0 to day 21 and grower feed from day 22 to day 42. Feed and water were provided ad libitum throughout the study via an automatic bell drinker and a 17-inch-diameter hanging tube. Each pen was dressed with built-up litter for bedding from day zero. Heating and lighting were controlled, with continuous lighting for 22 h and 2 h of darkness daily. The test facility, pens, and birds were observed twice daily for general conditions. All conditions were kept uniform for all three groups.
Table 1.
Compositions of basal starter and grower diets
| Ingredient | % (wt/wt) of amt fed |
|
|---|---|---|
| Starter diet | Grower diet | |
| Corn | 54.60 | 61.23 |
| Soybean meal (48%) | 36.50 | 31.50 |
| Corn oil | 4.80 | 3.65 |
| Salt | 0.45 | 0.45 |
| Limestone | 1.33 | 1.23 |
| Dicalcium phosphate | 1.76 | 1.54 |
| Vitamin-mineral premix | 0.25 | 0.20 |
| Mycosorb | 0.10 | 0.10 |
| dl-Methionine | 0.21 | 0.10 |
All poults were weighed and the values averaged by pen on days 0, 7, 14, 21, 28, 35, and 42, and feed intake was measured in order to assess feed conversion ratios and weight gain. The intact cecal pouches of 2 randomly caught birds per pen were removed immediately after euthanization on each weighing day, and the cecal contents were placed in sterile tubes containing sterilized 20% (wt/vol) maltodextrin on each day. The tubes were then flash frozen in liquid nitrogen, lyophilized, and stored at −80°C.
Total-DNA extraction and purification.
Cecal DNA was extracted as described previously (7). Briefly, 0.05 g of cecal contents was added to tubes containing 0.5 g of 0.1-mm-diameter glass beads and 0.5 g of 0.5-mm-diameter zirconia beads, to which hexadecyltrimethylammonium bromide (CTAB) extraction buffer (10% CTAB in 0.7 M NaCl mixed with equal volumes of 240 mM K2HPO4 [pH 8.0]) (Sigma-Aldrich, St. Louis, MO) was added. The resulting DNA was purified using a High Pure PCR product purification kit (Roche, Basel, Switzerland) according to the manufacturer's instructions and was eluted in a final volume of 50 μl. DNA was further diluted 1:10 with sterile Millipore water for subsequent analysis.
Community fingerprinting using bacterial ARISA.
The 16S-23S intergenic spacer region from the bacterial rRNA operon was amplified from cecal DNA using forward primer S-D-Bact-1522-b-S-20 (5′-TGCGGCTGGATCCCCTCCTT-3′) and reverse primer L-D-Bact-132-a-A-18 (5′-CCGGGTTTCCCCATTCGG-3′ (40). The 5′ end of the forward primer was labeled with 6-carboxyfluorescein (6-FAM) to allow for fluorescent detection of the amplicons by ARISA. PCR conditions and reagents were similar to those described previously, and a standard concentration of 20 ng of pooled cecal DNA was used in each reaction (51). PCR products were purified using a High Pure PCR product purification kit (Roche, Basel, Switzerland).
Samples for ARISA (10 μl) consisted of 1 μl of the fluorescently labeled PCR product, 0.5 μl of a GeneScan 1200 LIZ size standard (Applied Biosystems, Foster City, CA), and 8.5 μl of Hi-Di formamide (Applied Biosystems, Foster City, CA). The samples were denatured at 95°C for 3 min and were then immediately placed on ice for 5 min. An ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA) equipped with a 16-channel capillary system (36 cm long) filled with POP-7 polymer (Applied Biosystems, Foster City, CA) was used to analyze the samples as described in reference 50.
16S rRNA gene clone library construction.
PCR amplification for 16S rRNA gene clone library construction was carried out on pooled DNA samples (1 μl from each bird, on each sampling day, per treatment group) using the universal primer set 27F (5′-AGAGTTTGATCMTGGCTGAD-3′)-1492R (5′-TACGGYTACCTTGTTACGACTT-3′), which produced an amplicon of ∼1,500 bp (27). The final reaction mixtures included 100 ng of pooled DNA, 500 ng each primer, 2.5 mM MgCl2, 1× (NH4)2SO4 reaction buffer (pH 8.80), 500 μM each deoxynucleoside triphosphate (dNTP), 2 U Taq polymerase, and sterile water to a final reaction volume of 100 μl. PCR amplification was performed as follows. A 2-min denaturation step at 94°C was followed by 5 min at 85°C for the addition of Taq polymerase. After the addition of the polymerase, a total of 30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C were carried out. A final 10-min step at 72°C was employed to ensure that the remaining amplified products were fully extended. The reaction product was then cooled to 4°C prior to analysis on agarose gels. PCR products were purified using a High Pure PCR product purification kit, and the purified products were ligated into vector pCR 2.1 (Invitrogen, Carlsbad, CA) using the Quick-Stick ligase kit (Bioline, London, United Kingdom) according to the manufacturer's instructions. The ligation reaction was carried out at room temperature using a T4 DNA ligase. Escherichia coli strain INVαF′ competent cells were transformed with insert-containing vectors by heat shock (30 s at 42°C) according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). The pCR 2.1 vector contains an ampicillin resistance gene used in the selection and maintenance of E. coli and thus allowed for the identification of successfully transformed clones by growth on Luria-Bertani (LB) plates supplemented with 100 μg ml−1 ampicillin. The clones were also screened for α-complementation of β-galactosidase by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), allowing blue/white screening of putative transformants. Approximately 60 clones were selected for each library.
Plasmid extraction and sequencing.
Plasmid DNA was isolated from E. coli cells using the High Pure plasmid isolation kit according to the manufacturer's instructions (Roche, Basel, Switzerland), and plasmids were eluted in 50 μl of sterile deionized water. Sequencing was carried out by Macrogen Inc. (Seoul, South Korea) using M13 forward and reverse primers with an Applied Biosystems 3730xl DNA analyzer.
Analysis of DNA sequences.
Sequence and homology data were analyzed using the basic local alignment search tool (BLAST) (1) (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and putative identities were assigned to each clone. The phylogenetic associations of bacteria were determined using the classifier algorithm on the Ribosomal Database Project (RDP) website (http://rdp.cme.msu.edu/). Chimeric sequences were checked using Chimera Check on the same website. Phylogenetic trees were constructed using Molecular Evolutionary Genetics Analysis (MEGA) software, version 4.1 (http://www.megasoftware.net/) (49). The neighbor-joining method (41) was used to infer evolutionary history, and the Jukes-Cantor method (24) was used to compute evolutionary distances, for phylogenetic tree construction. A total of 500 replicates were used for bootstrap analysis (17).
Statistical differences in the library compositions of the floras were estimated by comparisons of the libraries using the LIBSHUFF utility (44). If the P value was ≤0.05, the libraries were considered significantly different from each other, indicating that the microbial compositions were different.
Statistical analysis.
One-way analysis of variance (ANOVA) was used to analyze data sets for the trial performance data (weight gain, feed consumption, and feed conversion ratios). ANOVA was performed using the MINITAB (Coventry, United Kingdom) statistical software package, version 15.0. The significance level was set to a P value of ≤0.05.
The similarity of ribotype profiles in ARISA, for each treatment group, was measured using the Bray-Curtis measure of similarity (6). Bacterial community matrices were analyzed using Primer software (Primer-E Ltd., United Kingdom) to create nonmetric multidimensional scaling (MDS) plots for each of the data sets. MDS is an ordination technique that represents the relationships inherent in multivariate data sets and attempts to preserve the ranked order of similarity of any two communities as an inverse function of the distance between the points representing those communities on the plot (25). The degree to which the plot matches the similarity matrix can be judged by examining the stress (Kruskal's stress): values less than 0.2 represent good ordination with little risk of misinterpretation of the data (11). MDS was performed using 50 random starting configurations of the sample points, and in all cases, 2-dimensional (2D) solutions are presented.
Permutational multivariate ANOVA (PERMANOVA) was used for testing the simultaneous response of one or more variables to one or more factors in an ANOVA experimental design on the basis of any distance measure, using permutation methods (2). Partitioning of the total sum of squares based on the distance matrix is performed according to the full experimental design as specified. A distance-based pseudo-F statistic is calculated for each term in the model based on the expectations of mean squares; this pseudo-F statistic is analogous to the F statistic for multifactorial univariate ANOVA (12). Permutations are then used to obtain P values for each term used. In the event of a small sample size, such that too few permutations are available, correct P values can also be obtained through Monte Carlo random draws (3). Pairwise comparisons among levels of factors can also be performed.
Nucleotide sequence accession numbers.
Representative DNA sequences were submitted to GenBank under accession numbers GUI171034 to GUI171170 and JF781593 to JF782043.
RESULTS
Performance data.
Bird performance data revealed that dietary supplementation had no effect on bird weights, feed consumption, or feed conversion ratios. The initial (day-of-hatch) body weights of chicks did not differ significantly for different treatments (P > 0.05) and averaged 42 g per bird per treatment. No significant differences in bird weight were noted between the control and supplemented groups on any day. Bird weights were continuously higher in the supplemented groups than in the control group on days 7, 14, 21, and 28 posthatch but were lower, though not significantly, in the supplemented groups on days 35 and 42 posthatch, as shown in Table S1a in the supplemental material. The average feed consumption per pen was measured on each sampling day and is shown in Table S1b in the supplemental material. The quantity of feed was maintained at 20 kg throughout the experiment, and the feed tubes were replenished as necessary upon inspection. Feed efficiency ratios (see Table S1c in the supplemental material) were calculated by dividing the feed consumption throughout a time period by the weight gained for that period. The averaged feed efficiency ratios varied with time and with diet, but no significant differences between groups were observed on any day.
ARISA.
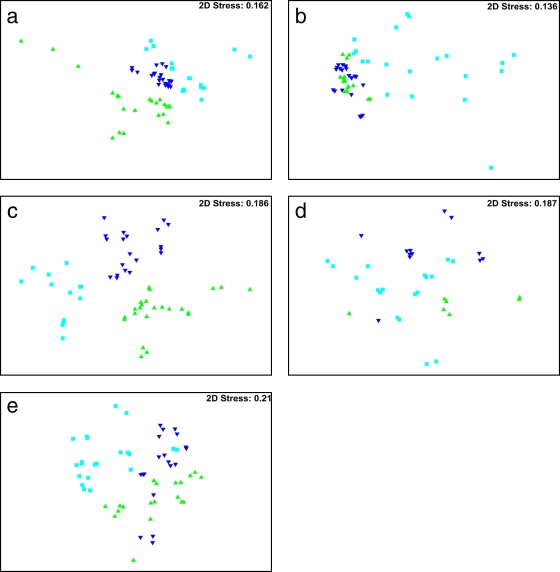
ARISA was used to generate bacterial community profiles, consisting of the individual ARISA amplicons present and their relative abundances, for each treatment group on every sampling day. Each amplicon detected in the profile indicated a unique ribotype. The majority of ribotypes detected lay in the range of 300 bp to 800 bp. Nonmetric multidimensional scaling (MDS) was used to determine whether the overall cecal bacterial community compositions of chickens whose diets were supplemented with MOS at either of two levels differed from those for chickens fed the control unsupplemented diet. MDS ordinations are typically interpreted based on the distance between ordinate points; where treatments appear close together, they can be regarded as having similar community compositions. Figure 1a to e show MDS ordinations based on Bray-Curtis similarities for all treatment groups on each sampling day. All plots show clear differentiation of the control group, the group receiving 1 kg MOS t−1, and the group receiving 2 kg MOS t−1 except on day 21 posthatch. The MDS plot for day 21 posthatch shows one well-defined group and two groups that are not easily distinguished from each other. These other two groups, however, are indeed different communities; they are in the same 2D plane but in separate 3D planes (this can be seen from a 3D plot, which cannot be shown on a 2D printout). On all days, the stress values on the ordination plot were below 0.2 (Fig. 1a to e), indicating that the observed plot is a reliable representation of the data. Overall, MDS ordination plots indicated that the ribotype composition of bacterial communities was affected by dietary supplementation with MOS at both the 1-kg t−1 and the 2-kg t−1 level and that the most marked differences from the control group were attributed to MOS supplementation at the 2-kg t−1 level.
Fig. 1.
Multidimensional scaling (MDS) plots demonstrating changes in bacterial community composition in the cecal contents of chickens whose diets were supplemented with MOS. ▴, control group; ▾, group receiving MOS supplementation at 1 kg t−1; ▪, group receiving MOS supplementation at 2 kg t−1. The Bray-Curtis measure of similarity was used to assess similarities in ribotype profiles in the data set. All samples were standardized by total abundance, and results were subjected to square root transformation. Eight samples were assessed for each group. (a) Day 14; (b) day 21; (c) day 28; (d) day 35; (e) day 42.
PERMANOVA was used to explore the statistical significance of the variation between the bacterial communities observed by MDS ordinations. PERMANOVA for each day analyzed (Table 2) confirmed that significant differences existed between all groups. Pairwise analysis of each grouping showed that significant differences (P ≤ 0.001) existed between all possible combinations of groups.
Table 2.
PERMANOVA of bacterial communities in cecal contents in response to dietary supplementation with MOSa
| Sample day and test | Statisticb |
No. of unique permutations | |||
|---|---|---|---|---|---|
| df | SS | F | P | ||
| Day 14 | |||||
| Overall test of diet | 2 | 23,178 | 8.8 | 0.001 | 998 |
| Pairwise tests | |||||
| Control vs 1 kg MOS t−1 | 0.001 | 998 | |||
| Control vs 2 kg MOS t−1 | 0.001 | 997 | |||
| 1 vs 2 kg MOS t−1 | 0.001 | 998 | |||
| Day 21 | |||||
| Overall test of diet | 2 | 31,612 | 8.02 | 0.001 | 998 |
| Pairwise tests | |||||
| Control vs 1 kg MOS t−1 | 0.001 | 992 | |||
| Control vs 2 kg MOS t−1 | 0.001 | 994 | |||
| 1 vs 2 kg MOS t−1 | 0.001 | 999 | |||
| Day 28 | |||||
| Overall test of diet | 2 | 33,641 | 10.28 | 0.001 | 998 |
| Pairwise tests | |||||
| Control vs 1 kg MOS t−1 | 0.001 | 998 | |||
| Control vs 2 kg MOS t−1 | 0.001 | 997 | |||
| 1 vs 2 kg MOS t−1 | 0.001 | 999 | |||
| Day 35 | |||||
| Overall test of diet | 2 | 19,640 | 4.83 | 0.001 | 999 |
| Pairwise tests | |||||
| Control vs 1 kg MOS t−1 | 0.001 | 996 | |||
| Control vs 2 kg MOS t−1 | 0.001 | 999 | |||
| 1 vs 2 kg MOS t−1 | 0.001 | 999 | |||
| Day 42 | |||||
| Overall test of diet | 2 | 30,594 | 8.82 | 0.001 | 999 |
| Pairwise tests | |||||
| Control vs 1 kg MOS t−1 | 0.001 | 999 | |||
| Control vs 2 kg MOS t−1 | 0.001 | 997 | |||
| 1 vs 2 kg MOS t−1 | 0.001 | 999 | |||
Analysis was based on abundances from square root-transformed Bray-Curtis similarities. P, ≤0.05; n, 8 for each group.
df, degrees of freedom; SS, sum of squares; F, variance of the group; P, probability.
16S rRNA gene sequence library analysis.
Phylogenetic analysis was carried out on cloned 16S rRNA gene amplicons from cecal DNA extracted from the control and mannan oligosaccharide-supplemented groups on each sampling day. Clones were assigned putative identities and individual accession numbers using BLAST and the GenBank database, available on the NCBI website. The similarity matches of sequenced clones with sequences in the GenBank database ranged from 89% to 100%. Approximately 10% of the clone sequences isolated displayed similarities of <97% to sequences already deposited in the BLAST database. These 10% represent bacterial sequences that have yet to be identified and may represent novel bacterial species. The isolated clones that were identified were highly diverse; many were related to sequences previously discovered in human feces, pig and chicken GI tracts, or the bovine rumen.
The phylogenetic classification of clones isolated from the control and supplemented groups is outlined in Table 3. A total of 594 partial 16S rRNA gene sequences were analyzed over the 5 sampling days, and three treatment groups with approximately 200 sequences per treatment were analyzed in total. Analysis showed that three major bacterial phyla dominated the broiler cecum: the low-G+C Gram-positive phylum Firmicutes and the Gram-negative phyla Bacteroidetes and Proteobacteria. The phylum Firmicutes was dominated by sequences related to Clostridia subclusters XIVa and IV, Bacilli, lactobacilli, and other Clostridia-related sequences. Clones related to the Clostridiaceae and the Ruminococcaceae dominated the bacterial communities of the broiler ceca in both the control and MOS-supplemented groups. Sequences belonging to the phylum Firmicutes were the most abundant in the cecum on almost all sampling days and in almost all treatment groups, making up 61% to 100% of all sequences except on day 35 in the control group, where they made up just 48% of all sequences detected.
Table 3.
Classification and abundances of 594 bacterial 16S rRNA gene clone sequences isolated from cecal contents of broilers from each diet group on each sampling day
| Phylum | Class | No. of bacterial 16S rRNA gene clone sequences for the indicated diet groupa on the following sample day: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 14 |
Day 21 |
Day 28 |
Day 35 |
Day 42 |
||||||||||||
| C |
M1 |
M2 |
C |
M1 |
M2 |
C |
M1 |
M2 |
C |
M1 |
M2 |
C |
M1 |
M2 |
||
| Firmicutes | Clostridia | |||||||||||||||
| Subcluster XIVa | 18 | 14 | 9 | 18 | 16 | 15 | 12 | 5 | 14 | 9 | 11 | 15 | 13 | 14 | 23 | |
| Subcluster IV | 17 | 19 | 16 | 14 | 13 | 16 | 11 | 7 | 13 | 2 | 11 | 7 | 10 | 14 | 8 | |
| Bacilli | 5 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 2 | |||||||
| Other | 8 | 5 | 1 | 5 | 5 | 5 | 4 | 7 | 14 | 2 | 3 | 6 | 5 | 5 | 2 | |
| Bacteroidetes | Bacteroidia | 1 | 8 | 1 | 14 | 12 | 15 | 14 | 6 | 7 | 11 | 8 | 10 | |||
| Proteobacteria | Gammaproteobacteria | 2 | 1 | 1 | 1 | 1 | 1 | 2 | ||||||||
| Total | 50 | 39 | 26 | 39 | 44 | 40 | 43 | 31 | 56 | 29 | 32 | 37 | 40 | 43 | 45 | |
C, control group; M1 and M2, groups receiving MOS supplementation at 1 and 2 kg t−1, respectively.
The Bacteroidetes were generally the second most abundant phylum detected within the cecum, and the sequences identified were closely related to Bacteroides spp. and Alistipes spp. Days 28, 35, and 42 posthatch saw the detection of a greater abundance of Bacteroidetes-related sequences, in both the control and MOS-supplemented groups, than days 14 and 21 posthatch. Proteobacteria represented the least abundant phylum detected within the cecum, and the sequences identified were related to the subphylum Gammaproteobacteria. The proteobacterial clones identified from both the control and supplemented groups were clustered with the genera Escherichia and Shigella.
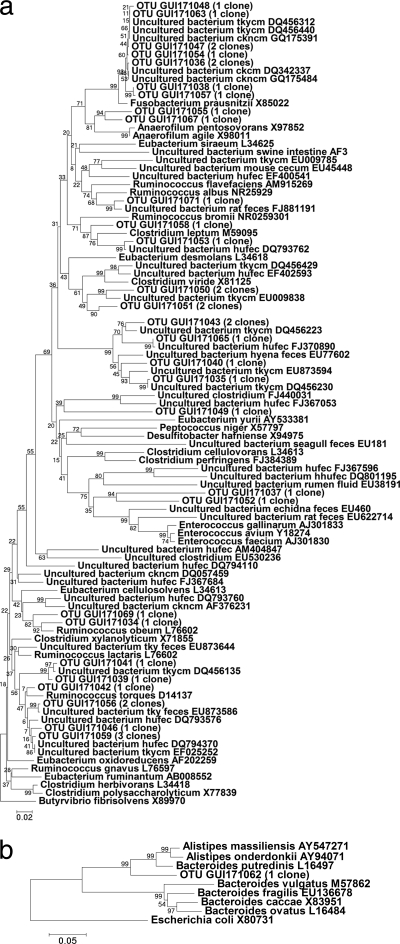
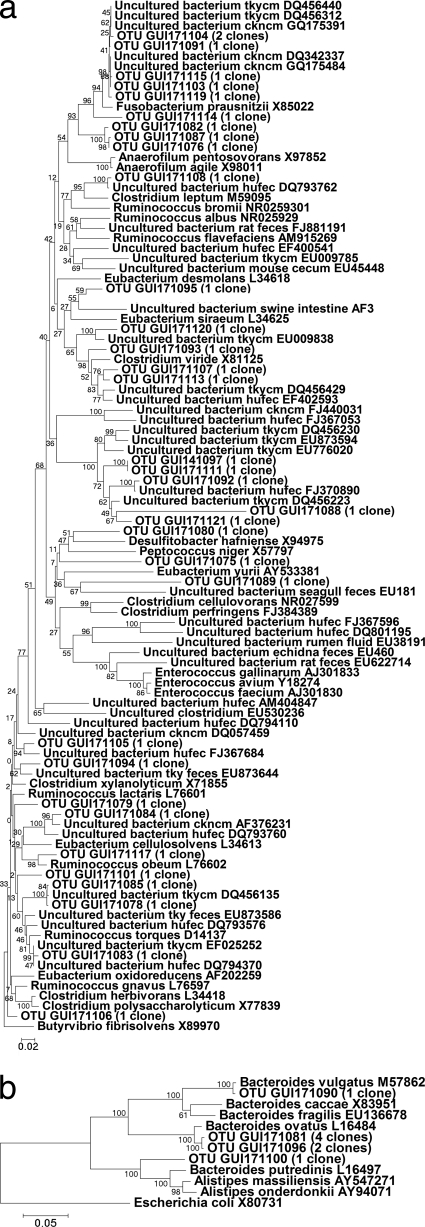
Representative phylogenetic trees for day 21 posthatch of this trial are shown in Fig. 2a and b, 3a and b, and 4a and b. No specific trend could be noted for MOS supplementation either specifically increasing or decreasing the abundance of any class of bacteria detected using phylogenetic trees or BLAST matching alone. In order to determine whether significant differences existed between the compositions of the clone libraries from the control and MOS-supplemented groups, pairwise comparisons of libraries were carried out using LIBSHUFF statistical analysis. Statistical evaluations comparing the control and MOS groups on all sampling days confirmed that the compositions of the clone libraries differed significantly (Table 4). The control group differed significantly from both MOS-supplemented groups on day 14, while on day 21 the control group differed significantly only from the group receiving 1 kg MOS t−1. On days 28 and 35 posthatch, both MOS-supplemented groups were significantly different from the control. A significant difference was also observed between the control group and the group receiving 2 kg MOS t−1, but not the group receiving 1 kg MOS t−1, on day 42 posthatch. Significant differences were also noted between the groups receiving 1 versus 2 kg MOS t−1 on all days except day 28 posthatch.
Fig. 2.
(a) Phylogenetic tree showing the relationships of Firmicutes sequences derived from the cloning of 16S rRNA gene fragments for the control group. Bootstrap support values (500 replicates) are given on the branches, and the sum of branch lengths for the optimal tree was 4.54. (b) Phylogenetic tree showing the relationships of Bacteroidetes sequences derived from the cloning of 16S rRNA gene fragments for the control group. Bootstrap support values (500 replicates) are given on the branches, and the sum of branch lengths for the optimal tree was 0.75.
Fig. 3.
(a) Phylogenetic tree showing the relationships of Firmicutes sequences derived from the cloning of 16S rRNA gene fragments for the group receiving 1 kg MOS t−1. Bootstrap support values (500 replicates) are given on the branches, and the sum of branch lengths for the optimal tree was 4.79. (b) Phylogenetic tree showing the relationships of Bacteroidetes sequences derived from the cloning of 16S rRNA gene fragments for the group receiving 1 kg MOS t−1. Bootstrap support values (500 replicates) are given on the branches, and the sum of branch lengths for the optimal tree was 0.77.
Fig. 4.
(a) Phylogenetic tree showing the relationships of Firmicutes sequences derived from the cloning of 16S rRNA gene fragments for the group receiving 2 kg MOS t−1. Bootstrap support values (500 replicates) are given on the branches, and the sum of branch lengths for the optimal tree was 4.71. (b) Phylogenetic tree showing the relationships of Bacteroidetes sequences derived from the cloning of 16S rRNA gene fragments for the group receiving 2 kg MOS t−1. Bootstrap support values (500 replicates) are given on the branches, and the sum of branch lengths for the optimal tree was 0.78.
Table 4.
Distribution of P values for comparison of 16S rRNA gene sequence libraries among samples from chickens receiving different dietsa
| Pairwise test of diets |
P value at sampling day: |
||||
|---|---|---|---|---|---|
| 14 | 21 | 28 | 35 | 42 | |
| Control vs 1 kg MOS t−1 | 0.053 | 0.008 | 0.001 | 0.023 | 0.319 |
| Control vs 2 kg MOS t−1 | 0.001 | 0.139 | 0.018 | 0.015 | 0.038 |
| 1 vs 2 kg MOS t−1 | 0.001 | 0.024 | 0.19 | 0.003 | 70.011 |
Differences were determined by pairwise comparisons of the clone libraries using LIBSHUFF. If the P value returned by LIBSHUFF is ≤0.05, the two libraries are significantly different in community composition, with a confidence level of 95%.
DISCUSSION
The development of culture-independent methods for the analysis of microbial communities has overcome the limitations of culture-based methods of analysis, allowing the examination of entire bacterial communities. This study examined the effects of dietary supplementation with MOS on broiler performance and bacterial community structure through the use of a number of different molecularly based methods. The results showed that dietary supplementation with MOS had a significant impact on changing bacterial community structure.
Mannan oligosaccharides have been shown quite widely to have positive effects on bird performance characteristics, including live weight gain, feed conversion efficiencies, and feed consumption (16, 35, 36, 43, 56; J. P. Blake et al., presented at Alltech's 22nd international symposium, Lexington, KY, 2005). The present study analyzed data from a 42-day broiler trial to evaluate birds for improvements in performance as a result of dietary supplementation with MOS. No significant differences were noted in broiler performance when live weight gain, feed consumption, or feed conversion efficiencies were compared. Similar results for broilers have also been noted in previous studies where such supplementation failed to convey a growth-promoting effect either through increased weight gain or through improved feed efficiencies (53, 55). It has been noted previously that when environmental stresses are not placed on broiler chickens, no positive responses to antimicrobials or antibiotic alternatives (MOS) are observed (22). Factors such as the broiler strain, housing type, litter age/sterility, feed composition, stocking density, temperature, presence of unfavorable organisms, and disease are possible stress inducers that may elicit an improved response to supplementation (19). These variables were highly controlled in the trial carried out in the present study, which may offer an explanation for the lack of response in the variables analyzed.
The molecular community approach ARISA was used to show the effects of dietary supplementation on total bacterial community structure. The internal transcribed spacer (ITS) region is variable in both length and sequence, making ARISA a useful technique for characterizing microbial communities. MDS ordinations and PERMANOVA indicated that supplementation of feed with MOS at both 1 kg t−1 and 2 kg t−1 invariably significantly altered bacterial community composition, even though no significant differences in bird performance characteristics were noted. These results demonstrate that MOS is capable of altering the microbial community structure of broiler cecal contents across a time scale and at different levels of dietary supplementation. Terminal restriction fragment length polymorphism (TRFLP), a similar microbial community fingerprinting technique, has been used previously to demonstrate the influence of dietary supplementation on the microbiota within the broiler cecum (19). The authors demonstrated that birds whose diets were supplemented with MOS had a microbial community structure significantly different from those of birds receiving another prebiotic (fructooligosaccharides [FOS]) or an antibiotic (zinc bacitracin) but not from that of the control group. In contrast, the broiler trial in this work displayed significantly altered bacterial communities in the supplemented groups versus the control group, a result that may highlight the higher sensitivity of ARISA than of other community analysis approaches. The composition of the intestinal bacterial community is associated with the performance of a bird; therefore, manipulation of the microbiota to produce a community most conducive to optimal performance will be one of the key considerations in designing prebiotic feeding strategies. The birds in this trial were housed in an environment that was atypical for a large broiler production facility. Further studies with birds housed in typical production environments and subjected to typical stresses may show that MOS has superior abilities to enhance bird performance.
The molecular community fingerprinting approach of ARISA is particularly useful as a means of rapidly obtaining broad-scale profiles of bacterial community structure and identifying sensitive changes due to a dietary variable. However, it lacks the ability to obtain specific phylogenetic information about the community profiles; therefore, other methods must be used in conjunction with ARISA to extract such information about a community (18, 39). 16S rRNA gene community libraries have been used previously to successfully estimate the phylogenetic diversity of bacteria present in the gastrointestinal tracts of many animals (5, 28, 30, 31, 34). They have also been used to monitor community changes in response to environmental variables such as time (8) and diet (15), indicating the suitability of clone libraries for identifying changes in poultry cecal contents in response to dietary augmentation with MOS in this trial.
The clone sequences identified from broilers in this study were diverse, and many were related to sequences previously isolated from the gastrointestinal tracts of other animal species. Numerous studies of the intestinal microbiota of humans, pigs, mice, cows, and chickens have contributed novel 16S rRNA gene sequences to the GenBank database, and distinct intestinal microbial communities have been observed for each species (20, 42, 48, 54). However, despite the large number of clone sequences isolated from the gastrointestinal tracts of various mammals and avian species, some clones isolated from this study still exhibited less than 97% similarity to any sequence deposited in GenBank. Since low numbers of previously unidentified clones were isolated from all groups of this trial, no trend could be specifically attributed to dietary supplementation. Similar results were found in previous clone library studies of microbial diversity within the ceca of broilers, where a number of clone sequences isolated were unique to the individual studies, showing less than 97% similarity to sequences deposited in the database (29, 58); Lu et al. and Zhu et al. suggested that these results might indicate that distinct bacterial species or genera inhabit the poultry intestinal tract. It is also possible that these unidentified bacterial clone sequences are unique to the distinct environments where the birds were raised, the strain of the birds used, or the use of different feed components (14).
Of the phyla isolated in the current study, Firmicutes were the most abundant in both the control and MOS-supplemented groups. This phylum was also the most abundant in broiler cecal contents analyzed in other trials (29, 58). Those previous trials found that low-G+C Gram-positive bacteria, including members of Clostridia subclusters XIVa and IV, were abundant in broiler cecal contents, in agreement with the results from this trial (29, 58). The effects of dietary supplementation with MOS on total Firmicutes or on total Clostridia have not been investigated previously. Interestingly, although this phylum is the most abundant in the gastrointestinal tracts of many species, little is known about the bacteria in it. Much of the gastrointestinal flora relating to this phylum remains uncultured, and the functionalities of these organisms can only be inferred phylogenetically. Much scope therefore exists for further elucidation of the bacteria within this phylum.
Bacilli represented a small proportion of the Firmicutes sequences and were mostly identified as Lactobacillus spp. (Table 3). It is not possible to say whether MOS affected these populations, since they were detected at low frequencies in this trial. In agreement with our observations, lactobacilli were also detected at low frequencies in broiler cecal contents in studies by Lu et al. and Zhu et al. (29, 58). Culture-dependent approaches that selectively enrich for Lactobacillus spp. have previously shown that MOS significantly increase their levels in cecal contents (4), although this result may be biased due to the selective enrichment step used. A molecular approach to quantifying total lactobacilli may be more appropriate for determining the effect on total Lactobacillus numbers. Interestingly, the MOS groups contained two previously identified Lactobacillus species that have important documented anti-Salmonella activities (Lactobacillus salivarius and Lactobacillus crispatus) as well as anti E. coli activities (L. crispatus) (10, 57).
The second most abundant phylum detected within the clone libraries of all three groups was Bacteroidetes, although they were detected at a lower frequency than the Firmicutes. This trend has also been observed in previous studies (28, 29, 58). The effect of MOS supplementation on the Bacteroidetes has not been studied previously, perhaps because this phylum is not of major concern with regard to avian-pathogenic diseases and because of the difficulties associated with culturing these bacteria. Bacteroidetes species are common bacteria in the gut, involved in many important metabolic activities, including the fermentation of carbohydrates, the utilization of nitrogenous substances, the biotransformation of bile acids, and the prevention of pathogen colonization (38).
The Gammaproteobacteria comprised the least frequently observed phylogenetic group identified in all of the clone libraries. Lu et al. (29) obtained similar results, detecting a low frequency of Gammaproteobacteria in cecal libraries from chickens. Some Gammaproteobacteria cause disease; these include E. coli, Salmonella spp., Helicobacter spp., Klebsiella spp., Shigella spp. and other enteric pathogens. However, the frequency of isolation of sequences related to this phylum was extremely low in this trial.
Clone library analysis demonstrates that the chicken cecum typically harbors a diverse microbiota dominated by obligate anaerobes. LIBSHUFF analysis indicated that statistically significant differences exist between the clone libraries from the control and MOS-supplemented groups and between those from the two different MOS groups. It is difficult to determine where these differences lie specifically by analyzing the sequences from BLAST matching and clone libraries alone, due to the lack of sensitivity of these techniques. It is likely that the significant differences observed between the clone libraries indicate different genera or species, which are not easily identifiable using these phylogenetic techniques. These results do, however, support the ARISA findings described herein.
The primary purpose of ecological analysis is the description of the microbiota in an ecosystem. The work presented here describes the effect of dietary manipulation with a prebiotic mannan oligosaccharide on the bacterial community structure within the broiler cecum throughout its life cycle. Comprehensive analysis by ARISA, followed by a detailed description of the bacteria present by use of 16S rRNA gene clone library analysis, has revealed that MOS can significantly alter the bacterial community structure of broiler cecal contents at both 1 kg t−1 and 2 kg t−1. The level of MOS provided also causes a significant difference between the communities. Although our data clearly show changes in the bacterial community composition as a result of dietary supplementation with MOS, deciphering these changes at the genus or species level and relating these changes to ecological function remain formidable challenges. Community analysis techniques such as ARISA and clone library analysis are useful means of rapidly obtaining broad-scale profiles of bacterial community structure in cecal contents that cannot be obtained by culture-dependent methods. A more focused approach is now necessary in order to determine whether these changes in bacterial community structure have a function and what, if any, health-promoting effects these changes produce. The poultry industry will benefit from the identification of bacterial species and microbial compositions that could enhance bird performance, maintain an optimal intestinal microbiota, or enhance immunity.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Anderson M. J. 2001. Permutation tests for univariate or multivariate analysis of variance and regression. Can. J. Fish. Aquat. Sci. 58:626–639 [Google Scholar]
- 3. Anderson M. J., Robinson J. 2003. Generalised discriminant analysis based on distances. Aust. N. Z. J. Stat. 45:301–318 [Google Scholar]
- 4. Baurhoo B., Letellier A., Zhao X., Ruiz-Feria C. A. 2007. Cecal populations of lactobacilli and bifidobacteria and Escherichia coli populations after in vivo Escherichia coli challenge in birds fed diets with purified lignin or mannanoligosaccharides. Poult. Sci. 86:2509–2516 [DOI] [PubMed] [Google Scholar]
- 5. Bjerrum L., et al. 2006. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques. Poult. Sci. 85:1151–1164 [DOI] [PubMed] [Google Scholar]
- 6. Bray J. R., Curtis J. T. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325–349 [Google Scholar]
- 7. Brodie E., Edwards S., Clipson N. 2002. Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb. Ecol. 44:260–270 [DOI] [PubMed] [Google Scholar]
- 8. Brown M. V., Schwalbach M. S., Hewson I., Fuhrman J. A. 2005. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ. Microbiol. 7:1466–1479 [DOI] [PubMed] [Google Scholar]
- 9. Chee S. H., Iji P. A., Choct M., Mikkelsen L. L., Kocher A. 2010. Characterisation and response of intestinal microflora and mucins to manno-oligosaccharide and antibiotic supplementation in broiler chickens. Br. Poult. Sci. 51:368–380 [DOI] [PubMed] [Google Scholar]
- 10. Chen X., et al. 2007. The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int. J. Food Microbiol. 115:307–312 [DOI] [PubMed] [Google Scholar]
- 11. Clarke K. R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. Ecol. 18:117–143 [Google Scholar]
- 12. Cornfield J., Tukey J. W. 1956. Average values of mean squares in factorials. Ann. Math. Stat. 27:907–949 [Google Scholar]
- 13. Dibner J. J., Richards J. D. 2005. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 84:634–643 [DOI] [PubMed] [Google Scholar]
- 14. Dibner J. J., Richards J. D., Knight C. D. 2008. Microbial imprinting in gut development and health. J. Appl. Poult. Res. 17:174–188 [Google Scholar]
- 15. Dumonceaux T. J., Hill J. E., Hemmingsen S. M., Van Kessel A. G. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eseceli H., Demir E., Degirmencioglu N., Bilgic M. 2010. The effects of Bio-Mos mannan oligosaccharide and antibiotic growth promoter performance of broilers. J. Anim. Vet. Adv. 9:392–395 [Google Scholar]
- 16a. European Union 18 October 2003. Regulation (EC) no. 1831/2003 of the European Parliament and of the Council of 22 September 2003 on additives for use in animal nutrition. Off. J. Eur. Union L 268/29-L 268/43. http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2003:268:0029:0043:EN:PDF
- 17. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 18. Fisher M. M., Triplett E. W. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geier M. S., Torok V. A., Allison G. E., Ophel-Keller K., Hughes R. J. 2009. Indigestible carbohydrates alter the intestinal microbiota but do not influence the performance of broiler chickens. J. Appl. Microbiol. 106:1540–1548 [DOI] [PubMed] [Google Scholar]
- 20. Hill J. E., et al. 2005. Comparison of ileum microflora of pigs fed corn-, wheat-, or barley-based diets by chaperonin-60 sequencing and quantitative PCR. Appl. Environ. Microbiol. 71:867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hooge D. M. 2004. Meta-analysis of broiler chicken pen trials evaluating dietary mannan oligosaccharides, 1993-2003. Int. J. Poult. Sci. 3:163–174 [Google Scholar]
- 22. Hooge D. M., Sims M. D., Sefton A. E., Connolly A., Spring P. 2003. Effect of dietary mannan oligosaccharide, with or without bacitracin or virginiamycin, on live performance of broiler chickens at relatively high stocking density on new litter. J. Appl. Poult. Res. 12:461–467 [Google Scholar]
- 23. Hugenholtz P., Huber T. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 53:289–293 [DOI] [PubMed] [Google Scholar]
- 24. Jukes T. H., Cantor C. R. 1969. Evolution of protein molecules, p. 21-132. In Munro H. N.(ed.), Mammalian protein metabolism. Academic Press, New York, NY [Google Scholar]
- 25. Kruskal J. 1964. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 29:1–27 [Google Scholar]
- 26. Lan Y., Verstegen M. W. A., Tamminga S., Williams B. A. 2005. The role of the commensal gut microbial community in broiler chickens. Worlds Poult. Sci. J. 61:95–104 [Google Scholar]
- 27. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In Stackebrandt E., Goodfellow M.(ed.), Nucleic acid techniques in bacterial systematics. John Wiley, Chichester, England [Google Scholar]
- 28. Lu J., Domingo J. 2008. Turkey fecal microbial community structure and functional gene diversity revealed by 16S rRNA gene and metagenomic sequences. J. Microbiol. 46:469–477 [DOI] [PubMed] [Google Scholar]
- 29. Lu J., et al. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu J., Santo Domingo J. W., Hill S., Edge T. A. 2009. Microbial diversity and host-specific sequences of Canada goose feces. Appl. Environ. Microbiol. 75:5919–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu J., Santo Domingo J. W., Lamendella R., Edge T., Hill S. 2008. Phylogenetic diversity and molecular detection of bacteria in gull feces. Appl. Environ. Microbiol. 74:3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manning T. S., Gibson G. R. 2004. Prebiotics. Best Pract. Res. Clin. Gastroenterol. 18:287–298 [DOI] [PubMed] [Google Scholar]
- 33. Mateo C. D., Jacques K. A., Harvey J. 2000. Organic chromium, mannan oligosaccharides and zinc bacitracin: effects on broiler performance and carcass characteristics. Poult. Sci. 79(Suppl. 1):116 [Google Scholar]
- 34. Matsui H., et al. 2010. Microbial diversity in ostrich ceca as revealed by 16S rRNA gene clone library and detection of novel Fibrobacter species. Anaerobe 16:83–93 [DOI] [PubMed] [Google Scholar]
- 35. Midilli M., et al. 2008. Effects of dietary probiotic and prebiotic supplementation on growth performance and serum IgG concentration of broilers. S. Afr. J. Anim. Sci. 38:21–27 [Google Scholar]
- 36. Parks C. W., Grimes J. L., Ferkett P. R. 2005. Effects of virginiamycin and a mannanoligosaccharide-virginiamycin shuttle program on the growth and performance of large white female turkeys. Poult. Sci. 84:1967–1973 [DOI] [PubMed] [Google Scholar]
- 37. Patterson J. A., Burkholder K. M. 2003. Application of prebiotics and probiotics in poultry production. Poult. Sci. 82:627–631 [DOI] [PubMed] [Google Scholar]
- 38. Phillips M. L. 2009. Gut reaction: environmental effects on the human microbiota. Environ. Health Perspect. 117:A198–A205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Popa R., et al. 2009. Limitations and benefits of ARISA intra-genomic diversity fingerprinting. J. Microbiol. Methods 78:111–118 [DOI] [PubMed] [Google Scholar]
- 40. Ranjard L., et al. 2001. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl. Environ. Microbiol. 67:4479–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 42. Salzman N. H., et al. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651–3660 [DOI] [PubMed] [Google Scholar]
- 43. Sims M. D., Dawson K. A., Newman K. E., Spring P., Hooge D. M. 2004. Effect of dietary mannan oligosaccharide, bacitracin methylene disalicylate, or both on the live performance and intestinal microbiology of turkeys. Poult. Sci. 83:1148–1154 [DOI] [PubMed] [Google Scholar]
- 44. Singleton D. R., Furlong M. A., Rathbun S. L., Whitman W. B. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sofos J. N. 2008. Challenges to meat safety in the 21st century. Meat Sci. 78:3–13 [DOI] [PubMed] [Google Scholar]
- 46. Spring P., Wenk C., Dawson K. A., Newman K. E. 2000. The effects of dietary mannanoligosaccharides on cecal parameters and the concentrations of enteric bacteria in the ceca of salmonella-challenged broiler chicks. Poult. Sci. 79:205–211 [DOI] [PubMed] [Google Scholar]
- 47. Suzuki M. T., Giovannoni S. J. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tajima K., et al. 1999. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol. Ecol. 29:159–169 [Google Scholar]
- 49. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software, version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 50. Thakuria D., Schmidt O., Liliensiek A.-K., Egan D., Doohan F. M. 2009. Field preservation and DNA extraction methods for intestinal microbial diversity analysis in earthworms. J. Microbiol. Methods 76:226–233 [DOI] [PubMed] [Google Scholar]
- 51. Thakuria D., Schmidt O., Mac Siúrtáin M., Egan D., Doohan F. M. 2008. Importance of DNA quality in comparative soil microbial community structure analyses. Soil Biol. Biochem. 40:1390–1403 [Google Scholar]
- 52. von Wintzingerode F., Göbel U. B., Stackebrandt E. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213–229 [DOI] [PubMed] [Google Scholar]
- 53. Waldroup P. W., Fritts C. A., Yan F. 2003. Utilization of Bio-Mos mannan oligosaccharide and Bioplex copper in broiler diets. Int. J. Poult. Sci. 2:44–52 [Google Scholar]
- 54. Wang M., Ahrné S., Jeppsson B., Molin G. 2005. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 54:219–231 [DOI] [PubMed] [Google Scholar]
- 55. Yang Y., Iji P. A., Kocher A., Mikkelsen L. L., Choct M. 2008. Effects of mannanoligosaccharide and fructooligosaccharide on the response of broilers to pathogenic Escherichia coli challenge. Br. Poult. Sci. 49:550–559 [DOI] [PubMed] [Google Scholar]
- 56. Zdunczyk Z., Juskiewicz J., Jankowski J., Biedrzycka E., Koncicki A. 2005. Metabolic response of the gastrointestinal tract of turkeys to diets with different levels of mannan-oligosaccharide. Poult. Sci. 84:903–909 [DOI] [PubMed] [Google Scholar]
- 57. Zhang G., Ma L., Doyle M. P. 2007. Salmonellae reduction in poultry by competitive exclusion bacteria Lactobacillus salivarius and Streptococcus cristatus. J. Food Prot. 70:874–878 [DOI] [PubMed] [Google Scholar]
- 58. Zhu X. Y., Zhong T., Pandya Y., Joerger R. D. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.