Abstract
The four-plasmid complement of the raw milk cheese isolate Lactococcus lactis subsp. lactis biovar diacetylactis DPC3901 was sequenced, and some genetic features were functionally analyzed. The complete sequences of pVF18 (18,977 bp), pVF21 (21,739 bp), pVF22 (22,166 bp), and pVF50 (53,876 bp) were obtained. Each plasmid contained genes not previously described for Lactococcus, in addition to genes associated with plant-derived lactococcal strains. Most of the novel genes were found on pVF18 and encoded functions typical of bacteria associated with plants, such as activities of plant cell wall modification (orf11 and orf25). In addition, a predicted high-affinity regulated system for the uptake of cobalt was identified (orf19 to orf21 [orf19-21]), which has a single database homolog on a plant-derived Leuconostoc plasmid and whose functionality was demonstrated following curing of pVF18. pVF21 and pVF22 encode additional metal transporters, which, along with orf19-21 of pVF18, could enhance host ability to uptake growth-limiting amounts of biologically essential ions within the soil. In addition, vast regions from pVF50 and pVF21 share significant homology with the plant-derived lactococcal plasmid pGdh442, which is indicative of extensive horizontal gene transfer and recombination between these plasmids and suggests a common plant niche for their hosts. Phenotypes associated with these regions include glutamate dehydrogenase activity and Na+ and K+ transport. The presence of numerous plant-associated markers in L. lactis DPC3901 suggests a plant origin for the raw milk cheese isolate and provides for the first time the genetic basis to support the concept of the plant-milk transition for Lactococcus strains.
INTRODUCTION
Lactococcus lactis strains are essential components of industrial starters for the manufacture of fermented dairy products. Their success in food fermentations is due largely to plasmid-encoded traits promoting efficient growth in milk (i.e., metabolism of lactose, milk proteins, citrate, and other complex nutrients), resistance to environmental stresses (e.g., bacteriophages, heavy metals, and temperature/osmotic stresses), and selective colonization advantages (i.e., production of bacteriocins and exopolysaccharides). Plasmids encoding these phenotypes are often mobilizable, which has greatly contributed to the genome plasticity of lactococci, enabling them to become adapted to and specialized for the dairy environment (25).
However, L. lactis is also naturally present on cows' skin and on plants, with the latter environment being the second most important ecosystem occupied by lactococci (21, 29). Plant strains are thought to be the natural source of dairy strains, since they can transfer from forage plants and meadow grasses to milk via cattle (37). Recent genotypic and phenotypic diversity analyses of large collections of dairy- and plant-derived L. lactis isolates have provided some evidence of such an origin (18) and showed that plant strains mostly belong to the subspecies lactis and exhibit a molecular diversity not observed in dairy strains (33). A subsequent genome-scale analysis confirmed that plant strains exhibit an array of metabolic phenotypes exceeding that of dairy strains and including traits associated with the adaptation to grow on diverse plant cell wall substrates but also genes for defense and stress response (43). Natural isolates often demonstrate greater tolerance of stress as a result of specific protection mechanisms developed to resist the numerous cytotoxic compounds and organisms (i.e., antibiotics, heavy metals, bacteriocins, and bacteriophages) present in the plant and animal environments (34, 42, 49). They also exhibit phenotypes of industrial interest that can be utilized in dairy and other food fermenting applications, such as amino acid conversion to flavor compounds (i.e., α-keto acid decarboxylase and glutamate dehydrogenase) or the production of broad-spectrum bacteriocins (21, 44, 45). Again, many of these phenotypes are often plasmid encoded, which reaffirms the crucial contribution of these extrachromosomal entities to the performance, fitness, and ability of lactococci to colonize specific biotopes.
The analysis of plasmid complement is therefore essential to evaluate the genetic potential and variability existing in dairy and natural lactococcal strains. Using this approach, novel genotypes useful to the dairy industry can be found, but also niche-specific traits determining the uniqueness and adaptive capabilities of the strain can be uncovered. Should the plant-milk transition theory be valid, strains recently transferred to milk and therefore potentially still carrying plant-specific traits could theoretically be retrieved in artisanal products such as raw milk cheeses, where the natural milk microflora, derived from the grass, remains intact (14, 37).
In this study, we have sequenced and functionally analyzed the four-plasmid complement of L. lactis subsp. lactis biovar diacetylactis DPC3901, a raw milk cheese isolate isolated and phenotypically characterized during previous studies aimed at identifying novel strains or genetic traits to be used in programs of starter improvement (6, 11). L. lactis DPC3901 plasmids contain a number of gene functions not previously described for Lactococcus and which are typical of bacteria associated with plants, as well as several other genes displaying homology only to single genes found on the plant-derived lactococcal plasmid pGdh442 or pKF147A (41, 47). Additionally, two L. lactis DPC3901 plasmids exhibited high levels of homology over many loci with pGdh442, which is indicative of extensive horizontal gene transfer (HGT) as well as plasmid recombination and implies that L. lactis DPC3901 may have originated from a plant niche.
MATERIALS AND METHODS
Lactococcal strains and media.
The natural strain L. lactis DPC3901 was isolated and partially characterized in previous studies (6, 11). Lactococcal strains (Table 1) were routinely propagated at 30°C in M17 medium (Oxoid, United Kingdom) supplemented with 0.5% (wt/vol) glucose (GM17) or lactose (LM17). Solid media contained 1.0% (wt/vol) bacteriological agar (Oxoid, United Kingdom). Bacterial strains were stocked in M17 containing 40% glycerol at −80°C. Working cultures were stored at 4°C and transferred periodically.
Table 1.
Bacterial strains used in this study
| Bacterial strain | Relevant characteristic(s) | Sourcea or reference |
|---|---|---|
| L. lactis subsp. lactis biovar diacetylactis DPC3901 | Raw milk cheese isolate; contains four plasmids of 18, 21, 22, and 50 kb | TFRC culture collection |
| L. lactis subsp. lactis biovar diacetylactis DPC3901-c1 | pVF18-cured derivative of L. lactis DPC3901 | This study |
| L. lactis subsp. lactis biovar diacetylactis DPC3901-c2 | pVF22-cured derivative of L. lactis DPC3901-c1 | This study |
| L. lactis subsp. cremoris MG1363 | Plasmid-free derivative of L. lactis subsp. cremoris 712 | 16 |
| L. lactis subsp. lactis biovar diacetylactis DRC3 | DNA size marker strain; contains eight plasmids with sizes from 2 to 78 kb | 24 |
| L. lactis subsp. lactis SK1 | Cheese starter | TFRC culture collection |
| L. lactis subsp. cremoris DPC4268 | Cheese starter | TFRC culture collection |
TFRC, Teagasc Food Research Centre, Moorepark, Fermoy, Cork, Ireland.
Classification of L. lactis DPC3901.
L. lactis DPC3901 was genotyped with the universal 16S rRNA primers CO1 (5′-AGTTTGATCCTGGCTCAG-3′) and CO2 (5′-TACCTTGTTACGACT-3′) and using the PCR conditions described elsewhere (2). Aliquots (5 μl) of the amplified products were subjected to electrophoresis in 1% agarose (Sigma) gels and stained with ethidium bromide. PCR products were purified using the QIAquick PCR purification kit (Qiagen GmbH, Germany) and sequenced by single-run primer extension (Beckman Coulter Genomics). Sequencing data were searched for homology against sequences available in public databases (GenBank, EMBL, DDBJ, and PDB) by using the nucleotide MegaBLAST algorithm of the Basic Local Alignment Search Tool (BLAST) (56). Confirmation of the genotypically predicted citrate-fermenting phenotype was obtained by streaking a freshly grown culture of L. lactis DPC3901 on KMK medium and checking for the appearance of blue colonies after 48 h of incubation at 30°C (9).
Plasmid DNA sequencing, assembly, and annotation.
Plasmid DNA was prepared as described elsewhere (32). Any residual genomic DNA was removed by treatment with Plasmid-Safe ATP-dependent DNase (Epicentre, Madison, WI), which selectively hydrolyzes linear double-stranded DNA to deoxynucleotides without affecting closed circular supercoiled or nicked circular double-stranded DNAs (dsDNAs). The purified plasmids were sequenced as a pool on a 454 Genome Sequencer FLX system after preparation of a sonication library, bar coding of the library with the Roche multiplex identifier system, and emulsion PCR (emPCR) amplification (Beckman Coulter Genomics). Bar coding of the library was necessary since pools of plasmids from different isolates were sequenced simultaneously. Automatic de novo assembly of data resulted in 56 large (>500 bp) contigs providing ca. 135 kb of sequence data (12× average coverage). Gap closure was performed by PCR using primers designed with the PrimerSelect v.8.0.2 software program (DNAStar), followed by sequencing of the PCR products (Beckman Coulter Genomics). Putative open reading frames (ORFs) were automatically predicted and annotated using the Glimmer 2.0 software program (7). Annotated DNA sequences were examined manually using the Artemis viewer (36), and BLASTX and BLASTP analyses (1) were used to examine intergenic regions for putative ORFs not identified by Glimmer, to identify putative frameshifts, and to refine the start codon assignments. The InterProScan software tool (www.ebi.ac.uk/Tools/InterProScan), the PFAM database (13), and the Conserved Domain Database (CDD) (23) were used to predict protein function. InterProScan predicts the occurrence of functional domains and motifs/signatures in a protein by combining 12 different databases and their relative protein signature recognition methods (55). Prediction of topology in putative alpha-helical membrane proteins, including a specification and the in/out orientation of the membrane-spanning segments, was obtained by using the TOPCONS hidden Markov model (http://topcons.cbr.su.se/index.php), which uses five different topology prediction algorithms (SCAMPI-seq, SCAMPI-msa, PRODIV-TMHMM, PRO-TMHMM, and OCTOPUS) to generate a consensus prediction for the protein (3). Putative ribosome binding sites (RBS) and terminator sequences were automatically identified by using the RBSFinder+TransTerm program (http://nbc11.biologie.uni-kl.de/). Manual identification of terminator sequences was achieved by following three main criteria: (i) a ΔG value of less than −6, (ii) a stem-loop structure rich in GC, and (iii) a T-rich sequence following the stem-loop structure.
Growth inhibition assays.
Bacterial sensitivity to cadmium (Cd2+), cobalt (Co2+), tetracycline, streptomycin, and kanamycin (Sigma) was assessed by using a microplate procedure. Two hundred microliters of GM17 or LM17 broth containing either CdCl2 (0 to 0.3 mM), CoCl2 (0 to 6 mM), tetracycline (0 to 10 μg/ml), streptomycin (10 to 500 μg/ml), or kanamycin (10 to 500 μg/ml) was inoculated at 1% with overnight cultures on 96-well microplates (Sarstedt, Newton, NC). Bacterial growth was assessed over 18 h at 30°C by measuring the absorbance at 600 nm with a GENios Plus reader (Tecan, Switzerland).
Determination of glutamate dehydrogenase (GDH) activity.
GDH activity was determined in cell-free extracts (CFEs) as previously described (20), with some modifications. Briefly, cell cultures (200 ml) growing in mid-log phase (optical density [OD] = 0.4) were centrifuged (4,100 × g, 15 min, 4°C) and washed twice with 50 mM triethanolamine buffer (TEA) (pH 7). Pellets were resuspended in 5 ml of TEA containing lysozyme (1.6 mg/ml) and saccharose (0.2 mg/ml), incubated at 30°C for 2 h, centrifuged, and resuspended in hypotonic buffer TEA to provoke spheroplast lysis. Cell debris was eliminated by centrifugation (20,000 × g for 30 min), and the supernatant was filtered (0.45-μm filter; Millipore) and used as CFE. The CFE protein concentration was determined by the Bradford method using bovine serum albumin as the standard (4). Determination of GDH activity in CFEs was based on a modification of a colorimetric glutamic acid assay (Boehringer Ingelheim GmbH, Germany) where the reduced cofactor (NADH or NADPH), produced by GDH-mediated oxidative deamination of glutamate, reacts with iodonitrotetrazolium in the presence of diaphorase to produce a colorimetric product (formazan) that is measured by absorbance at 492 nm. A 300-μl reaction mixture consisted of 40 μl of 50 mM potassium phosphate–50 mM TEA buffer (pH 9.0) containing 1% of Triton X-100 (Promega Corporation), 20 μl of 100 mM glutamic acid, 20 μl of 2 mM iodonitrotetrazolium, 20 μl of diaphorase (1.76 U/ml), 20 μl of NAD+ (17.33 mM) or NADP+ (13.8 mM), and 180 μl of water. The colorimetric reaction was started by adding 30 μl of CFEs to the reaction mixture, and the changes in the NADH concentration after 1 h of incubation at 37°C were determined by measuring the A492. A control test lacking glutamic acid was prepared for each strain in order to subtract for nonspecific reactions producing the reduced cofactors. The GDH activity was expressed by the increase in A492 per mg of protein of CFE and per min of reaction.
Plasmid curing.
Plasmid curing experiments were performed by growing strains at 37°C in LM17 lacking the buffering agent β-glycerophosphate (β-GP) for 50 generations before being transferred to fresh medium. The strain was plated on LM17 agar at each transfer in order to analyze plasmid profiles of individual colonies.
Measurement of bacterial growth rate and acidification capacity.
The growth rates and acidification capacities of L. lactis DPC3901 and of its pVF18/pVF22-cured derivative (L. lactis DPC3901-c2) were compared with those of cheese starter strains L. lactis subsp. cremoris DPC4268 and L. lactis subsp. lactis SK1 as previously described (12). Briefly, growth rate parameters (maximum growth rate, μmax, and generation time, G) were determined after growth in LM17 at 30°C, whereas the acidification capacities (starter activity) were evaluated, based on final pH, after growth in 11% reconstituted skim milk (RSM) over a 5-h incubation time through a simulated cheddar cheese-making temperature profile (32°C for 70 min, 40°C for 190 min, and 32°C for 40 min).
PCR analysis.
Detection of specific genes within the plasmid complement of L. lactis DPC3901 and its cured derivatives, DPC3901-c1 and DPC3901-c2, was obtained by PCR analysis using the primers listed in Table 2.
Table 2.
Primers used in this study to map gene features of L. lactis DPC3901 plasmids
| Primera | Sequence (5′–3′) | Product size (bp) | Target | Plasmid |
|---|---|---|---|---|
| orfs18-21 (F) | GAAAACATCAAGAAGAACGCAGAGT | 1,858 | orf18-21 | pVF18 |
| orfs18-21 (R) | TGGCAATCGAAGCATCAAGTAGAC | |||
| gdh (F) | GTGGGTCGACGATGCTGGTAAAGTT | 822 | gdh | pVF21 |
| gdh (R) | TGGACGCCGCTGGAAATCAATA | |||
| corA (F) | TTTATGTGTATGGTGACGAACGAGAAT | 812 | corA | pVF22 |
| corA (R) | CAGCACAGCAAAAACACCATAAGAAGAT | |||
| ldh (F) | GTGCTGCCAAATATAGATGCCCAACA | 484 | ldh | pVF50 |
| ldh (R) | ATGCCAAAGCGGTTAGATGATTATTACGA |
F, forward primer; R, reverse primer. Columns indicate the primer's name and sequence, expected size of the PCR product, target gene(s), and its plasmid location.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of plasmids pVF18, pVF21, pVF22, and pVF50 have been submitted to the EMBL/GenBank database and are available under accession numbers JN172910, JN172911, JN172912, and JN225497, respectively.
RESULTS
Plasmid DNA sequencing.
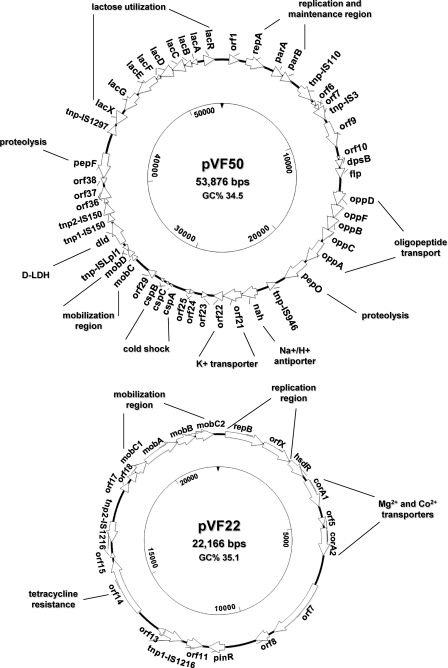
Sequencing of the 16S rRNA gene revealed a subsp. lactis genotype for L. lactis DPC3901, which was also classified in the biovar diacetylactis due to its ability to ferment citrate during growth on KMK medium (data not shown). L. lactis DPC3901 was previously found to harbor four plasmids greater than 10 kb in size and to be completely resistant to a number of 936- and c2-type phages (11). Consequently, the entire plasmid complement of L. lactis DPC3901 was sequenced to investigate its contribution to the technological capacities of the strain. By using a PCR-based gap closure strategy, the complete circular sequences of plasmids pVF50 (53,876 bp), pVF22 (22,166 bp), pVF21 (21,728 bp), and pVF18 (18,977 bp) were obtained (Fig. 1 ; see also Tables S1 to S4 in the supplemental material). The GC content of these plasmids ranged from 33.6 to 35.1%, which is similar to that of other lactococcal plasmids (42). Sequence analysis suggests that all plasmids could replicate according to a theta mechanism, since each one carries an origin of replication (ori), located upstream of the rep gene encoding the replication initiator protein, containing highly conserved structural motifs typical of theta replicons (data not shown) (39). The most significant genetic features of L. lactis DPC3901 plasmids are discussed below.
Fig. 1.
Physical and genetic maps of plasmids pVF50, pVF22, pVF21, and pVF18 of L. lactis DPC3901. Position and orientation of genes are indicated by arrows. Inner circles show the nucleotide numbering.
Genes from pVF18, pVF21, and pVF50 suggest a plant-based origin for L. lactis DPC3901.
pVF18 contains 25 genes encoding predicted proteins with no obvious dairy-associated functions, eight of which (orf8, orf11, orf18 to orf21, orf23, and orf25) have been rarely if ever described for Lactococcus. Some of these genes encode products whose functions are irrelevant to bacteria growing in milk but important to those associated with or striving to become established within the natural plant ecosystem. An example is orf11, which encodes a putative polysaccharide deacetylase (432 amino acids [aa]) belonging to a family of conserved proteins (PFAM01522) involved in hydrolysis of the plant cell wall. Members of this family include the chitooligosaccharide deacetylase NodB from Rhizobium (15), chitin deacetylases from Saccharomyces cerevisiae (27), and microbial endoxylanases (26). These last enzymes share sequence similarity with the NodB deacetylase (26), whose activity enables Rhizobium to associate symbiotically with leguminous plants, thus allowing them access to mineral nitrogen necessary for their growth. The nodB gene is plasmid encoded in Rhizobium sp. NGR234, and this was suggested to be the result of a recent HGT from Agrobacterium (15). Orf11-like proteins are highly conserved in Enterococcus faecium, Staphylococcus aureus, and some Ruminococcus and Clostridium members, but no homologues have been found to date in Lactococcus. Similarly, homologues of the product of orf25 can be found in various environmental subspecies of Streptococcus, Carnobacterium, Enterococcus, Corynebacterium, Bacillus, Geobacillus, and Clostridium but not in Lactococcus or other milk-derived bacteria. Orf25 is a 140-aa protein containing the conserved barrel domain typical of cupins (PFAM07883). The functions of these proteins range from isomerase and epimerase activities involved in the modification of cell wall carbohydrates in bacteria and plants to nonenzymatic germins and plant seed storage proteins, which provide the major nitrogen source for the developing plant (10).
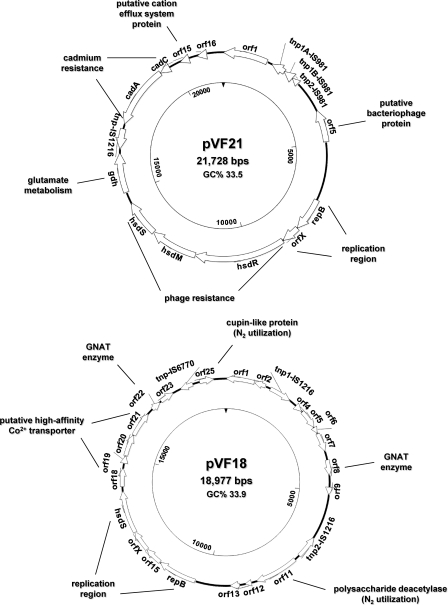
Traces of a putative plant origin can also be linked to pVF50 and pVF21. pVF50 is largely similar to the plant-derived lactococcal plasmid pGdh442 (47), with which it shares five gene modules spanning more than 30 kb of sequence and exhibiting 97 to 100% homology at both the DNA and protein levels. One of these shared gene blocks was so far exclusive to pGdh442 and is composed of three genes encoding membrane components of Na+ and K+ transport systems, which are virtually identical in pVF50 (nah, orf21, and orf22) and in pGdh442 (orf15 to orf17) but share only 41 to 45% identity to other homologues found in Clostridium (47). Also, pVF21 carries a region 5.6 kb long and containing the genes gdh, tnp-IS1216, cadA, and cadC, which was so far unique to pGdh442 (47). In particular, the glutamate dehydrogenase (GDH) activity encoded by the gdh gene is rare in L. lactis and so far has been identified only in strains of plant and animal origin (47, 48). Additionally, pVF21 carries a gene (orf15) predicted to encode a 105-aa hypothetical protein identical to its only database match, encoded by the plasmid pKF147A, isolated from an L. lactis plant strain (41). Finally, pVF21 encodes a hypothetical protein (Orf5) of 365 aa with no homologues in Lactococcus or other lactic acid bacteria (LAB) and with the most similar matches (27 to 32%) being proteins from two Bacillus cereus strains. Orf5 contains the conserved motifs (DGQHR, QR, and FxxxN) typical of the highly divergent DGQHR domain (TIGR03187), and although the function of proteins containing this domain is unknown, several members of this family have been annotated as putative bacteriophage proteins.
Functional Co2+ transport mechanisms in L. lactis DPC3901 plasmids.
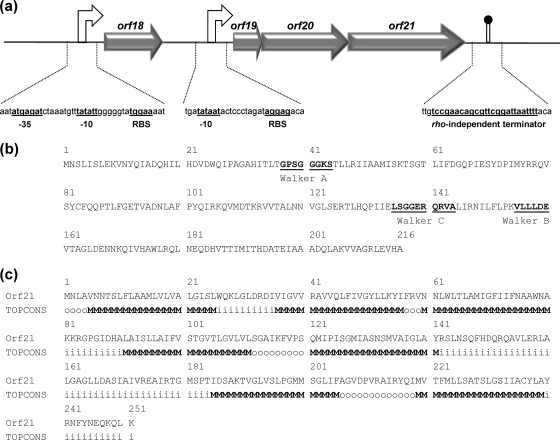
Cobalt (Co2+) is an essential component of many enzymes and must be transported into cells in appropriate amounts when needed (34). In L. lactis DPC3901, pVF22 carries two copies (corA1 and corA2) of the corA gene (COG0598), which encodes the principal transporter of Mg2+ in prokaryotes but also functions in the uptake of Co2+ (28). Additionally, pVF18 carries a putative polycistronic operon (orf19 to orf21 [orf19-21]) that may play a role in the high-affinity and regulated uptake of Co2+. Two 4-bp overlaps between orf20 and its neighbor genes, coupled with the presence of putative promoter sequences upstream of orf19 and of a strong rho-independent terminator (ΔG = −7.9 kcal/mol) downstream of orf21 (Fig. 2 a), suggest that orf19, orf20, and orf21 are transcribed as a single mRNA. This three-gene operon is uncommon in L. lactis and so far has been found only on the recently sequenced plasmid LkipL4726 from Leuconostoc kimchii IMSNU 11154 (30). orf19 encodes a small protein (75 aa) of unknown function exhibiting 92% identity to its only significant match, a hypothetical protein from LkipL4726 (30). orf20 and orf21 encode proteins containing features typical of the ATPase and permease components of ATP-binding cassette (ABC) transporters (Fig. 2b and c). Interestingly, homologs of Orf20 are widespread in Lactococcus, whereas Orf21 has a single lactococcal match on pGdh442 (47). A putative involvement in Co2+ uptake for the ABC transporter encoded by orf20-21 is suggested by the presence in Orf20 of a conserved domain (cd03225) found in the CbiO component of CbiMNQO, which is the most widespread transport system for the high-affinity uptake of Co2+ in prokaryotic genomes (34).
Fig. 2.
Sequence analysis of the putative polycistronic operon orf18-21. (a) Putative regulatory elements of the orf18-21 operon. The ribosome binding site (RBS), −10 and −35 promoter sequences, and rho-independent terminator signal are shown in bold and underlined. (b) Locations of the conserved domains typical of ABC transporter ATPases in Orf20: the Walker A (consensus GxxGxGKS/T, where x is any residue) and Walker B (consensus hhhhDE, where h is a hydrophobic residue) motifs involved in ATP binding and the Walker C (consensus LSGGQQ/R/KQR) motif involved in ATP hydrolysis are shown underlined. (c) TOPCONS consensus prediction of membrane topology for Orf21, showing seven transmembrane motifs (i, inside; M, transmembrane; o, outside) typical of ABC transporter permeases.
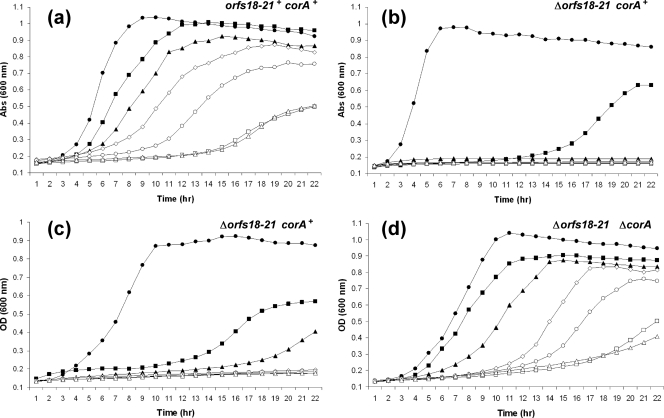
To verify the functionality of these mechanisms, the growth of L. lactis DPC3901 in the presence of increasing concentrations of Co2+ was compared to that of L. lactis MG1363, which lacks any high-affinity Co2+ uptake system but possesses a chromosomally encoded CorA transporter. L. lactis DPC3901 can grow relatively well in the presence of 4 mM Co2+ and tolerate concentrations as high as 6 mM (Fig. 3 a). In contrast, L. lactis MG1363 growth is affected by the presence of 1 mM Co2+ and is totally inhibited by concentrations of and above 2 mM (Fig. 3b). Despite its importance for bacterial metabolism, Co2+ uptake must be tightly regulated to avoid toxic effects (34). Results from the growth curves are indicative of the ability of L. lactis DPC3901 to maintain the intracellular Co2+ to levels that are compatible with cell life. To further confirm these indications, pVF18 and pVF22 were sequentially cured from L. lactis DPC3901 in order to first remove orf19-21 and then the corA genes. Growth curve analysis showed that the pVF18-cured derivative, named DPC3901-c1 (Δorf19-21 corA+), could not grow at Co2+ concentrations higher than 1 mM (Fig. 3c), whereas the derivative cured of both pVF18 and pVF22, DPC3901-c2 (Δorf19-21 ΔcorA), tolerated Co2+ in a fashion similar to that of the parent strain (Fig. 3d). These results confirm the involvement of the orf19-21 genes in mediating uptake and homeostasis of the potentially toxic metal Co2+ in L. lactis DPC3901.
Fig. 3.
Growth curves of L. lactis DPC3901 (a), MG1363 (b), DPC3901-c1 (c), and DPC3901-c2 (d) in medium containing 0 (•), 1 (▪), 2 (▴), 3 (⋄), 4 (○), 5 (□), or 6 mM (▵) cobalt chloride. The minus or plus superscript is indicative of the absence or presence, respectively, of the orf18-21 and corA genotypes in each strain or derivative. Abs, absorbance.
Antibiotic resistance in L. lactis DPC3901.
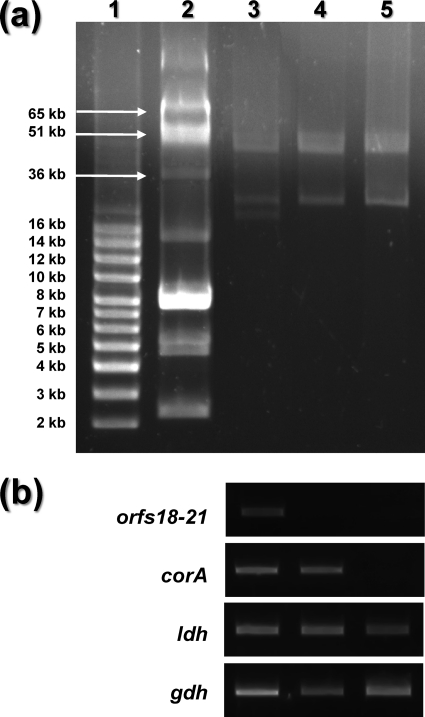
pVF22 carries a gene (orf14) encoding a TetS-like tetracycline resistance protein (cd04168) identical to those found on pKL0018 from the fish pathogen L. garvieae (22) and on pK214 from an L. lactis isolate from raw milk soft cheese (49). This protein functions through ribosomal protection, and its gene is typically found on mobile genetic elements, such as transposons or plasmids. In pVF22, orf14 is inserted within a genetic context resembling a transposon-like structure since it is flanked by two IS1216 transposases and by genes responsible for site-specific recombination (pinR and orf17) and for conjugal transfer (mobC1ABC2). The presence of this tet(S)-like gene on a conjugative plasmid would pose concerns regarding the use of L. lactis DPC3901 in foods for human consumption due to the potential spread of antibiotic resistance genes from food-borne LAB to pathogenic organisms (49). However, antibiotic resistance genes have been removed from probiotic strains by using methods that did not genetically modify the organism and retained the original probiotic characteristics (35). We took a similar approach and cured pVF22 from the pVF18-cured derivative of L. lactis DPC3901 (L. lactis DPC3901-c1) (Fig. 4 a). The strain lost pVF22 after ca. 100 generations but retained pVF50 and pVF21, as also confirmed by PCR (Fig. 4b). Compared to the parent strain, the new derivative (DPC3901-c2) exhibited unaltered acidification capacities and slightly better growth rates (Table 3), probably as a result of the loss of two metabolically burdening plasmids.
Fig. 4.
(a) Plasmid DNA profiles of L. lactis DPC3901 (lane 3) and its cured derivatives DPC3901-c1 (pVF18−; lane 4) and DPC3901-c2 (pVF18− pVF22−; lane 5). The plasmid profile of L. lactis DRC3 (lane 2) provides reference plasmids of large sizes (36, 51, and 65 kb) as a complement to the 2- to 16-kb molecular size ladder (lane 1). (b) PCR-based detection of specific gene features in the plasmid complement of L. lactis DPC3901, DPC3901-c1, and DPC3901-c2.
Table 3.
Comparison of growth rate parameters and acidification capacities of L. lactis DPC3901 and its pVF18/pVF22-cured derivative (DPC3901-c2) with those of selected lactococcal cheese startersa
| L. lactis strain | Growth rate |
Starter activity (pH) |
||
|---|---|---|---|---|
| μmaxb (h−1) | Generation time (h−1) | 11% RSM | GM17 | |
| DPC3901 | 0.90 (± 0.08) | 1.17 (± 0.10) | 5.37 (± 0.18) | 6.06 (± 0.04) |
| DPC3901-c2 | 0.99 (± 0.10) | 1.09 (± 0.12) | 5.25 (± 0.01) | 5.90 (± 0.05) |
| SK1 | 1.18 (± 0.01) | 0.85 (± 0.01) | 4.91 (± 0.24) | 5.67 (± 0.03) |
| DPC4268 | 1.10 (± 0.13) | 0.92 (± 0.11) | 4.78 (± 0.13) | 5.41 (± 0.01) |
Values are means ± SD.
μmax, maximum growth rate.
Putative determinants for antibiotic resistance were also found on pVF18, which contains two genes (orf8 and orf23) encoding putative GCN5-related N-acetyltransferases (GNAT) (cd04301). These enzymes catalyze the transfer of an acetyl group to the cognate substrate and are thought to be implicated in a variety of functions, ranging from antibiotic (aminoglycosides) resistance to regulation of cell growth and development (54). To test their possible involvement in antibiotic resistance, L. lactis DPC3901 and the sensitive strain L. lactis MG1363 were grown in the presence of increasing concentrations of streptomycin and kanamycin (data not shown). Both strains were similarly sensitive to both aminoglycosides, which suggests that the GNAT-like proteins encoded by pVF18 are not involved in resistance to these antibiotics. Interestingly, these proteins have rarely been found in Lactococcus but are conserved in Streptococcus, Clostridium, Enterococcus, and Staphylococcus.
pVF21 confers potential flavor-forming and phage resistance capability to DPC3901.
pVF21 carries a gdh gene encoding a 448-aa protein with homology to the NAD(P)-dependent glutamate dehydrogenase (GDH) enzyme of pGdh442 (46). GDH enzymes catalyze the reversible oxidative deamination of glutamate to α-ketoglutarate and ammonium by utilizing NAD, NADP, or both as a cofactor. Thus, we tested the functionality of the gdh gene of pVF21 by using L. lactis IL1403 as a GDH-negative reference strain. No GDH activity was registered for L. lactis IL1403, whereas L. lactis DPC3901 exhibited NAD+- and NADP+-dependent specific activities of 0.028 and 0.01, respectively, which are similar to those reported for an L. lactis subsp. cremoris strain used in the manufacture of Gouda and cheddar-type cheeses (20). These results suggest that pVF21 encodes a functional GDH enzyme providing L. lactis DPC3901 with the potential to produce aroma compounds from glutamate catabolism and/or to synthesize glutamate from α-ketoglutarate and ammonia.
pVF21 also carries three genes (hsdRMS) encoding a restriction-modification (R/M) system of type I, which is one of the many molecular mechanisms developed by L. lactis strains to defend themselves against bacteriophage (phage) attack (25). The hsdR gene encodes a restriction enzyme (1,025 aa) containing conserved helicases-related domains (cd00046; COG0610), while the 537-aa product of hsdM is a typical N6-adenine methyltransferase (pfam02384). Both proteins are virtually identical (99%) to the HsdR and HsdM enzymes encoded by genes on the lactococcal plasmid pAH82 and exhibiting low restriction efficiency (2 logs) against the small isometric-headed phage 712 (31). In contrast, the HsdS specificity protein (414 aa) of pVF21 shares only 38% identity with the homolog encoded by pAH82. Low similarities among HsdS subunits are common and reflect the large variability existing within their target recognition domains (TRDs), which impart target sequence specificity to both restriction and modification activities (25).
pVF50 contributes to adaptation of L. lactis DPC3901 to the dairy environment.
pVF50 shares over 30 kb of nearly identical sequence with pGdh442, which in the latter plasmid include the adjacent modules orf55-64 (orf55 to orf64) and orf1-3 (orf1 to orf3) and three flanking modules spanning orf15 to orf23 (47). pGdh442 contains 20 IS elements that have probably mediated the acquisition of its numerous gene modules (47) and can therefore promote their transfer to plasmids of a different organism. Adjacent modules in pVF50 exhibit a swapped organization compared to their arrangement in pGdh442, thus supporting the hypothesis that these gene blocks can be mobilized by the flanking IS elements (47).
Genes on pVF50 related to growth on milk components have certainly facilitated the adaptation of L. lactis DPC3901 to the dairy environment. The strain's ability to grow well in milk and to utilize lactose is afforded by the lactose phosphotransferase operon (lacXGEFDCBA), which is negatively regulated by the product of the divergently transcribed lacR gene (52, 53). The lacXGEFDCBAR region of pVF50 is 98 to 100% identical to that present on pSK11L at both the DNA and protein levels (42). A further contribution to lactose utilization in L. lactis DPC3901 might also be afforded by the dld gene (contig2.gene14), which is predicted to encode a d-lactate dehydrogenase (d-LDH) of 559 aa and might function in d-lactate utilization under aerobic conditions (42). pVF50 also encodes the Opp system for the uptake of oligopeptides generated from casein degradation by the cell wall proteinase (51) and the oligoendopeptidases PepO and PepF, which are able to hydrolyze oligopeptides of 5 to 35 residues (5). These genotypes should endow L. lactis DPC3901 with the ability to optimally utilize milk caseins as a nitrogen source. The opp and pepO genes are part of a region, containing the flp operon and flanked by transposases of the IS3 and IS6 families, that is 98% identical to the homologous region on pGdh442 (orf55-64) (47). The presence of these determinants led us to evaluate the strain's ability to grow and acidify milk under conditions resembling the manufacture of cheddar cheese. Growth rates and acidification capacity of L. lactis DPC3901 were slightly lower than those of the widely used cheese starters L. lactis DPC4268 and L. lactis SK1 but still sufficient for cheese manufacturing (Table 3).
pVF50 is the only L. lactis DPC3901 plasmid to contain a replication and maintenance region composed of a typical lactococcal theta-type ori site (39), a repA gene, and two partition genes (parA and parB) that are likely to provide pVF50 with a maintenance system counteracting plasmid loss at cell division. This region is identical to homologous regions present on the plant-derived lactococcal plasmids pKF147A (41) and pGdh442 (47) and on pSK08 from L. lactis ML3 (accession no. AF300944).
DISCUSSION
It is thought that plant-associated strains are the original source of many dairy strains since they can transfer from forage plants and meadow grasses to milk via cattle (37). Recently, some evidence of such an origin was suggested based on an analysis of chromosome length that grouped five L. lactis subsp. cremoris strains, including L. lactis MG1363, with plant strains, such as L. lactis KF147, and on the observation that remnants of genes for degradation and metabolism of plant-derived carbohydrates can also be found in the L. lactis MG1363 genome (18). The authors observed a reductive evolution in dairy strain genomes compared to those of the plant isolates, which they interpreted as part of the adaptation to growth in milk. This process involves the acquisition of genes that are mostly plasmid encoded.
Plasmids are capable of mediating extensive HGT and recombination, and this enables strains to acquire traits conferring a selective advantage to colonize specific biotopes (25). Some of these traits can be so specific to a particular niche as to be reasonably considered equivalent to traceability markers, thus enabling the plasmid complement to be used as a sort of bar-coding system of the organisms' origin. Retrieval of strains containing such traceable traits is feasible within raw milk cheeses, where the absence of pasteurization allows the transfer of plant-derived bacteria to cheese via the cow-milk network. In this study, the four-plasmid complement of the raw milk cheese isolate L. lactis subsp. lactis biovar diacetylactis DPC3901 was found to encode a number of gene functions not previously described for Lactococcus and generally typical of bacteria associated with the plant ecosystem (Table 4). These include activities of modification of the plant cell wall, a functional system for the high-affinity and regulated uptake of Co2+, and several other transporters with a confirmed or predicted role in the uptake of a variety of metal ions, which have single lactococcal homologues on plant-derived plasmids. These genotypes are irrelevant to bacteria growing in milk but important to those associated with or striving to become established within the natural plant ecosystem. Therefore, their presence in a dairy strain may be inferred to be a genetic trace of the previous niche inhabited by the organism, where they conferred on the host a real colonization advantage. The plant origin hypothesis is supported by a number of other observations. First, most of these plant-based origin gene markers are found on pVF18, which lacks any trait favoring colonization of the dairy environment and, interestingly, was the first plasmid to be cured from the host. Additionally, pVF50 and pVF21 share vast and highly homologous regions with the plant-derived lactococcal plasmid pGdh442 (47), which also encodes phenotypes (i.e., glutamate dehydrogenase and Na+ and K+ transporters) so far unique to pGdh442 in L. lactis (47, 48). This is indicative of HGT or recombination occurring between pVF50/pVF21 and pGdh442, which again suggests that L. lactis DPC3901 could have originally occupied a grass plant niche before transferring to the dairy environment. Also, the GDH enzyme of pGdh442 was shown to function mainly in glutamate biosynthesis, and this capacity has been suggested to be important for bacteria living on plants, since it allows more efficient elimination of the high levels of ammonia present in this environment (46). Therefore, the presence of this genotype further supports a plant origin for L. lactis DPC3901.
Table 4.
Novel genes not found before in L. lactis and rare genes with homologues on plant-associated lactococcal strains
| Plasmid | Gene(s) | Product(s) | Predicted function/role(s) | Best homolog found in: | % identity |
|---|---|---|---|---|---|
| Novel in L. lactis | |||||
| pVF18 | orf11 | Polysaccharide deacetylase | Hydrolysis of cell wall carbohydrates in plants; utilization of mineral nitrogen | Enterococcus faecium PC4.1 | 99 |
| pVF18 | orf19-orf20-orf21 | Hypothetical protein; ABC transporter, ATPase and permease components | Putative high-affinity Co2+ transporter; heavy metal homeostasis | Leuconostoc kimchii IMSNU 11154, LkipL4726 | 92–99 |
| pVF18 | orf25 | Cupin domain containing isomerase/epimerase or germin/plant storage protein | Modification of cell wall carbohydrates in plants; utilization of mineral nitrogen | Streptococcus uberis 0140J | 92 |
| pVF21 | orf1 | Primosomal protein N′ | DNA replication factor | Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 | 37 |
| pVF21 | orf5 | DGQHR domain-containing hypothetical protein | Putative bacteriophage protein | Bacillus cereus G9842 | 32 |
| pVF50 | tnp-IS110 | Transposase, IS110 family | DNA integration/recombination | Streptococcus pneumoniae SP19-BS75 | 75 |
| Rare in L. lactis (single hit) | |||||
| pVF18 | orf8, orf23 | GCN5-related N-acetyltransferase | Regulation of cell growth and development | Enterococcus faecium; Weissella paramesenteroides ATCC 33313 | 98; 84 |
| pVF18 | orf24 | Transposase, IS30 family | DNA integration/recombination | Enterococcus faecalis TUSoD Ef11 | 100 |
| pVF21 | gdh | Glutamate dehydrogenase | Reversible oxidative deamination of glutamate | Lactococcus lactis, pGdh442 | 99 |
| pVF21 | orf15 | Hypothetical protein LLKF_p0013 | Putative cation efflux system protein | Lactococcus lactis KF147, pKF147A | 99 |
| pVF22 | orf9 | Hypothetical protein | Putative glycopeptide antibiotic resistance protein | Streptococcus salivarius | 98 |
| pVF22 | orf13 | Hypothetical protein | Unknown | Lactococcus garvieae, pLK0018 | 99 |
| pVF50 | nah | Na+/H+ antiporter | Exchange of sodium/hydrogen ions | Lactococcus lactis, pGdh442 | 100 |
| pVF50 | orf21 | Putative K+ transporter | Uptake of potassium | Lactococcus lactis, pGdh442 | 99 |
| pVF50 | orf22 | Putative K+ transport system TrkA | Uptake of potassium | Lactococcus lactis, pGdh442 | 99 |
Computational and functional analyses showed that L. lactis DPC3901 carries a functional pVF18-encoded system for the regulated uptake of Co2+ in addition to a corA gene duplication on pVF22. Two CorA transporters are uncommon in L. lactis and are likely to enhance the strain's ability to take up Co2+ in environments (e.g., the soil) where its presence in trace amounts might be growth limiting for the strain (34). However, this also increases the strain's sensitivity to high concentrations of environmental Co2+. Our analyses showed that the presence of pVF18 and of its Orf19-21 transport system endows L. lactis DPC3901 with the ability to maintain Co2+ homeostasis by probably allowing a regulated efflux outside the cell. Sequence analysis suggests that Orf20 and Orf21 form an ABC transporter and that Orf20 contains a domain also found on CbiO from the high-affinity and widespread Co2+ transport system in prokaryotes, CbiMNQO (34). The integral membrane protein CbiM and the cobalt-binding periplasmic protein CbiN have been shown to be the minimal requirements for a functional CbiMNQO system (34). Thus, Orf20 could be the counterpart of the ATPase-like protein CbiO, whereas Orf21 functionality could resemble that of CbiM, which also contains seven transmembrane helices. Orf19 is probably a cytoplasmic protein but lacks any conserved Co2+ binding motifs (50), which discards the option that it might be a CbiN homolog. CbiMNQO transporters are generally controlled on the level of translation initiation by B12 riboswitch elements (34). In pVF18, expression of the orf19-21 operon could be controlled by orf18, which encodes a transcriptional regulator belonging to the XRE family (cd00093). Putative promoter sequences but no termination signal could be identified for orf18 (Fig. 3a), suggesting that it could be cotranscribed with orf19-21 and possibly regulate the activity of this operon, thus endowing L. lactis DPC3901 with the ability to maintain Co2+ homeostasis. To date, the orf19-21 operon has been found only on plasmids from Leuconostoc kimchii IMSNU 11154 (30) and L. lactis DPC3901 (this study). Interestingly, a recent and comprehensive comparative and functional analysis of Co2+ transporters in prokaryotic genomes retrieved no homologues of cbi genes in both Leuconostoc and Lactococcus (34). Thus, bacteria from these genera could have recently evolved a novel plasmid-encoded system dedicated to Co2+ uptake and homeostasis. Leuconostoc kimchii IMSNU 11154 was isolated from a traditional Korean fermented food made from cabbage (30), whereas L. lactis DPC3901 derives from a raw milk cheese, where the absence of pasteurization preserves the natural milk microflora, mostly derived from grass plants (37). LkipL4726 and pVF18 hosts could therefore have inhabited natural environments, where perhaps trace amounts of Co2+ have promoted the development of an appropriate high-affinity system. The observation that lactococcal homologs of the Orf20-21 transporter can be found only on the plant-derived pGdh442 (47) further supports the link between L. lactis DPC3901 and the plant environment.
Assuming that L. lactis DPC3901 originated from a plant niche, its transfer to the dairy niche probably occurred some time ago, since the strain carries an entire plasmid (pVF50) specialized for growing and becoming established in milk. The lac operon, as well as the Opp system and the oligoendopeptidases PepF and PepO, should endow L. lactis DPC3901 with the ability to optimally utilize lactose and milk caseins, respectively, thus resulting in a fast milk coagulation phenotype (51–53). This was confirmed, since L. lactis DPC3901 performed similarly to other starters under simulated cheddar cheese-making conditions (Table 3). pVF50 also carries a ParAB maintenance system that, in L. lactis, has so far been found only on pSK08, pCI2000, pGdh442, and pKF147A (17, 41, 47). These plasmids share with pVF50 a large size (over 50 kb) and a low copy number, which make them more at risk of being lost during cell division. The maintenance system presumably increases their stability within the host by ensuring their faithful segregation at each cell division. In the case of pVF50, this would allow the stable maintenance of lactose and peptide utilization abilities that are essential to L. lactis DPC3901 for a successful adaptation to the dairy environment. L. lactis DPC3901 is also well prepared to cope with attacks by lytic phages, which is one of the major threats for lactococci in milk and a major concern for the dairy industry since it affects strains' ability to perform efficiently in large-scale fermentations (25, 31). L. lactis DPC3901 was previously shown to be completely resistant to a number of 936- and c2-type phages (11), and the identification of a type I R/M system on pVF21 (this study) provides the genetic basis for this resistance. Type I R/M systems are particularly effective defense mechanisms, since the result is phage inactivation without compromising cell viability and also because novel specificities can be generated by recombinational shuffling of TRDs between different HsdS subunits, thus enabling the host to restrict a wider range of phages and to cope with the evolution of phage variants (31, 38). L. lactis DPC3901 also has the potential to produce aroma compounds from glutamate catabolism via a functional GDH enzyme encoded by pVF21. Indeed, this enzyme is identical to that encoded by pGdh442, which was shown to be capable of stimulating glutamate conversion to aroma compounds in L. lactis strains that had acquired the plasmid by conjugation (45). Finally, it is noteworthy that although L. lactis DPC3901 is capable of fermenting citrate, no citrate permease gene was found in its plasmid complement as has generally been reported for diacetylactis strains (40). It can be hypothesized that this genotype is chromosomally encoded in this strain, alongside the citrate lyase and other genes involved in citrate breakdown (19).
The capacities of phage resistance coupled with the starter and flavor-forming potential identified on the plasmid complement of L. lactis DPC3901 render this strain an all-round candidate for the preparation of starter formulations with enhanced robustness and novel technological attributes. However, the presence of tet(S) on the mobilizable pVF22 poses concerns regarding the use of this strain in foods for human consumption due to the potential transferability of antibiotic resistance genes from food-borne LAB to pathogenic organisms in the gut (49). Genes conferring resistance to tetracycline, erythromycin, and vancomycin are being detected more frequently in lactococci, enterococci, and lactobacilli isolated from fermented meat and milk products, probably as a consequence of improper veterinary practices (8, 14, 49). We have thus created an antibiotic resistance gene-free derivative of L. lactis DPC3901 via plasmid curing, which retains all the important technological properties of the parent strain and can be safely employed to generate novel starter formulations with improved protection against phage attack and the ability to diversify and widen the flavor profile of the final product.
To conclude, we have found plasmid-encoded markers that potentially trace L. lactis subsp. lactis biovar diacetylactis DPC3901 back to a plant origin and provide for the first time the genetic basis to support the concept of the plant-milk transition for L. lactis strains.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by Irish Dairy Levy. V.F. received a grant from the Irish Research Council for Science, Engineering and Technology (IRCSET) under the Embark initiative.
We thank C. Guinane for assistance with submission of DNA sequences.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Altschul S., et al. 1998. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. FASEB J. 12:A1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beresford T., Condon S. 1991. Cloning and partial characterization of genes for ribosomal ribonucleic-acid in Lactococcus lactis subsp. lactis. FEMS Microbiol. Lett. 78:319–324 [DOI] [PubMed] [Google Scholar]
- 3. Bernsel A., Viklund H., Hennerdal A., Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37:W465–W468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 5. Christensen J. E., Dudley E. G., Pederson J. A., Steele J. L. 1999. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 76:217–246 [PubMed] [Google Scholar]
- 6. Cogan T. M., et al. 1997. Characterization of the lactic acid bacteria in artisanal dairy products. J. Dairy Res. 64:409–421 [Google Scholar]
- 7. Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delgado S., Mayo B. 2004. Phenotypic and genetic diversity of Lactococcus lactis and Enterococcus spp. strains isolated from Northern Spain starter-free farmhouse cheeses. Int. J. Food Microbiol. 90:309–319 [DOI] [PubMed] [Google Scholar]
- 9. Drici H., Gilbert C., Kihal M., Atlan D. 2010. Atypical citrate-fermenting Lactococcus lactis strains isolated from dromedary's milk. J. Appl. Microbiol. 108:647–657 [DOI] [PubMed] [Google Scholar]
- 10. Dunwell J. M. 1998. Cupins: a new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol. Genet. Eng. Rev. 15:1–32 [DOI] [PubMed] [Google Scholar]
- 11. Fallico V. 2011. Plasmid biology of natural Lactococcus lactis strains and molecular mechanisms of bacteriophage-host interaction. Ph.D. thesis. University College Cork, Cork, Ireland [Google Scholar]
- 12. Fallico V., McAuliffe O., Fitzgerald G. F., Hill C., Ross R. P. 2009. The presence of pMRC01 promotes greater cell permeability and autolysis in lactococcal starter cultures. Int. J. Food Microbiol. 133:217–224 [DOI] [PubMed] [Google Scholar]
- 13. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Florez A. B., Ammor M. S., Mayo B. 2008. Identification of tet(M) in two Lactococcus lactis strains isolated from a Spanish traditional starter-free cheese made of raw milk and conjugative transfer of tetracycline resistance to lactococci and enterococci. Int. J. Food Microbiol. 121:189–194 [DOI] [PubMed] [Google Scholar]
- 15. Freiberg C., et al. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394–401 [DOI] [PubMed] [Google Scholar]
- 16. Gasson M. J. 1983. Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kearney K., Fitzgerald G. F., Seegers J. F. M. L. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelly W. J., Ward L. J. H., Leahy S. C. 2010. Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. Evol. 2:729–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kempler G. M., Mckay L. L. 1981. Biochemistry and genetics of citrate utilization in Streptococcus lactis ssp. diacetylactis. J. Dairy Sci. 64:1527–1539 [Google Scholar]
- 20. Kieronczyk A., Skeie S., Langsrud T., Yvon M. 2003. Cooperation between Lactococcus lactis and nonstarter lactobacilli in the formation of cheese aroma from amino acids. Appl. Environ. Microbiol. 69:734–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klijn N., Weerkamp A. H., Devos W. M. 1995. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl. Environ. Microbiol. 61:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maki T., Hirono I., Kondo H., Aoki T. 2008. Drug resistance mechanism of the fish-pathogenic bacterium Lactococcus garvieae. J. Fish Dis. 31:461–468 [DOI] [PubMed] [Google Scholar]
- 23. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKay L. L., Baldwin K. A. 1984. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl. Environ. Microbiol. 47:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mills S., McAuliffe O. E., Coffey A., Fitzgerald G. F., Ross R. P. 2006. Plasmids of lactococci: genetic accessories or genetic necessities? FEMS Microbiol. Rev. 30:243–273 [DOI] [PubMed] [Google Scholar]
- 26. Millward-Sadler S. J., et al. 1994. Evidence for a general role for high-affinity noncatalytic cellulose-binding domains in microbial plant-cell wall hydrolases. Mol. Microbiol. 11:375–382 [DOI] [PubMed] [Google Scholar]
- 27. Mishra C., et al. 1997. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast 13:327–336 [DOI] [PubMed] [Google Scholar]
- 28. Niegowski D., Eshaghi S. 2007. The CorA family: structure and function revisited. Cell. Mol. Life Sci. 64:2564–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nomura M., Kobayashi M., Narita T., Kimoto-Nira H., Okamoto T. 2006. Phenotypic and molecular characterization of Lactococcus lactis from milk and plants. J. Appl. Microbiol. 101:396–405 [DOI] [PubMed] [Google Scholar]
- 30. Oh H. M., et al. 2010. Complete genome sequence analysis of Leuconostoc kimchii IMSNU 11154. J. Bacteriol. 192:3844–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Sullivan D., et al. 2000. Novel type I restriction specificities through domain shuffling of HsdS subunits in Lactococcus lactis. Mol. Microbiol. 36:866–875 [DOI] [PubMed] [Google Scholar]
- 32. O'Sullivan D. J., Klaenhammer T. R. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rademaker J. L. W., et al. 2007. Diversity analysis of dairy and nondairy Lactococcus lactis isolates, using a novel multilocus sequence analysis scheme and (GTG)5-PCR fingerprinting. Appl. Environ. Microbiol. 73:7128–7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodionov D. A., Hebbeln P., Gelfand M. S., Eitinger T. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188:317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosander A., Connolly E., Roos S. 2008. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 74:6032–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rutherford K., et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 37. Salama M. S., Musafijajeknic T., Sandine W. E., Giovannoni S. J. 1995. An ecological study of lactic acid bacteria: isolation of new strains of Lactococcus including Lactococcus lactis subspecies cremoris. J. Dairy Sci. 78:1004–1017 [Google Scholar]
- 38. Schouler C., Gautier M., Ehrlich S. D., Chopin M. C. 1998. Combinational variation of restriction modification specificities in Lactococcus lactis. Mol. Microbiol. 28:169–178 [DOI] [PubMed] [Google Scholar]
- 39. Seegers J. F. M. L., Bron S., Franke C. M., Venema G., Kiewiet R. 1994. The majority of lactococcal plasmids carry a highly related replicon. Microbiology 140:1291–1300 [DOI] [PubMed] [Google Scholar]
- 40. Sesma F., Gardiol D., Holgado A. P. D., Demendoza D. 1990. Cloning of the citrate permease gene of Lactococcus lactis subsp. lactis biovar diacetylactis and expression in Escherichia coli. Appl. Environ. Microbiol. 56:2099–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Siezen R. J., et al. 2010. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J. Bacteriol. 192:2649–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siezen R. J., et al. 2005. Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl. Environ. Microbiol. 71:8371–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siezen R. J., et al. 2008. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 74:424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smit B. A., et al. 2005. Identification, cloning, and characterization of a Lactococcus lactis branched-chain alpha-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 71:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanous C., Chambellon E., Le Bars D., Delespaul G., Yvon M. 2006. Glutamate dehydrogenase activity can be transmitted naturally to Lactococcus lactis strains to stimulate amino acid conversion to aroma compounds. Appl. Environ. Microbiol. 72:1402–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tanous C., Chambellon E., Sepulchre A. M., Yvon M. 2005. The gene encoding the glutamate dehydrogenase in Lactococcus lactis is part of a remnant Tn3 transposon carried by a large plasmid. J. Bacteriol. 187:5019–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tanous C., Chambellon E., Yvon M. 2007. Sequence analysis of the mobilizable lactococcal plasmid pGdh442 encoding glutamate dehydrogenase activity. Microbiology 153:1664–1675 [DOI] [PubMed] [Google Scholar]
- 48. Tanous C., Kieronczyk A., Helinck S., Chambellon E., Yvon M. 2002. Glutamate dehydrogenase activity: a major criterion for the selection of flavour-producing lactic acid bacteria strains. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 82:271–278 [PubMed] [Google Scholar]
- 49. Teuber M., Meile L., Schwarz F. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 76:115–137 [PubMed] [Google Scholar]
- 50. Thilakaraj R., Raghunathan K., Anishetty S., Pennathur G. 2007. In silico identification of putative metal binding motifs. Bioinformatics 23:267–271 [DOI] [PubMed] [Google Scholar]
- 51. Tynkkynen S., et al. 1993. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J. Bacteriol. 175:7523–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van Rooijen R. J., Devos W. M. 1990. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J. Biol. Chem. 265:18499–18503 [PubMed] [Google Scholar]
- 53. van Rooijen R. J., Vanschalkwijk S., Devos W. M. 1991. Molecular cloning, characterization, and nucleotide sequence of the tagatose 6-phosphate pathway gene cluster of the lactose operon of Lactococcus lactis. J. Biol. Chem. 266:7176–7181 [PubMed] [Google Scholar]
- 54. Vetting M. W., et al. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433:212–226 [DOI] [PubMed] [Google Scholar]
- 55. Zdobnov E. M., Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
- 56. Zhang Z., Schwartz S., Wagner L., Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7:203–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.