Abstract
The chromosome of Thermotoga maritima strain MSB8 was found to have an 8,870-bp region that is not present in its published sequence. The isolate that was sequenced by The Institute for Genomic Research (TIGR) in 1999 is apparently a laboratory variant of the isolate deposited at the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM 3109) in 1986. This newly sequenced region from the DSMZ culture was located between TM1848 (cbp, cellobiose phosphorylase) and TM1847 (the 3′ end of a truncated ROK regulator). The new region contained seven genes: a beta glucosidase gene (bglA), three trehalose ABC transporter genes (treEFG), three xylose ABC transporter genes (xylE2F2K2), and the 5′ end of a gene encoding the ROK regulator TM1847. We present a new differential scanning fluorimetry method using a low pH that was necessary to screen potential ligands of these exceptionally thermostable periplasmic substrate-binding proteins. This method showed that trehalose, sucrose, and glucose stabilized TreE, and their binding was confirmed by measuring changes in intrinsic fluorescence upon ligand binding. Binding constants of 0.024 μM, 0.300 μM, and 56.78 μM at 60°C, respectively, were measured. XylE2 ligands were similarly determined and xylose, glucose, and fucose bound with Kd (dissociation constant) values of 0.042 μM, 0.059 μM, and 1.436 μM, respectively. Since there is no discernible phenotypic difference between the TIGR isolate and the DSMZ isolate despite the variance in their genomes, we propose that they be called genomovars: T. maritima MSB8 genomovar TIGR and T. maritima MSB8 genomovar DSM 3109, respectively.
INTRODUCTION
In the course of examining the evolution of maltose ABC transporters among members of the order Thermotogales, we noticed that many of the species for which genome sequences were determined have three members of this family (16). The genome of the first sequenced species, Thermotoga maritima strain MSB8, was exceptional in having only two mal operons, mal1 and mal2. The arrangement of genes adjacent to the mal3 orthologs in several Thermotoga species is similar to that near TM1848 in the T. maritima genome, but there is no Mal3 transporter in that region of the sequenced T. maritima genome. In 1994, Liebl et al. (8) published the sequence of a portion of a malE gene that they found adjacent to a beta glucosidase gene (bglA) that they cloned and characterized in the same study. When we searched GenBank for homologs of the MalE3 proteins of the Thermotoga species, we found that Liebl et al.'s MalE was a strong match, more so than the T. maritima MalE1 and MalE2. A bglA gene is upstream of the malE3 in all the other Thermotoga species, so we speculated that the region containing the T. maritima mal3 ortholog had been deleted during cultivation of that strain (16). This report confirms that hypothesis and demonstrates that the isolate that was sequenced was a laboratory variant of Thermotoga maritima MSB8 and should be designated as genomovar TIGR. The original isolate deposited at the DSMZ culture collection, genomovar DSM 3109, contains the complete third mal transporter operon. Another ABC transporter operon was also found, as was Liebl et al.'s bglA gene, which was also missing from the T. maritima MSB8 genomovar TIGR genome sequence published in 1999 (14).
To examine the functions of these two new transporters, we sought a method to screen the binding properties of their periplasmic substrate-binding proteins. Differential scanning fluorimetry (DSF) can provide a rapid screening assay, but these highly thermostable proteins do not denature at temperatures achievable by commonly used instruments. We present here a modification of typical DSF methods that can be applied to binding studies of extremely thermostable proteins. This assay allowed us to more easily screen many sugars and to narrow the number of ligands needed to measure ligand-binding affinities using intrinsic fluorescence. These results expand our knowledge of the physiological capabilities of this organism and also provide a cautionary tale for genome-sequencing projects in that cultivation can create genome variants that may confuse investigations of metabolic function.
MATERIALS AND METHODS
Strains.
Thermotoga maritima MSB8 genomovar DSM 3109, Thermotoga petrophila RKU-1 (DSM 13995), and Thermotoga naphthophila RKU-10 (DSM 13996) were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). Thermotoga maritima MSB8 genomovar TIGR and Thermotoga sp. strain RQ2 were kindly provided by Karl O. Stetter. Thermotoga neapolitana NS-E was provided by the late Holger W. Jannasch.
PCR amplification and DNA sequencing.
The region between TM1848 and TM1847 was amplified by PCR using the IDUltra Taq DNA polymerase enzyme kit (ID Labs Biotechnology, Inc.) using the primers listed in Table S1 in the supplemental material, and the products were amplified using an MJ Mini thermocycler.
The 10-kb PCR product from T. maritima MSB8 genomovar DSM 3109 was sequenced using a primer-walking method. After amplification as described above, the purified PCR product was sequenced using the primers listed in Table S2 in the supplemental material. To increase the reliability of the sequences, at least two identical sequences were obtained and sequencing was performed on both DNA strands. All sequencing was done by capillary electrophoresis on a 3130xl genetic analyzer (Applied Biosystems).
Fragments (1,478 bp) of the 16S rRNA genes from both T. maritima MSB8 genomovars were sequenced using two sets of primers (see Table S1 in the supplemental material) and the same PCR conditions used above but with IDproof Taq DNA polymerase (ID Labs Biotechnology, Inc.). The purified PCR products (QiaQuick PCR purification kit; Qiagen) were used as templates in sequencing reactions containing a single primer (see Table S1) and BigDye Terminator v3.1 (Applied Biosystems).
Prediction software.
Open reading frames (ORFs) were predicted using GeneMark (10) and ORF Finder. The operons were predicted by FGENESB. The transcription and translation terminators were predicted by FindTerm and RibEX (1). Protein transmembrane helices were predicted by the TMHMM Server v2.0 (7).
Cloning, expression, and purification of TreE and XylE2.
The putative substrate-binding protein-encoding genes treE and xylE2 from T. maritima MSB8 genomovar DSM 3109 were amplified using the IDProof Taq DNA polymerase enzyme kit (ID Labs Biotechnology, Inc.) with the primers listed in Table S1 in the supplemental material. The genes were cloned into the pET15b vector and the pET15b-treE and pET15b-xylE2 plasmids were transformed in Escherichia coli BL21-CodonPlus (DE3)-RIPL competent cells (Stratagene) for expression. All clones were validated by sequencing. The periplasmic substrate-binding protein genes malE1 and malE2 were cloned as previously described (13).
The recombinant proteins were extracted and purified according to the following protocol for the ligand screening. Cells were grown in an LB medium containing ampicillin (50 μg/ml) for 4 to 5 h and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 16 h at 24°C. In a typical experiment, cell pellets of 0.1 g to 0.5 g were washed with 5 ml of phosphate buffer and lysed using the B-PER bacterial protein extraction kit (Pierce), and the overexpressed His-tagged proteins were purified with a nickel-Sepharose high-performance column (GE) according to the manufacturer's protocol. The recombinant TreE tended to aggregate, especially at 4°C. This phenomenon was also observed for the related trehalose/maltose binding protein (TMBP) from Thermococcus litoralis (5). To prevent this, the cell lysate containing TreE was treated with RNase A (Qiagen) and DNase I (Pierce) (5). The proteins were dialyzed against 2 liters of buffer (20 mM sodium phosphate, pH 7.4) at room temperature.
Differential scanning fluorimetry screening of ligand binding.
The samples were mixed to a final volume of 20 μl, according to the following preparation unless otherwise indicated: 2.5 μg protein, 4 μl 100 mM citric acid buffer (pH 3.50 to 3.75), 0.032 μl 5,000× Sypro orange (Invitrogen), 1.2 μl 2.5 M NaCl, and 2 μl ligand, typically with a final concentration of 10 mM ligand. The final concentrations of cellobiose, mannan, pullulan, and sorbitol were 25 μM, 0.225% (wt/vol), 0.5% (wt/vol), and 0.1% (wt/vol), respectively. The fluorescence intensities were measured using an iCycler IQ real-time detection system with excitation at 490 nm and emission at 530 nm. The samples were heated from 25 to 94°C with a heating rate of 0.5°C per min. All the assays were done in triplicate.
The midpoint temperature of the unfolding transition (Tm) was obtained with the program Gnuplot from curve fitting to a Boltzmann equation (15) as follows:
where L, U, and a are the minimum and maximum fluorescence intensities and the slope of the curve, respectively. The ΔTm of the protein for a specific ligand was calculated as the difference between the Tm values of the ligand-bound and ligand-free proteins.
Sugar purity.
The purity of the sugars was at least 98% unless noted otherwise. Arabinose, melibiose, raffinose, cellobiose, sorbitol, tagatose, lactose, ribose, trehalose, maltose, l-fucose, xylose, fructose, galactose, l-rhamnose, myo-inositol, glucose, sucrose, mannan, mannose, α-1,4-maltotetraose (95%), and pullulan were obtained from Sigma-Aldrich. The xyloglucan oligosaccharide (95%), β-1,4-mannotriose (95%), and β-1,4-mannotetraose (95%) were supplied from Megazyme, and the α-1,4-maltotriose (97%) was from ICN.
Intrinsic fluorescence spectroscopy.
All fluorescence measurements were performed using an SLM Aminco-Bowman 2 spectrofluorimeter. The protein samples were incubated at 60°C and the temperature of the cuvette with the sample was equilibrated at 60°C for 5 min. Fluorescence emission spectra were measured at an excitation wavelength of 280 nm for XylE2 and 295 nm for TreE, and the emission intensities were measured over the wavelength range of 300 to 400 nm. The dissociation constants were measured by adding increasing amounts of selected carbohydrates into a stirred cuvette at 60°C. In the experiments, 3-μl portions of different stock solutions of the carbohydrate were added to 1.5 ml of 0.45 μM or 0.08 μM protein solution (20 mM sodium phosphate, pH 7.4). After the addition of the sugar, the sample was stirred for 2 min to reach equilibrium and then the fluorescence intensity at the predetermined emission wavelength was recorded for 45 s.
The Kd values of TreE for trehalose and XylE2 for glucose and xylose were obtained from curve fitting using KaleidaGraph to the equation accounting for ligand depletion as follows: F=F0 + ΔF/2Pt [(Kd + Pt + Lt) − (Kd + Pt + Lt)2. − 4Pt Lt]1/2, where F is the measured fluorescence at ligand concentration Lt and Pt is the total protein concentration (3, 19). This formula was used since the protein concentration was greater than the measured Kd. Other Kd values were obtained from curve fitting using GraphPad Prism to the following equation: F=(Fmax·L)/(Kd + L), where F is the measured fluorescence at ligand concentration L.
Nucleotide sequence accession number.
The genomic sequences between TM1848 and TM1847 of genomovar DSM 3109 were deposited in GenBank as JF907620.
RESULTS AND DISCUSSION
Identification of the two genomovars of Thermotoga maritima MSB8.
PCR amplifications with oligonucleotides that hybridized to the loci TM1848 (cbp) and TM1847 (ROK) were performed using DNA isolated from the T. maritima strain used to prepare DNA for the TIGR sequencing project (14) and from the strain obtained from the DMSZ collection (DSM 3109). The amplification from the inoculum used for the TIGR sequencing project gave a fragment of 1,216 bp, as predicted from the genome sequence, while the amplification from the DMSZ strain gave a fragment of approximately 10 kb (see Fig. S1 in the supplemental material). Other Thermotogales species gave fragments of sizes predicted by their reference genome sequences (see Fig. S1).
To ensure that both cultures were genuinely T. maritima MSB8, the 16S rRNA gene of each culture was sequenced. Both sequences were identical and matched with 100% identity to the sequence of the 16S rRNA gene deposited in 1987 as Thermotoga maritima MSB8 DSM 3109 (M21774.1). This demonstrated that these two cultures represent genomic variants or genomovars of T. maritima MSB8. We propose to designate the strain deposited at the DSMZ collection as Thermotoga maritima MSB8 genomovar DSM 3109 while the variant used by TIGR for its genome-sequencing project will be referred as Thermotoga maritima MSB8 genomovar TIGR.
Seven ORFs in the region between TM1848 and TM1847 deleted from genomovar TIGR.
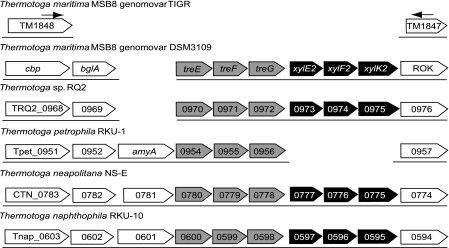
The sequence of the deleted region was completed by primer walking using the 10-kb PCR product from T. maritima MSB8 genomovar DSM 3109 as the template. This genomovar was found to have an additional 8,870 bp of genomic DNA between the loci TM1848 (cbp) and TM1847 (ROK). This region is missing from the genome of the genomovar TIGR between the positions 1,827,804 and 1,827,805. The new sequence has been deposited in GenBank as JF907620. This additional genomic DNA contains seven ORFs as predicted by GeneMark.hmm 2.4. A comparison of this region with the syntenic regions of other Thermotoga species shows that they all have similar genes at this location (Fig. 1), suggesting that genomovar DSM 3109 lost 8,870 bp of genomic DNA, giving rise to genomovar TIGR during laboratory cultivation.
Fig. 1.
Gene organization in syntenic regions of Thermotoga species in the vicinity of the TM1848 and TM1847 orthologs. The arrows represent the primer annealing sites used for PCR amplification of the 10-kb region. The tre operon is represented by gray boxes, while the xyl2 operon is represented by black boxes.
The first ORF encodes a beta glucosidase (BglA) that was first characterized in 1994, before publication of the genome sequence (8). The sequence matches with 100% identity to the sequence in GenBank deposited by W. Liebl (gi395290). The region downstream of bglA contains a palindromic sequence that suggests the presence of a rho-independent transcription terminator.
The next three ORFs encode a putative member of the maltose ABC transporter family and will be designated as a trehalose ABC transporter system (Tre). T. maritima has two known maltose operons, mal1 and mal2, that were previously characterized (13). As we demonstrate below, the newly discovered mal3 operon encodes a transporter with a binding preference for trehalose, so we propose that this be called the tre operon. It encodes a periplasmic substrate-binding protein (TreE) and two transmembrane proteins (TreF and TreG) (Fig. 1). Like the mal1 and mal2 operons and several other ABC transporter operons in T. maritima and other organisms, such as the E. coli malEFG operon, the tre operon contains no gene encoding an ATP binding protein. The newly discovered tre operon is only distantly related to the two mal T. maritima operons and is more closely related to the mal3 operons previously found in most other Thermotogales species, which are also related to the archaeal Thermococcus litoralis and Pyrococcus furiosus trehalose/maltose transporters (16) (data not shown). Unlike those archaeal trehalose/maltose binding proteins, TreE does not contain a lipid binding site at its N terminus that anchors it to the membrane (5). Both TreF and TreG proteins have six putative transmembrane helices.
The fourth, fifth, and sixth ORFs encode a second putative ABC transporter system containing a periplasmic substrate-binding protein, a transmembrane protein with eight putative transmembrane helices, and an ATP-binding protein with two nucleotide-binding domains (Fig. 1). The substrate-binding protein of this ABC transporter system is homologous to TM0114 (XylE1), a substrate-binding protein that binds both glucose and xylose (12, 20). This suggests that this ABC transporter operon should be named xyl2.
TM1847 encodes a putative transcription factor that belongs to the ROK (Repressor, ORF, Kinase) family. However, the 5′ portion of the gene is deleted in genomovar TIGR. As in other Thermotoga species, genomovar DSM 3109 contains a full ORF likely to encode a functional protein (Fig. 1) that could possibly regulate the Xyl2 transporter.
Binding properties of TreE and XylE2. (i) Thermal stabilities.
To identify the ligands that interact with the substrate-binding proteins encoded in this region, we used differential scanning fluorimetry (DSF). DSF analysis of ligand binding is based on the concept that proteins are stabilized by their ligands, and the difference between the denaturation temperatures of the ligand-bound and ligand-free forms, their ΔTm, can be measured (9, 11, 15, 17). A ΔTm close to zero indicates no thermal stability change in the presence of a ligand, while a positive value points to an interaction between the protein and the ligand. DSF can be performed in a real-time PCR thermocycler that records the increase of fluorescence intensity of the dye Sypro orange as the dye binds to hydrophobic residues that are revealed as the protein unfolds (15).
The substrate-binding proteins from T. maritima are extremely thermostable, and their unfolding temperatures (Tm) are typically above 94°C, the temperature limit for the PCR thermocycler (2, 20, 23). The Tm can be lowered using mildly denaturing conditions. We determined that an acidic buffer was the best denaturant for these thermophilic substrate-binding proteins. Using a citric acid buffer (pH 3.50 to 3.75), the Tm of MalE1 was linearly correlated with the pH, with an R2 of 0.99 (see Fig. S2 in the supplemental material).
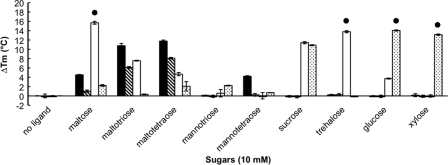
Prior to using this technique with the newly discovered substrate-binding proteins, we tested the ligand-induced stability of recombinant MalE1 and MalE2 from T. maritima. These proteins were previously characterized and their apparent dissociation constants (Kd) were determined using fluorescence spectroscopy (13, 23). MalE1 binds maltotriose, maltose, and mannotetraose with respective Kd values of 0.008 μM, 24 μM, and 38 μM (13). At 10 mM sugar, we observed that maltotriose, maltose, and mannotetraose increased the thermal stability of MalE1 with ΔTm values of 10.77°C, 4.52°C, and 4.22°C, respectively (Fig. 2; also see Table S3 in the supplemental material). MalE2 binds maltose, maltotriose, and trehalose with respective Kd values of 8.4 μM, 11 μM, and 9.5 μM, but mannotetraose was not a ligand (13). At 10 mM sugar, we observed that maltose and maltotriose increased the thermal stability of MalE2 with ΔTm values of 1.02°C and 6.85°C, respectively, while the ΔTm value in the presence of 10 mM mannotetraose was 0.24°C (Fig. 2; also see Table S3). Although trehalose binds to MalE2 (13), it did not increase the thermal stability of MalE2 (ΔTm=−0.05°C) (Fig. 2; also see Table S3). Trehalose is stable at high temperatures and low pHs (4), and so hydrolysis does not account for our observation. Instead, it is likely that the tertiary structure of MalE2 did not offer sufficient contacts between the sugar and the binding pocket to increase its stability in the presence of trehalose. Trehalose could have remained bound to the unfolded MalE2, leading to a lower ΔTm value (9). Interestingly, maltotetraose, a sugar that was not previously tested, stabilized both MalE1 and MalE2 with ΔTm values of 11.75°C and 8.67°C, respectively (Fig. 2; also see Table S3). No change in the thermal stability of MalE1 and MalE2 was recorded in the presence of other sugars (see Table S3). Thus, our ligand-screening assay gave no false-positive results and one false-negative result and revealed one potentially new ligand for these two previously studied proteins.
Fig. 2.
Thermal stability of the following recombinant proteins of T. maritima determined by differential scanning fluorimetry: MalE1 (black), MalE2 (crosshatched), TreE (white), and XylE2 (stippled). •, actual ΔTm is greater than or equal to the value indicated.
To determine the protein-ligand interactions of the newly identified substrate-binding proteins, the genes encoding TreE and XylE2 were expressed in E. coli. We screened 26 sugars with each protein by using DSF (see Table S3 in the supplemental material). We observed that the thermal stability of TreE was increased the most in the presence of 10 mM trehalose and maltose (Fig. 2; also see Tables S3 and S4). The upper values of the unfolding transition curves for both proteins were above 94°C, so their exact ΔTm values could not be determined. TreE was also stabilized with 10 mM sucrose, maltotriose, maltotetraose, and glucose with ΔTm values of 11.41°C, 7.57°C, 4.67°C, and 3.72°C, respectively (see Table S3). XylE2 was best stabilized by 10 mM glucose and xylose (Fig. 2; also see Tables S3 and S5) and was also stabilized with 10 mM sucrose, 10 mM l-fucose, 2.5 mM cellobiose, and 10 mM myo-inositol with ΔTm values of 10.89°C, 8.82°C, 6.03°C, and 5.41°C, respectively (see Table S3).
(ii) Intrinsic fluorescence spectroscopy.
Fluorescence spectroscopy was used to verify that the substrate-binding proteins bind the sugars identified with DSF and to measure their binding affinities. The Kd values were determined at 60°C, the highest temperature that the instrument could reach. TreE binds trehalose, sucrose, and glucose with Kd values of 0.024 μM, 0.300 μM, and 56.78 μM, respectively (Table 1). Fluorescence emission spectra were measured at an excitation wavelength of 280 nm and 295 nm to monitor aromatic residues and tryptophan, respectively. Despite this, no fluorescence changes were observed with maltose and maltotriose (data not shown). TreE's unfolding temperature (ΔTm) was dependent on maltose concentration (see Table S4 in the supplemental material), and phylogenetic analyses show that its closest relatives transport maltose (data not shown). Taken together, these results suggest that TreE binds maltose. Changes in intrinsic fluorescence cannot always be observed during binding as found with glucose binding by T. maritima XylE1 (12, 20).
Table 1.
Apparent Kd values at 60°C of TreE and XylE2 for different sugars as measured by changes in intrinsic fluorescence upon ligand binding
| Protein | Sugar | Kd value at 60°C (μM) |
|---|---|---|
| TreE | Trehalose | 0.024 |
| Sucrose | 0.300 | |
| Glucose | 56.78 | |
| XylE2 | Glucose | 0.059 |
| Xylose | 0.042 | |
| l-Fucose | 1.436 |
XylE2 binds glucose, xylose, and l-fucose with Kd values of 0.059 μM, 0.042 μM, and 1.436 μM, respectively (Table 1). Although sucrose, myo-inositol, and cellobiose stabilized XylE2 to heat denaturation, the Kd values for these ligands were relatively high at 116.4 μM, 56.47 μM, and 3.955 μM, respectively. It is likely these Kd values resulted from binding by the high-affinity ligand glucose contained as minor impurities in the sucrose, myo-inositol, and cellobiose rather than binding by those compounds.
T. maritima strain MSB8 genomovars.
The term genomovar was introduced by R. Rosselló in 1991 to describe different variants, or genomic groups, of Pseudomonas stutzeri (18). The term genomovar can be used for those strains exhibiting genomic variations yet with no detectable phenotypic differences that would allow them to be classified as separate species (21, 22). We demonstrated that the T. maritima isolate used for the genome-sequencing project published in 1999 (14) is a variant that lost 8,870 bp of genomic DNA between the loci TM1848 and TM1847 when compared to the isolate deposited in the DSMZ strain collection in 1986 (6). Both genomovars of T. maritima MSB8 have the same 16S rRNA gene sequence and no discernible phenotypic differences. We propose to designate the type strain (DSM 3109) as T. maritima MSB8 genomovar DSM 3109 and the sequenced variant as T. maritima MSB8 genomovar TIGR. The 8,870-bp region is largely syntenic with regions from other Thermotoga species that also have a bglA gene and a putative tre operon, suggesting that T. maritima MSB8 genomovar TIGR lost these genes.
A sequencing project for T. maritima genomovar DSM 3109 is in progress at the Joint Genome Institute (JGI). Upon its completion, the genome of the original isolate T. maritima MSB8 genomovar DSM 3109, deposited at the DSMZ culture collection, will be available in GenBank. The sequence of the genomovar DSM 3109 will likely provide new insights into the genetics, physiology, and metabolism of Thermotoga maritima MSB8.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from the NASA Astrobiology Program (NNX08AQ10G), the U.S. Department of Energy Office of Biological and Environmental Research (DE-PS02-08ER08-12), and the National Science Foundation Assembling the Tree of Life program (DEB0830024).
We thank Pascal Lapierre at the University of Connecticut Biotechnology Center for computational support, Carolyn Teschke for consultation about and the use of the fluorescence spectroscope, and Kristen Swithers and Chaman Ranjit for preliminary examinations of genomic DNAs.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Abreu-Goodger C., Merino E. 2005. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 33: W690–W692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cuneo M. J., Beese L. S., Hellinga H. W. 2008. Ligand-induced conformational changes in a thermophilic ribose-binding protein. BMC Struct. Biol. 8: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldstein A., Barrett R. W. 1987. Ligand dissociation constants from competition binding assays: errors associated with ligand depletion. Mol. Pharmacol. 31: 603–609 [PubMed] [Google Scholar]
- 4. Higashiyama T. 2002. Novel functions and applications of trehalose. Pure Appl. Chem. 74: 1263–1269 [Google Scholar]
- 5. Horlacher R., et al. 1998. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 180: 680–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huber R., et al. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144: 324–333 [Google Scholar]
- 7. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- 8. Liebl W., Gabelsberger J., Schleifer K. H. 1994. Comparative amino acid sequence analysis of Thermotoga maritima beta-glucosidase (BglA) deduced from the nucleotide sequence of the gene indicates distant relationship between beta-glucosidases of the BGA family and other families of beta-1,4-glycosyl hydrolases. Mol. Gen. Genet. 242: 111–115 [DOI] [PubMed] [Google Scholar]
- 9. Lo M. C., et al. 2004. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 332: 153–159 [DOI] [PubMed] [Google Scholar]
- 10. Lukashin A. V., Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26: 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matulis D., Kranz J. K., Salemme F. R., Todd M. J. 2005. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 44: 5258–5266 [DOI] [PubMed] [Google Scholar]
- 12. Nanavati D. M., Thirangoon K., Noll K. M. 2006. Several archaeal homologs of putative oligopeptide-binding proteins encoded by Thermotoga maritima bind sugars. Appl. Environ. Microbiol. 72: 1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nanavati D. M., Nguyen T. N., Noll K. M. 2005. Substrate specificities and expression patterns reflect the evolutionary divergence of maltose ABC transporters in Thermotoga maritima. J. Bacteriol. 187: 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nelson K. E., et al. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399: 323–329 [DOI] [PubMed] [Google Scholar]
- 15. Niesen F. H., Berglund H., Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2: 2212–2221 [DOI] [PubMed] [Google Scholar]
- 16. Noll K. M., Lapierre P., Gogarten J. P., Nanavati D. M. 2008. Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales. BMC Evol. Biol. 8: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pantoliano M. W., et al. 2001. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6: 429–440 [DOI] [PubMed] [Google Scholar]
- 18. Rosselló R., Garcia-Valdes E., Lalucat J., Ursing J. 1991. Genotypic and phenotypic diversity of Pseudomonas stutzeri. Syst. Appl. Microbiol. 14: 150–157 [Google Scholar]
- 19. Swillens S. 1995. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol. Pharmacol. 47: 1197–1203 [PubMed] [Google Scholar]
- 20. Tian Y., et al. 2007. Structure-based design of robust glucose biosensors using a Thermotoga maritima periplasmic glucose-binding protein. Protein Sci. 16: 2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ursing J., Aleksic S. 1995. Yersinia frederiksenii, a genotypically heterogeneous species with few differential characteristics. Contrib. Microbiol. Immunol. 13: 112–116 [PubMed] [Google Scholar]
- 22. Vandamme P., et al. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wassenberg D., Liebl W., Jaenicke R. 2000. Maltose-binding protein from the hyperthermophilic bacterium Thermotoga maritima: stability and binding properties. J. Mol. Biol. 295: 279–288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.