Abstract
Phasins (PhaP) are proteins normally associated with granules of poly(3-hydroxybutyrate) (PHB), a biodegradable polymer accumulated by many bacteria as a reserve molecule. These proteins enhance growth and polymer production in natural and recombinant PHB producers. It has been shown that the production of PHB causes stress in recombinant Escherichia coli, revealed by an increase in the concentrations of several heat stress proteins. In this work, quantitative reverse transcription (qRT)-PCR analysis was used to study the effect of PHB accumulation, and that of PhaP from Azotobacter sp. strain FA8, on the expression of stress-related genes in PHB-producing E. coli. While PHB accumulation was found to increase the transcription of dnaK and ibpA, the expression of these genes and of groES, groEL, rpoH, dps, and yfiD was reduced, when PhaP was coexpressed, to levels even lower than those detected in the non-PHB-accumulating control. These results demonstrated the protective role of PhaP in PHB-synthesizing E. coli and linked the effects of the protein to the expression of stress-related genes, especially ibpA. The effect of PhaP was also analyzed in non-PHB-synthesizing strains, showing that expression of this heterologous protein has an unexpected protective effect in E. coli, under both normal and stress conditions, resulting in increased growth and higher resistance to both heat shock and superoxide stress by paraquat. In addition, PhaP expression was shown to reduce RpoH protein levels during heat shock, probably by reducing or titrating the levels of misfolded proteins.
INTRODUCTION
Escherichia coli is used for the production of many heterologous products, such as proteins (42), biofuels (8), and bioplastics (26). High growth rates and the availability of different tools to manipulate its metabolism make E. coli an ideal candidate as a host for the synthesis of these compounds. However, the accumulation of high levels of recombinant products is known to produce stress in cells (16, 18, 38).
The heat shock stress response can be defined as the response of the cell to high temperatures, during which proteins are no longer able to fold properly (12). In E. coli, the heat shock response is regulated by the sigma factor RpoH and results in a transient increase in the expression of cytoplasmic chaperones, such as GroEL, IbpA, and DnaK, and proteases, such as Lon and FtsH (12). IbpA and IbpB are small heat shock proteins that bind to inclusion bodies formed by misfolded proteins, thereby preventing irreversible aggregation and aiding in folding (1, 24). It has been shown that the production of heterologous proteins (18) and biofuels, such as ethanol (11) and butanol (38), causes an increase in the expression of several stress-related proteins, in particular, heat shock proteins, such as GroEL, GroES, DnaK, IbpA, and IbpB.
Poly(3-hydroxybutyrate) (PHB), the best known polyhydroxyalkanoate, is a biodegradable polymer accumulated by many bacteria as a reserve material and has thermoplastic properties that make it suitable for the replacement of traditional plastics (26). Recombinant PHB-synthesizing E. coli strains carrying pha genes from different bacterial sources have been constructed (21). One such strain carrying PHB synthesis genes from Azotobacter sp. strain FA8, phaBAC (34), accumulates the polymer constitutively from different carbon sources, including agroindustrial by-products (30). Natural PHB producers accumulate the polymer in granules and have several proteins involved in their formation and regulation (33, 36). Among these are phasins (PhaP), small PHB granule-associated proteins that positively affect polymer synthesis and the number and size of PHB granules (35, 43).
Proteome analysis of PHB-producing recombinant E. coli strains compared to nonproducing strains revealed an increase in heat shock proteins, suggesting that PHB accumulation causes stress in cells (16). In accordance with this, the small heat shock proteins IbpAB and DnaK have been found associated with PHB granules in E. coli (14). There is experimental evidence that suggests that the presence of phasins may relieve this stress, as expression of PhaP1 from Cupriavidus necator (formerly Ralstonia eutropha) enhances growth and PHB synthesis both in natural producers and in recombinant E. coli (36, 45).
The growth-enhancing effect of PhaP observed in PHB-accumulating E. coli is not well understood. It has been proposed that PhaP could help prevent contact between cellular components and PHB granules (16), thus reducing the stress caused by the presence of the polymer. In PHB granule proteome experiments, heat shock proteins were observed to associate with PHB granules in recombinant E. coli in the absence of PhaP, but not when PhaP1 from C. necator was present (14, 40). Whether this altered binding of heat shock proteins is due to changes in transcription of heat shock genes, to changes in protein concentrations, or simply to protein displacement is unknown. In previous work (33), we observed that PhaP from Azotobacter sp. FA8, a 20.38-kDa phasin, enhances growth and PHB accumulation in recombinant E. coli, as a strain (K24KP) that constitutively expresses phaBAC and phaP achieves higher growth rates and cell densities and accumulates more PHB than the control strain without PhaP (9).
The aims of this work were to analyze the effect of PHB accumulation on gene transcription in recombinant E. coli and to characterize the enhancing effect of PhaP on PHB production and on cell growth. Based on the results obtained with both PHB-accumulating and nonaccumulating strains, a relationship between heat shock proteins and PhaP was proposed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All E. coli strains and plasmids used in this work are listed in Table 1. All pha genes used were those of Azotobacter sp. FA8.
Table 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| K1060a | F−fadE62 lacI60 tyrT58(AS) fabB5 mel-1 | 32 |
| KQ1 | Same as K1060, carrying pQEKm and pBBR1MCS; Kmr Cmr | This work |
| K24K1 | Same as K1060, carrying pJP24K and pBBR1MCS; Kmr Cmr | 9 |
| K24KP | Same as K1060, carrying pJP24K and pADP; Kmr Cmr | 9 |
| MC4100a | araD139 Δ(argF-lac)U19 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 6 |
| ADA100 | Same as MC4100, but λφ(ibp::lacZ) | 3 |
| ADAQ1 | Same as ADA100, carrying pQEKm and pBBR1MCS; Kmr Cmr | This work |
| ADAK1 | Same as ADA100, carrying pJP24K and pBBR1MCS; Kmr Cmr | This work |
| ADAKP | Same as ADA100, carrying pJP24K and pADP; Kmr Cmr | This work |
| ADA1001 | Same as ADA100, carrying pBBR1MCS; Cmr | This work |
| ADA100P | Same as ADA100, carrying pADP; Cmr | This work |
| Plasmids | ||
| pQEKm | Expression vector; ColE1 ori; Apr Kmr | 30 |
| pJP24K | pQEKm derivative containing the phaBAC genes from Azotobacter sp. strain FA8 under the control of a T5 promoter/lac operator element; Kmr | 30 |
| pBBR1MCS | Broad host range; lacPOZ′ mob RP4; Cmr | 23 |
| pADP | pBBR1MCS carrying phaP from Azotobacter sp. FA8; Cmr | 9 |
Strain obtained through the E. coli Genetic Stock Center, Yale University, New Haven, CT.
Growth conditions.
For strains KQ1, K24K1, and K24KP, MYAG medium was used, containing (per liter of deionized water) 6.0 g Na2HPO4, 3.0 g KH2PO4, 1.4 g (NH4)2SO4, 0.5 g NaCl, 0.2 g MgSO4·7H2O, 10.0 g yeast extract, 5.0 g casein amino acids, and 30.0 g glycerol. For plasmid maintenance, 50 μg·ml−1 kanamycin and 20 μg·ml−1 chloramphenicol were added. For strains ADA1001 and ADA100P, LB medium was used, supplemented with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). All cultures were grown at 37°C in 500-ml Erlenmeyer flasks containing 50 ml of medium and shaken at 250 rpm.
RNA extraction and quantitative reverse transcription (qRT)-PCR analysis.
Total RNA was extracted from 0.2 ml of pellets of 12-h cultures of strains KQ1, K24K1, and K24KP using the phenol-chloroform protocol, as previously described (37). cDNA was first synthesized from 5 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. A tube for each sample without transcriptase was made to check for genomic DNA. To remove the remaining RNA, samples were treated with RNase H for 20 min at 37°C. The resulting cDNA was diluted 1/10 before use in the quantitative PCR (qPCR) assays. Total RNA and DNA were quantified using a Nanodrop 3000 (Thermo Fisher Scientific Inc.). qPCR amplification of the cDNAs (95°C for 10 min, 95°C for 15 s, and 60°C for 1 min; 40 cycles) was performed with the Brilliant Sybr green qPCR master mix (Stratagene, Santa Clara, CA) plus 1 μM forward and reverse primers. The primers used are detailed in Table 2. The CT (cycle threshold) method was used for calculations. The CT is the minimum number of cycles necessary for the amplification product to be detected. CT values were normalized to the rrsB gene. The results were expressed as the log2 of the fold change, comparing the expression of K24K1 or K24KP to the expression of the same gene in the control strain KQ1 or the expression of K24KP with that of K24K1.
Table 2.
Primers used for qRT-PCR analysis
| Gene | Primer pair | Description | Reference |
|---|---|---|---|
| groEL | 5′-GTAACGACGCTCGTGTGAAA-3′ | Chaperone; its expression is regulated by RpoH. | 12 |
| 5′-GCAACGGAAACACCATCTTT-3′ | |||
| groES | 5′-AATGGCCGTATCCTTGAAAA-3′ | Chaperone; its expression is regulated by RpoH. | 12 |
| 5′-AATTGCCAGAATGTCGCTTT-3′ | |||
| dnaK | 5′-GAACCCGCAAAACACTCTGT-3′ | Chaperone; its expression is regulated by RpoH. | 12 |
| 5′-GTGCCATTTTCTGGCCTTTA-3′ | |||
| ibpA | 5′-AATCTGCTGGTGGTGAAAGG-3′ | Small heat shock protein; its expression is regulated by RpoH. | 1 |
| 5′-ACCAGGTTAGCACCACGAAC-3′ | |||
| rpoH | 5′-GGCTGAAAAGCTGCATTACC-3′ | Alternative sigma factor σH; it controls the heat shock response. | 12 |
| 5′-ATTACCATGGCGATCTGGAA-3′ | |||
| rpoE | 5′-GTCGTCCACCTTCCAGTGAT-3′ | Alternative sigma factor σE; it controls the responses to heat shock and other stresses on membrane and periplasmic proteins. | 27 |
| 5′-TAAATCTTCCGGGAGGGACT-3′ | |||
| rpoS | 5′-CGGAGTTGAGGTTTTTGACG-3′ | Alternative sigma factor σS; master regulator of the general stress response | 17 |
| 5′-GGCCGTTAACAGTGGTGAAT-3′ | |||
| dps | 5′-TGCTTTATACCCGCAACGAT-3′ | Protects against different types of stresses; its expression is regulated by RpoS. | 2 |
| 5′-GAAGCCATCCAGCATTTCAT-3′ | |||
| poxB | 5′-ATCGCCAAAACACTCGAATC-3′ | Pyruvate oxidase; its expression is regulated by RpoS. | 7 |
| 5′-CTCCGCTAAGTTGTGCTTCA-3′ | |||
| yfiD | 5′-AACTCTTTCTGGCTGCTGGA-3′ | Stress-induced alternate pyruvate formate-lyase subunit | 44 |
| 5′-ACCTTCAACGCGAACTTCTG-3′ | |||
| rrsB | 5′-GTGCTACAATGGCGCATACA-3′ | 16S rRNA | 29 |
| 5′-GGCATTCTGATCCACGATTA-3′ |
Immunoblot analysis.
For immunoblot experiments, cells from three independent cultures were pelleted and sonicated in 1 ml lysis buffer (50 mM Tris-Cl, pH 8, 1 mM EDTA, pH 8, 100 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). The soluble and insoluble fractions were separated by centrifugation (10 min at 10,000 × g). The protein concentration was measured according to the Bradford method (4). Equal amounts (30 μg) of total protein were used for Western immunoblot assay. Proteins were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (BA85; Schleicher & Schuell). The membrane was probed with rabbit anti-RpoH (31) at 1:5,000, followed by horseradish peroxidase-conjugated anti-rabbit antibody (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:5,000. The blots were developed with the ECL Western Blotting analysis system (Amersham-Pharmacia, Uppsala, Sweden) following the manufacturer's instructions. To estimate the intracellular content of RpoH, membranes were scanned with the luminescent image analyzer LAS-1000 plus and quantified with Image Gauge View 3.12 (Fuji Photo Film Co., Ltd.).
Biomass determination.
Samples were immediately chilled at 0°C by placement on ice. The cell dry weight (CDW) concentration, defined as grams (CDW) per liter of culture broth, was determined by placing an accurately measured volume of culture (10 ml) into a previously dried and weighed 15-ml centrifuge tube. After centrifugation at 10,000 × g for 10 min at 4°C, the cell pellet was washed twice with deionized water by resuspension and centrifugation. After decanting the supernatant, the centrifuge tube was dried in an oven at 85°C for 36 h and then cooled in a desiccator and weighed.
Analysis of PHB production.
The PHB content was quantitatively determined by gas chromatography using a slight modification of the method described by Braunegg et al. (5). About 10 mg freeze-dried biomass (0.1 mbar; 8 h) was accurately weighed and placed in Teflon-stoppered vials. Methanolysis was carried out by adding 2 ml CH3OH-H2SO4 (85:15 [vol/vol]) and 2 ml CHCl3 to the samples, which were incubated at 100°C for 140 min and cooled to room temperature. The aqueous phase was discarded, and the organic phase was extracted twice with deionized water. After settling of the layers, the organic phase was dried over anhydrous Na2SO4. The methyl esters of 3-hydroxybutyrate were quantified in a Hewlett Packard HP 5890 gas chromatograph equipped with a Hewlett-Packard FFAP (Hewlett-Packard Co., Palo Alto, CA). Pure PHB from C. necator was used as a standard. The PHB concentration was defined as grams of polymer per liter of culture broth. The PHB content was defined as a percentage of CDW (wt/wt).
Treatments with heat, paraquat, and hydrogen peroxide.
For heat stress experiments, bacterial cultures, grown in LB medium to an optical density at 600 nm (OD600) of 0.5, were transferred from 30 to 45 or 52°C and then kept at the transferred temperature for 30 min. For oxidative-stress experiments, 1.5 mM paraquat was added to cultures grown in LB medium at 37°C to an OD600 of 0.5. The viability of cells for each culture was determined by the colony count method, using LB plates. Survival was expressed as a percentage of the number of CFU at time zero, which was taken as 100%. Sensitivity to H2O2 was measured using sterile Whatman No. 1 filter disks (6 mm) impregnated with 5 μl of 30% H2O2 (Merck, Darmstadt, Germany) placed on top of bacterium-seeded plates. Zones of inhibition were measured after incubation at 37°C for 24 h.
Analysis of ibpA expression by measurement of β-galactosidase activity.
Expression of the ibpA-lacZ fusion was determined by means of β-galactosidase assays. Strain ADA100, transformed with different plasmids, was grown aerobically in LB medium, and 0.2-ml samples were taken at different times during cultivation. β-Galactosidase activity measurements were performed as previously described (41) from at least three independent experiments. Values are given in Miller units (28).
Comparison between PhaP and IbpA protein sequences.
Sequence alignment was performed using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with the default settings option.
RESULTS
Biomass, polymer accumulation, and protein production are higher in the presence of PhaP.
E. coli strains K24K1 (PHB producer), K24KP (PHB and PhaP producer), and KQ1 (a control strain that does not produce PHB or PhaP) were grown in shaken flask cultures using MYAG medium. In accordance with previous results (9), strain K24KP produced more PHB and biomass than the strain without PhaP (Table 3). To better analyze the metabolic state of the cells and to compare the results obtained in this work with previous publications, the concentration of soluble proteins related to total biomass was also determined after 12 h of growth (early stationary phase). Soluble-protein levels in the PHB producer (K24K1) were half those observed for the control strain; however, the presence of PhaP (K24KP) restored soluble protein to the control strain levels (Table 3).
Table 3.
Biomass, PHB, and protein concentrations for 12-h shaken flask cultures of strains KQ1 (control strain), K24K1 (PHB-producing strain), and K24KP (PHB- and PhaP-producing strain)
| E. coli strain | CDWa (g·liter −1) | PHB (g·liter−1) | % PHB (wt/wt)b | Soluble protein concnc (g·liter−1) | Soluble protein concn/CDW |
|---|---|---|---|---|---|
| KQ1 | 9.20 ± 0.07 | NDd | ND | 2.7 ± 0.6 | 0.29 ± 0.13 |
| K24K1 | 11.60 ± 0.06 | 2.4 ± 0.1 | 21 ± 1 | 1.7 ± 0.3 | 0.15 ± 0.09 |
| K24KP | 12.23 ± 0.08 | 3.0 ± 0.3 | 27 ± 2 | 3.5 ± 0.5 | 0.28 ± 0.13 |
CDW, cell dry weight.
The amount of PHB is given as a percentage of the CDW (average ± standard deviation).
Soluble proteins were extracted by sonication and centrifugation, and the amounts of proteins were determined by the Bradford assay as described in Materials and Methods.
ND, not determined.
The expression of stress-related genes is increased in the presence of PHB.
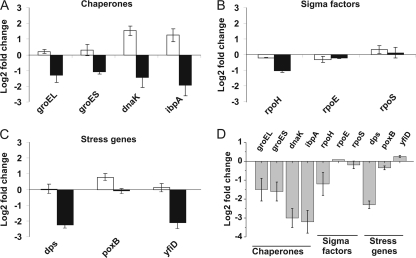
A previous proteomic analysis showed an increase in proteins related to the heat shock response in PHB-producing recombinant E. coli compared to the non-PHB-producing strain (16). To analyze whether this increase is due to changes in transcription, several stress-related genes were chosen for expression analysis by qRT-PCR. The genes and corresponding primers are detailed in Table 2. The expression of each gene in the PHB-producing strain (K24K1) was compared to its expression in the control strain (KQ1) (Fig. 1A to C, white bars). The most significant increases in gene expression in K24K1 were for the chaperones dnaK and ibpA (approximately 3- and 2.4-fold, respectively), with only slight increases observed for the chaperone genes groEL and groES. Minor changes were observed in the expression of the sigma factor genes rpoH, rpoE, and rpoS, which regulate the heat shock, envelope, and stationary-phase stress responses, respectively. Finally, only a slight increase was observed for poxB, which encodes a pyruvate oxidase (7) induced by RpoS.
Fig. 1.
Effects of PHB and PhaP accumulation on the expression of selected genes by qRT-PCR. The expression of chaperones (A), sigma factors (B), and stress-related genes (C) in strains K24K1 (PHB producer; white bars) and K24KP (PHB and PhaP producer; black bars) was compared to the expression in the control strain, KQ1. (D) The expression of all selected genes in strain K24KP was compared to the expression in K24K1. The results represent the mean values ± standard deviations of duplicate measurements from three independent experiments.
The expression of stress-related genes decreases in the presence of PhaP.
The effect of PhaP on the expression of the stress-related genes during PHB production was examined, comparing K24KP to K24K1. This revealed dramatic decreases in expression for most of the genes tested in the presence of PhaP. The greatest changes were observed for the chaperone genes ibpA and dnaK, followed by the RpoS-controlled stress genes dps and yfiD, the chaperone genes groEL and groES, and rpoH (Fig. 1D). No changes were observed for rpoE and rpoS. Unexpectedly, the presence of PhaP in PHB-producing cells (K24KP) reduced the expression of 7 out of 10 stress-related genes to levels even below those observed in the control strain that does not accumulate PHB (KQ1) (Fig. 1A to C, black bars), demonstrating the magnitude of the PhaP effect.
ibpA encodes a small heat shock protein previously found associated with PHB granules only in the absence of PhaP. Since ibpA transcription was the most affected by PhaP expression, we confirmed our qRT-PCR results using a fusion of the ibpA promoter to the reporter gene lacZ (3). After 12 h of growth in MYAG, the expression levels for the ibpA-lacZ fusion in ADAK1, the PHB-producing derivative, were two times higher than in ADAQ1, the control strain without PHB (153 ± 25 and 74 ± 11 Miller units for strains ADAK1 and ADAQ1, respectively). ibpA-lacZ expression levels for the PHB- and PhaP-producing strain were 2.5 times lower than in the PHB-producing strain (61 ± 17 for ADAKP), in accordance with the qRT-PCR results.
PhaP enhances growth in non-PHB-producing E. coli and reduces the expression of ibpA.
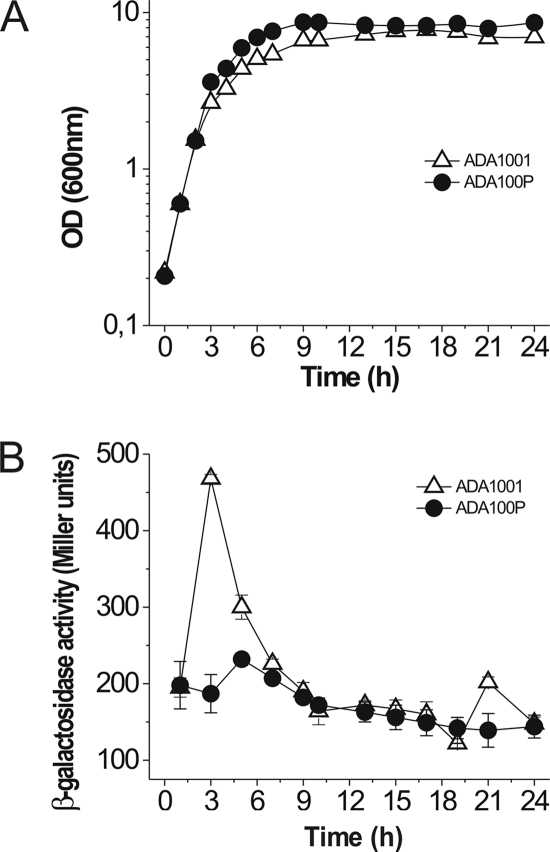
Our qRT-PCR results suggested that PhaP expression had significant effects on E. coli even in the absence of PHB expression. We tested this hypothesis by comparing growth and ibpA-lacZ expression in control (ADA1001) and PhaP-expressing (ADA100P) strains. In flask cultures at 37°C, the two strains displayed similar initial growth rates (Fig. 2A); however, they differed after 3 h, reaching maximum growth rates (μmax) of 0.79 ± 0.06 for the PhaP strain compared to 0.69 ± 0.05 for the control, resulting in a 38% higher final biomass for the PhaP-bearing strain than for the control (3.47 ± 0.12 and 2.50 ± 0.05 g·liter−1, respectively). The expression levels of ibpA-lacZ were similar for both strains during most of the time but increased dramatically in the control strain around the third hour of growth, forming a sharp peak (Fig. 2B). Interestingly, this peak occurred at the same time that differences in the growth rates between the two strains began to appear.
Fig. 2.
Growth and ibpA expression in the presence and absence of PhaP. Growth (A) and ibpA expression (B) were measured for strains ADA1001 (control strain) and ADA100P (PhaP producer) in cultures grown at 37°C. The expression of ibpA was calculated by measuring the β-galactosidase activity of the ibpA-lacZ fusion present in strains ADA1001 and ADA100P. The results shown represent the mean values for each parameter ± standard deviations of duplicate measurements from three independent cultures.
PhaP protects E. coli against heat and oxidative stresses.
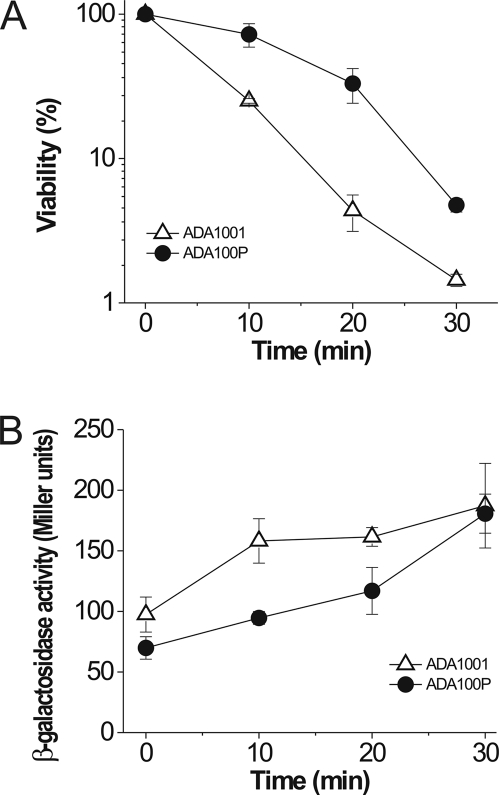
Our results demonstrated that PhaP affects the expression of several stress-related genes, suggesting that the protein may confer stress protection on the cells. This was tested by monitoring the effects of PhaP expression on cell viability and ibpA expression in response to heat and oxidative stresses. Survival of lethal heat shock was assessed by transferring control (ADA1001) and PhaP-expressing (ADA100P) strains from 30°C to 52°C and monitoring viability. PhaP expression conferred increased resistance to heat stress, with 72% survival after 10 min at 52°C compared to 25% in the absence of PhaP (Fig. 3A). In addition, expression of the ibpA-lacZ fusion was much lower in the PhaP-expressing strain (Fig. 3B). After 30 min of exposure to 52°C, the two strains showed similar expression levels.
Fig. 3.
Heat resistance in the presence and absence of PhaP. Viability (A) and ibpA expression (B) were measured in strains ADA1001 (control strain) and ADA100P (PhaP producer) after heat shock. Cells were grown at 30°C to an OD600 of 0.5 and then transferred to 52°C for 30 min. The results represent the mean values ± standard deviations of duplicate measurements from three independent experiments.
Cells were also subjected to oxidative stress using 1.5 mM paraquat. Similar to what was observed during heat shock, PhaP expression conferred increased resistance, with 72% survival after 10 min compared to 33% for the control strain. Sensitivity to H2O2 was analyzed using impregnated filter disks, but in this case, the results were similar for both strains (the inhibition halos were 1.70 ± 0.06 cm for ADA1001 and 1.77 ± 0.05 cm for ADA100P).
Since these results were similar to those obtained in cells overexpressing ibpA (22), the possibility that PhaP could be structurally related to IbpA was analyzed. The amino acid sequences of both proteins were compared using ClustalW, but no significant similarities were found.
RpoH levels are reduced in the presence of PhaP.
Our results demonstrated that the presence of PhaP decreases expression of several heat shock genes, such as ibpA, that are regulated by the sigma factor RpoH, suggesting lower RpoH activity. In addition, rpoH transcription was also observed to decrease in the presence of PhaP in the PHB-producing cells (Fig. 1B). Since RpoH is mostly regulated at posttranscriptional levels (13), we tested the effect of PhaP on the RpoH concentration using Western blot analysis both before and after heat shock at 45°C. Samples containing equal amounts of proteins from cultures of control and PhaP-expressing strains (ADA1001 and ADA100P) were used before and 5 and 10 min after shifting to 45°C. The levels of RpoH both before and after heat shock were higher in ADA1001 (Fig. 4). There was a slight decrease in the concentration of RpoH 5 and 10 min after heat shock for both strains, but the control strain continued to have higher RpoH levels than ADA100P at all times tested (Fig. 4), demonstrating that the presence of PhaP decreases RpoH levels.
Fig. 4.
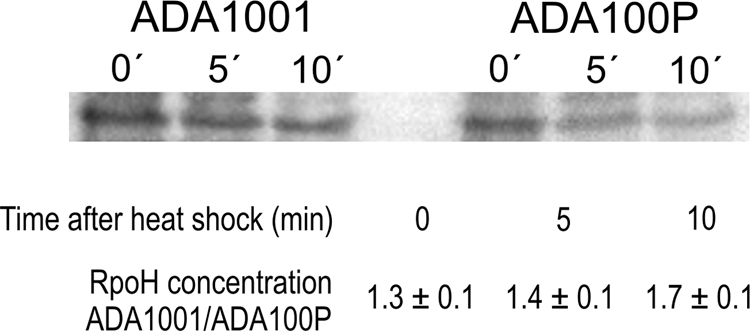
Immunoblots of RpoH in E. coli in the presence and absence of PhaP. Strains ADA1001 and ADA100P were subjected to heat shock, and samples were extracted 0, 5, and 10 min after transfer to 45°C. Equal amounts (30 μg) of protein were loaded in each lane. The ratios between the RpoH concentrations (quantified from the immunoblots) of the two strains are shown below.
DISCUSSION
Previous studies on the effect produced by PHB accumulation in E. coli were based on proteomics analysis and showed increased levels of several stress-related proteins (14, 16). The analysis of the transcription of several stress-related genes in PHB-producing strains performed in the present work shed light on the effects of PHB accumulation in recombinant E. coli, and also on the effect of PhaP, in both PHB-accumulating and nonaccumulating strains. The increased expression of dnaK and ibpA in the PHB-producing strain suggests that the accumulation of PHB in E. coli generates stress in the cells. This is in accordance with whole-cell proteomic studies performed in PHB-producing E. coli (16) that reported an increase in DnaK protein levels, in addition to increased amounts of GroEL, GroES, and Dps. A previous study that analyzed pgi mutants, which have high levels of reduced cofactors, reported an increase in the transcription of the stress-related genes rpoH, dnaK, groES, ibpA, and ibpB when the cells accumulated PHB (20). In the present work, no significant differences between the expression levels of rpoH, groEL, and groES were observed when comparing the PHB-producing strain with its control strain. The discrepancies between the studies are probably due to the different genetic backgrounds, and possibly also to other factors, such as the culture age and carbon source. The stress produced by the synthesis of PHB can be compared to the response observed during the production of heterologous proteins in E. coli, which generates an increase in the transcription (10, 19) and concentrations (18) of several chaperones, such as GroEL, GroES, DnaK, IbpA, and IbpB.
In C. necator mutant strains that do not produce PhaP1, the heat shock chaperones DnaK and GroEL were found in the granules (35), and in recombinant E. coli, IbpAB and GroEL are also associated with PHB granules (14, 40). It is likely that PHB production induces the heat shock response by titrating available cytoplasmic chaperones onto the surfaces of the PHB granules. Since free chaperones bind and inactivate available RpoH (13), titration of these proteins by PHB would release RpoH, thereby inducing expression of heat shock genes. This hypothesis is supported by the increase in the expression of several of these chaperones observed in our transcriptional analysis.
The concentration of soluble proteins in the PHB-producing strain (K24K1) was half that of the control strain without PHB (KQ1), in accordance with results obtained in E. coli strains expressing the phb genes from C. necator (16). This effect has been attributed to competition for substrates required for the synthesis of amino acids and other cellular metabolites, such as acetyl-coenzyme A (CoA) and/or NAD(P)H, which are used for polymer accumulation (25). Our results showed that the presence of PhaP from Azotobacter sp. FA8 restored the concentration of soluble proteins in the PHB-producing strain (K24KP) to levels similar to those in the control strain (KQ1). Phasins, such as PhaP1 from C. necator and PhaP from Azotobacter sp. FA8, have been shown to enhance not only polymer accumulation, but also cellular growth in E. coli (9, 45). As K24KP accumulated even more PHB than K24K1, this suggests that the lower protein concentration observed for K24K1 is not only due to the lower availability of substrates used for PHB synthesis, but also to other factors attenuated by the presence of PhaP. A possible explanation, suggested by the presence of the elongation factor Ef-Tu in isolated PHB granules in recombinant E. coli (14), could be that in the absence of PhaP the polymer might interact with components of the protein synthesis machinery, thereby affecting protein production.
Previous work proposed that the stress induced in PHB-producing strains is partly due to the interaction of PHB with other cellular components (14, 16). In view of this, it has been suggested that this effect could be attenuated by the presence of phasins, which form a barrier between the polymer and the cytoplasm (16, 40). Proteomic studies of purified PHB granules from E. coli showed that IbpA was the most abundant protein found in the granules, but only in the absence of PhaP (40). This could be solely due to competition between IbpA and PhaP for granule surface binding or to a reduced concentration of IbpA in the phaP-expressing recombinants. Our transcription analysis demonstrated that expression of ibpA, together with the other chaperones, is decreased in PHB-accumulating recombinants in the presence of PhaP. If PhaP prevents the titration of chaperones by the PHB granules, the increased level of free chaperones could bind to and inactivate RpoH, thereby decreasing the heat shock response and, consequently, the expression of these chaperones.
Significantly, our results demonstrated that PhaP expression also has a strong beneficial effect in E. coli independent of PHB accumulation. This was first suggested by the dramatic decreases in expression of the chaperone genes ibpA, groEL, groES, and dnaK in strain K24KP to levels significantly below those observed in non-PHB-producing cells, indicating that expression of PhaP significantly decreases cytoplasmic stress during cellular growth. This result was quite unexpected, since the expression of heterologous proteins is known to induce the heat shock response instead of reducing it (42). Further experiments showed that expressing PhaP in non-PHB-producing cells resulted in increased growth rates and cell density at 37°C compared to control cells. Interestingly, the peak in ibpA expression in control versus PhaP-expressing cells coincided with the beginning of differences in growth rates between the two strains. This could be due to a relative increase in the concentration of misfolded proteins in the control cells, which slows down growth and triggers the expression of ibpA. The presence of PhaP alleviates this stress, resulting in higher growth rates and final cell densities.
Our results also demonstrated that PhaP expression in non-PHB-producing E. coli provides a protective effect against stresses: increased resistance to both heat shock and superoxide stress by paraquat, but not to hydrogen peroxide, was observed. Interestingly, the protective effect of PhaP is similar to that observed in E. coli overexpressing ibpA and ibpB (22), suggesting an overlap in function between PhaP and IbpA. This was further reinforced by our finding that ibpA expression was reduced in PhaP-expressing strains, both during normal growth at 37°C and in response to heat shock, and the previous observation that IbpA was no longer present in PHB granules when PhaP was expressed (40). We note, however, that there are no sequence similarities between PhaP and IbpA, suggesting no common origin of the two proteins. Since the overexpression of IbpA and IbpB has been shown to positively influence recombinant protein production in E. coli (15), the similarities observed between the effects of IbpA and PhaP suggest that PhaP could also enhance the production of recombinant proteins in E. coli.
The primary signal that results in the induction of the heat shock response is the appearance of protein misfolding due to an increase in temperature. The amounts of misfolded proteins relative to those of chaperones and proteases regulate the heat shock response by affecting RpoH levels (13). Taking this into account, the reduced heat shock protein transcription observed in this work in strains expressing PhaP, together with reduced expression of rpoH during PHB production and RpoH protein levels during heat shock, suggests that PhaP may affect protein folding similarly to IbpA (Fig. 5).
Fig. 5.
Hypothetical model of the effect of PhaP on the heat shock response. When misfolded proteins appear in the cell as a result of heat or other kinds of stress, chaperones bind to them, and RpoH freed from chaperones activates the heat shock response (gray arrows). In this hypothetical model, PhaP could decrease the amount of misfolded proteins by an unknown mechanism (dashed arrows), thus reducing the recruitment of chaperones, which are available to bind to RpoH, inactivating it.
We note that in our experiments, levels of RpoH decreased slightly after 5 and 10 min of heat shock in both the presence and absence of PhaP. During heat shock, RpoH protein levels transiently increase before decreasing due to degradation (39). It is likely that in our experiments the transient increase peaked before our 5-min time point.
This work has demonstrated the protective role of PhaP in PHB-synthesizing E. coli and linked the effects of the protein to the expression of stress-related genes, especially ibpA. Furthermore, the effect of PhaP was analyzed independently of the production of PHB, showing that the protein has an unexpected protective effect in E. coli under both normal and stress conditions. The results obtained in this work open the door for novel biotechnological applications of this protein, for example, in the production of recombinant proteins and other heterologous products in E. coli.
ACKNOWLEDGMENTS
We thank F. Narberhaus for providing us with the RpoH antiserum, F. Baneyx for strain ADA100, E. Gogol for help in the qRT-PCR experiments, and S. Ruzal and M. Allievi for help during the quantification of RpoH.
This work was partially supported by grants from the National Institutes of Health (grant GM036278 to C.A.G.) and from the Universidad de Buenos Aires (project UBACyT X173). M.J.P. is a career investigator from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina). A.D.A. holds a postdoctoral fellowship from CONICET. While working at the University of California, San Francisco, A.D.A. was supported by fellowships from the American Society for Microbiology and the Fulbright Association. M.V.C. holds a graduate student fellowship from Agencia Nacional de Promoción Científica y Tecnológica (ANCyPT, Argentina).
Footnotes
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Allen S. P., Polazzi J. O., Gierse J. K., Easton A. M. 1992. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J. Bacteriol. 174:6938–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Almirón M., Link A. J., Furlong D., Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646–2654 [DOI] [PubMed] [Google Scholar]
- 3. Bianchi A. A., Baneyx F. 1999. Hyperosmotic shock induces the sigma32 and sigmaE stress regulons of Escherichia coli. Mol. Microbiol. 34:1029–1038 [DOI] [PubMed] [Google Scholar]
- 4. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 5. Braunegg G., Sonnleitner B., Lafferty R. M. 1978. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in bacterial biomass. Eur. J. Appl. Microbiol. Biotechnol. 6:29–37 [Google Scholar]
- 6. Casadaban M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541–555 [DOI] [PubMed] [Google Scholar]
- 7. Chang Y. Y., Wang A. Y., Cronan J. E., Jr 1994. Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS(katF) gene. Mol. Microbiol. 11:1019–1028 [DOI] [PubMed] [Google Scholar]
- 8. Clomburg J. M., Gonzalez R. 2010. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 86:419–434 [DOI] [PubMed] [Google Scholar]
- 9. de Almeida A., Nikel P. I., Giordano A. M., Pettinari M. J. 2007. Effects of granule-associated protein PhaP on glycerol-dependent growth and polymer production in poly(3-hydroxybutyrate)-producing Escherichia coli. Appl. Environ. Microbiol. 73:7912–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gill R. T., Valdes J. J., Bentley W. E. 2000. A comparative study of global stress gene regulation in response to overexpression of recombinant proteins in Escherichia coli. Metab. Eng. 2:178–189 [DOI] [PubMed] [Google Scholar]
- 11. Gonzalez R., et al. 2003. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant). Biotechnol. Prog. 19:612–623 [DOI] [PubMed] [Google Scholar]
- 12. Gross C. A. 1996. Function and regulation of the heat shock proteins. Escherichia coli and Salmonella, p. 1382–1399 ASM Press, Washington DC [Google Scholar]
- 13. Guisbert E., Yura T., Rhodius V. A., Gross C. A. 2008. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol. Mol. Biol. Rev. 72:545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han M. J., et al. 2006. Analysis of poly(3-hydroxybutyrate) granule-associated proteome in recombinant Escherichia coli. J. Microbiol. Biotechnol. 16:901–910 [Google Scholar]
- 15. Han M. J., Park S. J., Park T. J., Lee S. Y. 2004. Roles and applications of small heat shock proteins in the production of recombinant proteins in Escherichia coli. Biotechnol. Bioeng. 88:426–436 [DOI] [PubMed] [Google Scholar]
- 16. Han M. J., Yoon S. S., Lee S. Y. 2001. Proteome analysis of metabolically engineered Escherichia coli producing poly(3-hydroxybutyrate). J. Bacteriol. 183:301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoffmann F., Rinas U. 2004. Stress induced by recombinant protein production in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 89:73–92 [DOI] [PubMed] [Google Scholar]
- 19. Jürgen B., et al. 2000. Monitoring of genes that respond to overproduction of an insoluble recombinant protein in Escherichia coli glucose-limited fed-batch fermentations. Biotechnol. Bioeng. 70:217–224 [DOI] [PubMed] [Google Scholar]
- 20. Kabir M. M., Shimizu K. 2003. Fermentation characteristics and protein expression patterns in a recombinant Escherichia coli mutant lacking phosphoglucose isomerase for poly(3-hydroxybutyrate) production. Appl. Microbiol. Biotechnol. 62:244–255 [DOI] [PubMed] [Google Scholar]
- 21. Khanna S., Srivastava A. K. 2005. Recent advances in microbial polyhydroxyalkanoates. Process. Biochem. 40:607–619 [Google Scholar]
- 22. Kitagawa M., Matsumura Y., Tsuchido T. 2000. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol. Lett. 184:165–171 [DOI] [PubMed] [Google Scholar]
- 23. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 24. Kuczyńska-Wi´nik D., et al. 2002. The Escherichia coli small heat-shock proteins IbpA and IbpB prevent the aggregation of endogenous proteins denatured in vivo during extreme heat shock. Microbiology 148:1757–1765 [DOI] [PubMed] [Google Scholar]
- 25. Lee S. Y., Lee Y. K., Chang H. N. 1995. Stimulatory effects of amino acids and oleic acid on poly(3-hydroxybutyric acid) synthesis by recombinant Escherichia coli. J. Ferment. Bioeng. 79:177–180 [Google Scholar]
- 26. Madison L. L., Huisman G. W. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mecsas J., Rouviere P. E., Erickson J. W., Donohue T. J., Gross C. A. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618–2628 [DOI] [PubMed] [Google Scholar]
- 28. Miller J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 29. Neidhardt F. C., et al. (ed.), 1996. Escherichia coli and Salmonella, cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC [Google Scholar]
- 30. Nikel P. I., de Almeida A., Melillo E. C., Galvagno M. A., Pettinari M. J. 2006. New recombinant Escherichia coli strain tailored for the production of poly(3-hydroxybutyrate) from agroindustrial by-products. Appl. Environ. Microbiol. 72:3949–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Obrist M., Narberhaus F. 2005. Identification of a turnover element in region 2.1 of Escherichia coli sigma32 by a bacterial one-hybrid approach. J. Bacteriol. 187:3807–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Overath P., Schairer H. U., Stoffel W. 1970. Correlation of in vivo and in vitro phase transitions of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 67:606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pettinari J. M., Chaneton L., Vázquez G., Steinbüchel A., Méndez B. S. 2003. Insertion sequence-like elements associated with putative polyhydroxybutyrate regulatory genes in Azotobacter sp. FA8. Plasmid 50:36–44 [DOI] [PubMed] [Google Scholar]
- 34. Pettinari M. J., et al. 2001. Poly(3-hydroxybutyrate) synthesis genes in Azotobacter sp. strain FA8. Appl. Environ. Microbiol. 67:5331–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pötter M., et al. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301–2311 [DOI] [PubMed] [Google Scholar]
- 36. Pötter M., Steinbüchel A. 2005. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules 6:552–560 [DOI] [PubMed] [Google Scholar]
- 37. Rhodius V. A., Wade J. T. 2009. Technical considerations in using DNA microarrays to define regulons. Methods 47:63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rutherford B. J., et al. 2010. Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl. Environ. Microbiol. 76:1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Straus D. B., Walter W. A., Gross C. A. 1987. The heat shock response of E. coli is regulated by changes in the concentration of sigma 32. Nature 329:348–351 [DOI] [PubMed] [Google Scholar]
- 40. Tessmer N., et al. 2007. Heat-shock protein HspA mimics the function of phasins sensu stricto in recombinant strains of Escherichia coli accumulating polythioesters or polyhydroxyalkanoates. Microbiology 153:366–374 [DOI] [PubMed] [Google Scholar]
- 41. Tetsch L., Koller C., Haneburger I., Jung K. 2008. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol. Microbiol. 67:570–583 [DOI] [PubMed] [Google Scholar]
- 42. Valdez-Cruz N. A., Caspeta L., Perez N. O., Ramirez O. T., Trujillo-Roldan M. A. 2010. Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters. Microb. Cell Fact. 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wieczorek R., Pries A., Steinbüchel A., Mayer F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wyborn N. R., et al. 2002. Expression of the Escherichia coli yfiD gene responds to intracellular pH and reduces the accumulation of acidic metabolic end products. Microbiology 148:1015–1026 [DOI] [PubMed] [Google Scholar]
- 45. York G. M., Stubbe J., Sinskey A. J. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]