Abstract
Contamination of foods, especially produce, with Salmonella spp. is a major concern for public health. Several methods are available for the detection of Salmonella in produce, but their relative efficiency for detecting Salmonella in commonly consumed vegetables, often associated with outbreaks of food poisoning, needs to be confirmed. In this study, the effectiveness of three molecular methods for detection of Salmonella in six produce matrices was evaluated and compared to the FDA microbiological detection method. Samples of cilantro (coriander leaves), lettuce, parsley, spinach, tomato, and jalapeno pepper were inoculated with Salmonella serovars at two different levels (105 and <101 CFU/25 g of produce). The inoculated produce was assayed by the FDA Salmonella culture method (Bacteriological Analytical Manual) and by three molecular methods: quantitative real-time PCR (qPCR), quantitative reverse transcriptase real-time PCR (RT-qPCR), and loop-mediated isothermal amplification (LAMP). Comparable results were obtained by these four methods, which all detected as little as 2 CFU of Salmonella cells/25 g of produce. All control samples (not inoculated) were negative by the four methods. RT-qPCR detects only live Salmonella cells, obviating the danger of false-positive results from nonviable cells. False negatives (inhibition of either qPCR or RT-qPCR) were avoided by the use of either a DNA or an RNA amplification internal control (IAC). Compared to the conventional culture method, the qPCR, RT-qPCR, and LAMP assays allowed faster and equally accurate detection of Salmonella spp. in six high-risk produce commodities.
INTRODUCTION
Salmonella spp. are important food-borne pathogens of significant concern for public health both domestically and internationally (20, 39, 42). According to the latest CDC report, millions of people are infected by Salmonella every year in the United States alone. Out of the total U.S. food-borne diseases caused by all known food-borne pathogens, Salmonella accounts for 11%, 35%, and 28% of illnesses, hospitalizations, and deaths, respectively (39). Salmonella transmission to humans has been linked to numerous sources, especially undercooked food. Fresh vegetables and fruits can be contaminated with Salmonella during the often lengthy farm-to-fork process. Consumption of fresh fruits and vegetables in the United States has increased by almost 50% since the 1970s (5) and was steady from 2000 to 2009 (3). Owing to this historical trend and the continued improvement of detection methods, it is sometimes difficult to discern whether perceived increases in food-borne outbreaks (1) are the result of these modern trends or other factors, such as increasing populations or closer proximity of livestock and human sludge in agricultural areas, that might play a role in large outbreaks such as the jalapeno-borne Salmonella outbreak in 2008 (21, 29). Additionally, this increase in Salmonella outbreaks could be due to the implementation of better surveillance systems, such as PulseNet (http://www.cdc.gov/pulsenet), which allows Salmonella cases to be grouped into an outbreak that otherwise would have been considered as sporadic cases.
Traditional culture methods for Salmonella detection in foods consist of a series of steps that include nonselective enrichment, selective enrichment, and selective/differential plating and, finally, biochemical and serological confirmation. The traditional microbiological method for Salmonella isolation is described in detail in chapter 5 of the Bacteriological Analytical Manual (BAM) (2, 14) by the U.S. Food and Drug Administration (FDA). The method is labor-intensive and requires a minimum of 5 days to complete the analysis (14). Consequently, there is a need to develop and validate faster screening and detection methods for this pathogen in produce.
In recent years, molecular methods designed for targeting Salmonella DNA have focused on genes such as invA, fimC, the ttrRSBCA locus, phoP, and other genome markers. Moreover, these genes have been targeted by conventional and quantitative real-time PCR (qPCR) technologies (22, 28, 45, 46). Most of these molecular methods have the potential to reduce detection time to <3 days. qPCR is faster and more sensitive than conventional PCR and provides real-time data without the use of gels (43). Another promising molecular method introduced recently for Salmonella detection is the loop-mediated isothermal amplification (LAMP) assay (15). LAMP is a technique that amplifies nucleic acid using Bst DNA polymerase under isothermal conditions with high levels of specificity, efficiency, and speed (30, 32). LAMP differs from PCR in that four or six primers are used for the amplification of a single target gene. The amplification uses a single temperature step from 63°C to 65°C and is maintained at 65°C for ∼60 min. Many amplicons with various structural conformations are produced in LAMP reactions. These resultant fragments can be detected by simple turbidity or fluorescence, the latter of which is correlated with production of magnesium pyrophosphate, a by-product of the reaction (32). LAMP has several advantages over PCR, including the use of simple and cost-effective equipment, as well as high levels of specificity and amplification efficiency. Two LAMP assays for the detection of Salmonella spp. have been developed recently; one is based on detection of the invA gene (15, 26, 44) and the other targets the phoP gene (24). Other LAMP assays for Salmonella detection target mainly target specific serovars or specific O groups (35, 36, 47).
Several studies have shown that the invA gene and its mRNA are good targets for detection of Salmonella spp. by qPCR in environmental samples (4, 27, 31, 33, 37, 46), LAMP (15, 34), quantitative real-time reverse transcriptase PCR (RT-qPCR) (11, 18, 41), or conventional reverse transcriptase PCR (RT-PCR) (18). Both qPCR and LAMP have the potential to detect nonviable cells, particularly because bacterial DNA is more stable than bacterial RNA and can persist in a sample long after the target organism has died (9). invA mRNA production depends heavily on the physiological stage of the cells, and an important disadvantage of RT-qPCR is the need for production of sufficient invA mRNA to be detected (13). In nonhost environments, Salmonella is most likely to persist in a starved and highly stressed state, which might not produce invA mRNA and would therefore not be detected using this methodology. For nearly all methods, Salmonella spp. detection in foods is usually achieved after food samples are enriched overnight (10). This step increases cell number and subsequent mRNA production.
In this study, we evaluated the effectiveness of three molecular methods (qPCR, RT-qPCR, and LAMP) for Salmonella detection targeting the invA gene or its mRNA from artificially contaminated cilantro, lettuce, parsley, spinach, tomatoes, and jalapeno peppers. The results were compared to the standard culture detection methodology (BAM) (2). These types of produce were selected because of their vulnerability to contamination with Salmonella and because of their having been implicated in numerous Salmonella outbreaks and contamination events in the last decade.
MATERIALS AND METHODS
Bacterial strains and media.
Four strains belonging to four Salmonella enterica serovars (S. enterica serovar Enteritidis strain SARB18, S. Typhimurium strain SARA9, S. Newport strain 1240H, and S. Saintpaul strain 1358H) obtained from the FDA, Center for Food Safety and Applied Nutrition (CFSAN), Division of Microbiology were used in this study for artificial contamination of produce. Strains were grown overnight in Luria-Bertani (LB) medium at 35°C with shaking (250 rpm). Inclusivity is defined here as the ability of the assay to detect the intended pathogen target in a wide range of strains belonging to the same bacterial species (i.e., Salmonella strains only). Inclusivity of the real-time PCR (qPCR) assay for Salmonella spp. invA used in the present study was demonstrated earlier for 96 Salmonella serotypes (13). An additional 81 strains belonging to the Systems and Assays for Food Examination (SAFE) collection encompassing strains of other Salmonella enterica subspecies (other than subspecies I enterica from the FDA collection) were tested further for inclusivity of qPCR and LAMP assays (see Table S1 in the supplemental material). Exclusivity is defined here as the lack of signal or a negative reaction with closely related non-Salmonella strains. Exclusivity of the qPCR and invA Salmonella LAMP assays were demonstrated earlier (13, 15).
Preparation of Salmonella inocula.
Salmonella inocula for artificial contamination of produce were prepared as described previously (13). Briefly, Salmonella cultures in the exponential growth stage were serially diluted (10-fold) in Butterfield's phosphate buffer (BPB). Dilutions containing approximately 105 CFU/ml (high inoculation level) and <10 CFU/ml (low inoculation level) were used for artificial contamination studies. CFU counts were determined in triplicate by spreading 100 μl of 1/107, 1/108, and 1/109 dilutions of each Salmonella strain onto tryptic soy agar (TSA [Difco]). The CFU count reported is the average CFU on a plate with 25 to 300 colonies.
Sample processing and artificial contamination.
Cilantro (coriander [Coriandrum sativum] leaves), lettuce (Lactuca sativa leaves), parsley (Petroselinum crispum leaves), spinach (Spinacia oleracea leaves), tomato (Solanum lycopersicum fruit), and jalapeno peppers (Capsicum annuum fruit) were obtained from local supermarkets in College Park, Maryland (Table 1). These matrices were processed essentially as described previously (13) but with some modifications. Briefly, for each isolate and produce, three 25-g portions of food were each placed aseptically into a sterile Seward stomacher bag (Seward, United Kingdom). The three 25-g portions were designated as follows: A for no inoculation, B for high-level inoculation (105 CFU/ml), and C for low-level inoculation (<101 CFU/ml). Tomatoes and peppers were chopped aseptically in a blender into sizes similar to what is present in regular, chunky salsa and then weighed before being placed into preenrichment bags. The bags were massaged gently by hand for 1 min and kept at 4°C for 2 h before enrichment with 225 ml of universal enrichment broth (Difco, BD, Sparks, MD), shaken vigorously by hand for 30 s, and incubated (without shaking) at 35 ± 1°C for 24 ± 1 h.
Table 1.
Detection of Salmonella enterica by invA qPCR, invA mRNA RT-qPCR, invA LAMP assays, and BAM in artificially contaminated produce after 24 h of enrichment
| Produce | Salmonella strain | Minimum inoculation level detected (CFU/25 g) | Positivity for presence of Salmonella spp. by: |
qPCR (Cq) DNA presenceb | Salmonella enrichment (cells/ml)c | |||
|---|---|---|---|---|---|---|---|---|
| qPCR (Cq)a | LAMP | RT-qPCR (Cq)a | BAM | |||||
| Cilantro | S. Enteritidis SARB18 | 2 | + (23.90 ± 0.42) | + | + (28.26 ± 0.40) | + | − | 8.23E7 |
| S. Newport 1240H | 3 | + (28.57 ± 0.30) | + | + (29.76 ± 0.82) | + | − | 3.77E6 | |
| S. Saintpaul 1358H | 10 | + (32.93 ± 1.17) | + | + (34.13 ± 0.36) | + | − | 2.17E5 | |
| S. Typhimurium SARA9 | 20 | + (24.24 ± 0.15) | + | + (26.85 ± 0.82) | + | − | 6.58E7 | |
| Lettuce | S. Enteritidis SARB18 | 2 | + (21.84 ± 0.16) | + | + (24.42 ± 0.42) | + | − | 3.20E8 |
| S. Newport 1240H | 14 | + (24.40 ± 0.75) | + | + (24.55 ± 0.41) | + | − | 5.92E7 | |
| S. Saintpaul 1358H | 1 | + (22.92 ± 0.32) | + | + (24.45 ± 0.02) | + | − | 1.57E8 | |
| S. Typhimurium SARA9 | 2 | + (27.18 ± 1.26) | + | + (24.79 ± 0.22) | + | − | 9.45E6 | |
| Parsley | S. Enteritidis SARB18 | 8 | + (21.62 ± 0.04) | + | + (30.16 ± 0.24) | + | − | 3.71E8 |
| S. Newport 1240H | 2 | + (24.57 ± 0.82) | + | + (32.96 ± 0.08) | + | − | 5.29E7 | |
| S. Saintpaul 1358H | 10 | + (24.33 ± 0.14) | + | + (26.30 ± 0.41) | + | − | 6.20E7 | |
| S. Typhimurium SARA9 | 2 | + (22.78 ± 0.05) | + | + (30.43 ± 0.63) | + | − | 1.72E8 | |
| Spinach | S. Enteritidis SARB18 | 1 | + (27.96 ± 0.16) | + | + (28.96 ± 1.08) | + | − | 5.65E6 |
| S. Newport 1240H | 10 | + (29.75 ± 1.07) | + | + (28.12 ± 0.39) | + | − | 1.73E6 | |
| S. Saintpaul 1358H | 1 | + (28.85 ± 0.15) | + | + (24.43 ± 0.15) | + | − | 3.14E6 | |
| S. Typhimurium SARA9 | 1 | + (24.73 ± 0.06) | + | + (22.60 ± 0.34) | + | − | 4.76E7 | |
| Tomato (round) | S. Enteritidis SARB18 | 19 | + (18.06 ± 0.10) | + | + (27.70 ± 0.12) | + | + (34.36) | 3.88E9 |
| S. Newport 1240H | 15 | + (19.27 ± 0.32) | + | + (23.30 ± 0.11) | + | − | 1.75E9 | |
| S. Saintpaul 1358H | 2 | + (17.10 ± 0.14) | + | + (25.35 ± 0.41) | + | − | 7.32E9 | |
| S. Typhimurium SARA9 | 15 | + (19.36 ± 0.08) | + | + (25.04 ± 0.30) | + | − | 1.65E9 | |
| Jalapeno pepper | S. Enteritidis SARB18 | 2 | + (26.16 ± 0.59) | + | + (25.75 ± 1.86) | + | − | 1.85E7 |
| S. Newport 1240H | 2 | + (28.33 ± 0.40) | + | + (30.51 ± 0.10) | + | − | 4.51E6 | |
| S. Saintpaul 1358H | 2 | + (24.10 ± 0.15) | + | + (33.60 ± 0.21) | + | − | 7.21E7 | |
| S. Typhimurium SARA9 | 2 | + (21.38 ± 0.13) | + | + (20.58 ± 0.46) | + | + (34.40) | 4.34E8 | |
Cq, cycle threshold was set at 1,000 fluorescent units (FU, dR) in a Stratagene Mx3005P real-time PCR machine. Cq values are given in parentheses. +, Salmonella positive by the method. −, Salmonella negative by the method.
Same RNA samples amplified by qPCR to test for DNA contamination.
The number of Salmonella cells/ml was assumed to be equivalent to the number of invA copies/ml of enrichment as calculated by qPCR.
Two 1-ml samples were taken from each bag (A, B, and C) for DNA and RNA analysis. For DNA testing, a 1-ml sample was heated at 100°C for 12 min and then centrifuged for 2 min at 16,000 × g (Eppendorf, New York). The supernatants were recovered and used for qPCR and LAMP assays. For RNA analysis, 2 volumes (2 ml) of RNA Protect reagent (Qiagen, Valencia, CA) was added to each 1-ml sample and processed as recommended by the manufacturer. This reagent is used for RNA stabilization and protection to avoid the activity of endogenous RNases that can degrade RNA during storage and extraction procedures. Samples were stored at −70°C.
Food samples were analyzed as described previously (2, 13). Briefly, after enrichment for 24 ± 1 h (day 2), 0.1-ml and 1-ml aliquots from each sample were transferred to 10 ml of Rappaport-Vassiliadis (RV) medium and to 10 ml of tetrathionate (TT) broth (Difco), respectively. RV broth samples were incubated for 24 ± 1 h at 42 ± 0.2°C, and TT broth samples were incubated for 24 ± 1 h at 43 ± 0.2°C. On day 3, tube contents were vortexed for 10 s, and 10-μl portions of the TT and RV media were streaked on bismuth sulfite (BS) agar, xylose lysine deoxycholate (XLD) agar, and Hektoen enteric (HE) agar and incubated at 35 ± 2°C for 24 ± 1 h. On day 4, the plates were examined for the presence of typical Salmonella colonies. Typical colonies were confirmed as Salmonella as described in chapter 5 of the BAM (2).
Nucleic acid extraction.
RNA extraction was done with an RNeasy minikit (Qiagen) according to the manufacturer's instructions. This kit was chosen because it has been shown to be superior to other commercial RNA extraction kits for obtaining DNA-free RNA from Salmonella cells (38). For RNA extraction, lysis was done with 100 μl of lysozyme (1 mg/ml) at room temperature for 5 min. Treatment with DNase I (Qiagen) was done at room temperature for 30 min.
Standards for qPCR and RT-qPCR.
DNA standards and RNA standards for invA were generated as described previously (13). The number of copies of the invA DNA and RNA standards was calculated by assuming an average molecular mass of 680 Da for one nucleotide of double-stranded DNA and 340 Da for one nucleotide of single-stranded RNA. The calculation was done as follows: copies per ng = (NL × 10−9)/nm where n is the length of the standard (in base pairs or nucleotides), m is the molecular mass/nucleotide (in Da), and NL is Avogadro's constant (6.02 × 1023 molecules/mol).
invA qPCR and data analysis.
All primers and probes used in this study were purchased from IDT (Coralville, IA) and are given in Table 2, and qPCRs were done with Platinum Quantitative PCR SuperMix-uracil DNA glycosylase (UDG) according to the manufacturer's instructions (Invitrogen Carlsbad, CA). This kit is a ready-to-use cocktail consisting of a 2× reaction mix (Platinum Taq polymerase, 40 mM Tris-HCl, 100 mM KCl, 6 mM MgCl2, 0.4 mM each deoxynucleoside triphosphate [dNTP], 0.8 mM dUTP, UDG, and stabilizers). This kit contains dUTP instead of dTTP and ensures that any amplified DNA will contain uracil. The UDG enzyme removes uracil residues from single- and double-stranded DNA, preventing dUTP-containing DNA amplicons from serving as a template in future PCRs (9, 25). Reaction mixtures were scaled down to a final volume of 20 μl. MgCl2 was added to the master mix to a final concentration of 5 mM. Additional Platinum Taq polymerase was supplied to give a final content of 2.5 units/reaction. The final concentrations of primers in the qPCR mix were 200 nM for the invA gene and 100 nM for the internal amplification control (IAC). The IAC is described in detail below. Each probe was added to a final concentration of 150 nM. qPCR and data analysis were done with an Mx3005P (Agilent Technologies, Inc., Santa Clara, CA) real-time PCR system. A 2-μl portion of each boiled supernatant enrichment sample was added to each qPCR tube. The qPCR conditions were as follows: 2 min at 50°C for UDG incubation, 2 min at 95°C to activate the hot-start Taq polymerase, 35 cycles of denaturation for 15 s at 95°C and, finally, primer annealing and extension for 30 s at 60°C (the acquisition of both Cy5 and Texas Red dyes was done at the end of this cycle). The term Cq is equivalent to the original CT (threshold cycle) terminology according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (6, 7).
Table 2.
Primers and probes used in this study to detect invA DNA and RNA by qPCR, RT-qPCR, and LAMP
| Method | Target | Primers and probesb | Sequence (5′–>3′) | Position on genec | Product size (bp) | Accession no.a | Reference |
|---|---|---|---|---|---|---|---|
| qPCR/RT-qPCR | invA | invA_176F | CAACGTTTCCTGCGGTACTGT | 176 | 116 | DQ644631 | 13 |
| invA_291R | CCCGAACGTGGCGATAATT | 291 | |||||
| invA-Tx_208 | TX-CTCTTTCGTCTGGCATTATCGATCAGTACCA-BHQ2 | 208 | |||||
| IAC | IACf | CTAACCTTCGTGATGAGCAATCG | 1 | 198 | FJ357008 | 8 | |
| IACr | GATCAGCTACGTGAGGTCCTAC | 198 | |||||
| IAC-Cy5 | Cy5-AGCTAGTCGATGCACTCCAGTCCTCCT-Iowa BlackRQ-Sp | 62 | |||||
| LAMP | invA | FIP | GACGACTGGTACTGATCGATAGTTTTTCAACGTTTCCTGCGG | 272 and 343 | Various sizes | M90846 | 15 |
| BIP | CCGGTGAAATTATCGCCACACAAAACCCACCGCCAGG | 364 and 442 | |||||
| F3 | GGCGATATTGGTGTTTATGGGG | 224 | |||||
| B3 | AACGATAAACTGGACCACGG | 483 | |||||
| Loop F | GACGAAAGAGCGTGGTAATTAAC | 319 | |||||
| Loop B | GGGCAATTCGTTATTGGCGATAG | 392 |
Accession number at the NCBI (http://www.ncbi.nih.gov/).
TX, Texas Red. IAC primers and probe used for the internal control (either in DNA or RNA format).
Positions on genes are given according to the accession numbers.
invA LAMP and data analysis.
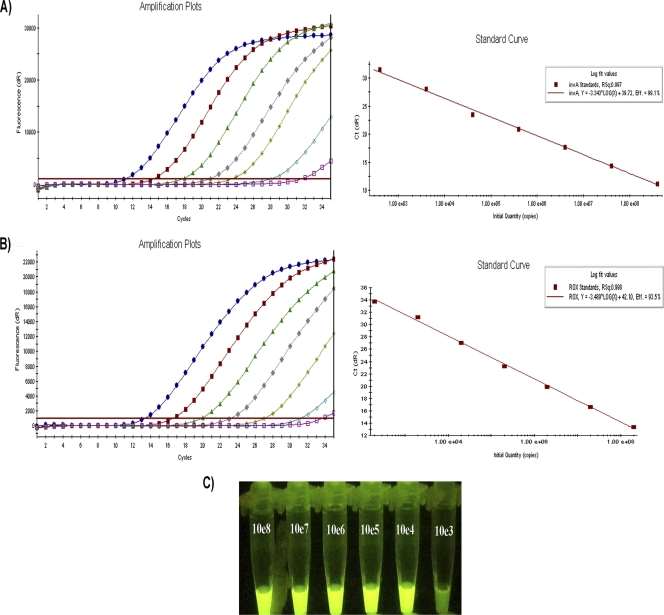
The LAMP reaction was done with a Loopamp DNA amplification kit (Eiken Chemical, Japan). The LAMP reaction mixture was prepared as described previously (15) and contained primers for Salmonella detection (Table 2), Bst polymerase (1 μl), Loopamp florescent detection reagent (1 μl), and 2 μl of template DNA solution (supernatant of the boiled enrichment sample). The reaction mixture was incubated for 1 h at 65°C in a thermal cycler (Bio-Rad, Hercules, CA) followed by 2 min at 80°C to inactivate the Bst polymerase. LAMP amplicons were detected by exposure to UV light, where positive reactions were visualized as bright green fluorescence (Fig. 1 C).
Fig. 1.
Determination of the detection limit of the RT-qPCR, qPCR, and LAMP assays. Calibration curves generated using 10-fold dilutions of DNA or RNA standards for invA (triplicates). The Cq values are plotted against the concentration of the nucleic acid target as copies/reaction for both DNA and RNA (instrument default view; Mx3005P QPCR system). To determine the reaction efficiency (E), the slope of each calibration curve was determined from the following equation: E = 10−1/slope −1. (A) Tenfold invA RNA standard dilutions amplified by RT-qPCR, including RNA IAC in each reaction tube. (B) Tenfold invA DNA standard dilutions amplified by qPCR, including DNA IAC in each reaction tube. The E and R2 values are shown. (C) LAMP amplification of 10-fold invA DNA standard dilutions: bright green, positive; light green, negative.
One-step RT-qPCR and data analysis.
RT-qPCRs were done with the master mix as described previously (13). Final concentrations of primers and probes were as indicated for qPCR. RT-qPCR and data analysis were done with an Mx3005P QPCR system real-time PCR instrument (Agilent Technologies, Inc.). The same RNA samples were not supplemented with reverse transcriptase to detect DNA contamination by qPCR. The interpretation of the results in the case of DNA contamination was as stated previously (13). Samples with differences between the Cq values for RT-qPCR and qPCR >4 cycles were considered a positive result. For values below this range, the sample was considered negative for the presence of live Salmonella cells.
Exogenous DNA and RNA internal controls.
An exogenous DNA or RNA IAC was incorporated into the qPCR and the RT-qPCR assay, respectively. Two-microliter portions of IAC DNA (1.1 pg/μl) and IAC RNA (0.5 pg/μl) were added to the qPCR and RT-qPCRs, respectively. DNA and IAC RNA were generated and processed as described previously (8). This IAC was used to assess inhibition of the qPCR or RT-qPCRs.
RESULTS AND DISCUSSION
Evaluation of the invA qPCR, RT-qPCR, and LAMP methods.
Primers and probes for qPCR and RT-qPCR protocols used in the present study for comparison of Salmonella detection in produce were as described previously (Table 2) (13). However, numerous parameters were changed: (i) a different real-time PCR instrument was used in the present study, (ii) an IAC was added to the invA qPCR, and (iii) the number of cycles for the protocols was reduced from 40 to 35. This helped to avoid artifactual signal arising from nontarget background bacteria that might have grown to high concentrations, subsequently inducing weak cross-reaction signals (12) and/or autohydrolysis of the TaqMan probe (reference 19 and our personal observation). All these changes required retesting both methods in order to determine the efficiency and detection limit of qPCR and RT-qPCR under the new conditions.
invA DNA standards generated by PCR from genomic DNA and invA RNA standards generated by in vitro transcription of invA PCR products were evaluated by qPCR and RT-qPCR (Fig. 1). Despite all the changes mentioned above, the performances of both invA qPCR and RT-qPCR assays in the Mx3005P QPCR system real-time PCR instrument (Agilent Technologies, Inc.) were similar to what was observed for the Rotor-Gene 3000 (Corbett) real-time PCR instrument (13), with linear calibration curves having a correlation coefficient (R2) of ≥0.99 and linear ranges of ≥7 orders of magnitude for both invA DNA and RNA (Fig. 1). However, owing to the reduction of cycles from 40 to 35, the detection limit was hindered 10-fold, with detection limits of 100 and 400 copies for invA DNA and RNA, respectively. The efficiencies of the RT-qPCR and qPCR were 0.99 and 0.94, respectively. This latter property was different from what was observed with the Rotor-Gene, where qPCR had a higher efficiency (0.96) whereas that of RT-qPCR was lower. These differences might be related to the fact that an IAC was included in both types of assays this time.
The robustness of DNA and RNA IAC was observed for all dilutions tested (see Fig. S1 in the supplemental material). The decreased detection limit of qPCR and RT-qPCR assays compared to previous findings (13) was caused by the reduction from 40 to 35 reaction cycles for both assays. The inclusion of an IAC in qPCR or RT-qPCRs has become obligatory to rule out the presence of PCR inhibitors that can cause false-negative results for Salmonella-positive samples (16, 17). Regardless of these results, both assays retained an efficiency of >0.90 and can be considered equivalent within the range recommended for a good quantitative qPCR (6, 7).
The six primers targeting the invA gene and assay protocol in the LAMP assay were as described previously (15); however, the assay used a real-time turbidimeter. In this study, LAMP products were visualized by fluorescence, which might result in a reduced detection limit for the assay; therefore, we tested the detection limit of the LAMP protocol under our laboratory conditions. Using the same six primers for LAMP (Table 2), we found that the detection limit was in effect lower than that reported previously (15). In our case, the invA LAMP detection limit was 104 CFU/reaction instead of 2 CFU/reaction as reported previously (15) (Fig. 1C). This reduction of the detection limit could be the result of using visual determination by fluorescence instead of turbidimetry. The LAMP assay detection limit was lower than those of the two qPCR methods, but we compared its performance in detecting Salmonella in produce with the other methods because the testing was done with enriched samples that usually contain Salmonella at levels much higher than the detection limit of the LAMP assay.
Specificity of the invA qPCR TaqMan and invA LAMP assay.
The specificity of the invA qPCR TaqMan has been reported previously (13); nonetheless, we tested its specificity by including a panel of 81 additional Salmonella strains (representing the seven Salmonella enterica subspecies and another Salmonella species, Salmonella bongori) (see Table S1 in the supplemental material). The earlier tests included only a few strains of other S. enterica subspecies (II [three strains], IIIa [one strain], IIIb [one strain], and IV [one strain], along with one S. bongori strain) (13). The positive qPCR results obtained with these additional strains, in our opinion, reenforce the conclusions in the earlier publication and further demonstrate the broad spectrum of inclusivity of our invA qPCR assay. For the LAMP assay, we tested 191 Salmonella strains, including 110 previously tested by qPCR (13), and an additional 81 from this study (see Table S1). invA LAMP inclusivity was evaluated earlier with only 39 Salmonella serovars belonging to Salmonella subsp. enterica (I) and seven isolates from Salmonella enterica subsp. arizonae (IIIa) (15). In this study, we increased assay inclusivity to 89 serovars as well as all other Salmonella enterica subspecies. All Salmonella strains were identified correctly by invA qPCR (100% inclusivity), and all but two were detected as positive by invA LAMP (99% inclusivity) (see Table S1). These two strains are likely to possess some nucleotide changes in the target region of the LAMP primers that preclude the amplification of that region. They were positive by qPCR using primers and probe targeting another region of the invA gene. The exclusivity of the LAMP assay was examined by testing 48 non-Salmonella species (13), and this examination did not produce any positive signal (data not shown).
Application of invA qPCR, RT-qPCR, and LAMP to Salmonella detection in six different produce commodities.
To our knowledge, the invA LAMP assay has been used for Salmonella detection in eggs (15, 34) and pork (40), whereas phoP LAMP (using the phoP gene as the target) was used for detection of Salmonella in milk and minced pork (24). However, the phoP gene is present in most Enterobacteriaceae, and unspecific amplification might occur when testing environmental samples. By contrast, the invA gene is a target present only in Salmonella spp. (23) and, therefore, is more specific than the phoP gene. Thus, invA LAMP was chosen for testing and comparison instead of phoP LAMP.
In order to assess the performance of the qPCR, RT-qPCR, and LAMP detection assays in this study, six different produce commodities were artificially contaminated with four different Salmonella enterica strains (representing four serovars) at levels of 105 and <10 CFU/25 g (Table 1). After enrichment for 24 h, the samples were analyzed for the presence of Salmonella using the qPCR, RT-qPCR, LAMP, and BAM methods (14). Samples of each produce commodity that were not inoculated were used as negative controls. All samples artificially contaminated at a high level were positive for Salmonella using the four methods. Only the lower level of inoculation is shown in order to compare the performance of the three molecular detection methods with the culture method (Table 1). Salmonella levels as low as 1 CFU/25 g (e.g., lettuce) were detected after enrichment for 24 ± 2 h. The absence of qPCR and RT-qPCR inhibitors was demonstrated by amplification of the DNA or RNA IACs in all samples, respectively (data not shown). Although the LAMP assay detection limit was low in pure culture (104 invA CFU/tube), it was able to successfully detect Salmonella in all the samples that were positive by both qPCR and RT-qPCR assays.
In all RT-qPCR assays, the same RNA samples were amplified by qPCR to test for DNA contamination. Most of the samples had no amplification; however, some samples showed a high Cq value indicative of the presence of small amounts of DNA contamination (e.g., tomato for S. Enteritidis SARB18). These data indicate that DNA contamination was negligible throughout this study. Rather than performing replicates of several inoculations using the same strain, we opted to spike the six produce matrices with four different Salmonella serovars, which is a more powerful approach. The ultimate goal of the assay was to detect numerous disparate serovars. Therefore, increasing the biocomplexity of the test provided a more rigorous challenge to the ability of each method to detect Salmonella.
Detection of Salmonella spp. in foods is usually done after food samples have been enriched overnight at 37°C (10), and the methods compared here are intended for an initial screening for the presence of Salmonella in produce after enrichment for 24 h. However, the main drawback of qPCR and LAMP methodologies is the reliance on the detection of DNA (9), which can be done even after the target cells are dead: the FDA cannot take regulatory action unless the Salmonella cells are shown to be viable, but the BAM protocol can take several days to complete. Using invA RT-qPCR reduces the testing time, and the recovery of isolates can be improved because only positive samples are analyzed further. The LAMP assay contains no IAC and, therefore, the presence of inhibitors in the samples cannot be monitored. By contrast, qPCR and RT-qPCR both have an IAC in the reaction.
Conclusions.
The results showed that Salmonella spp. were detected successfully on the six items of produce tested (cilantro, lettuce, parsley, spinach, tomato, and jalapeno pepper) by the molecular detection methods qPCR, RT-qPCR, and LAMP. The efficiency of these methods was equivalent to that of the conventional BAM method. RT-qPCR has several advantages over the other two molecular methods. All three molecular methods have the potential to be used as an initial screening step, but only RT-qPCR has the potential to be used as a preliminary screening tool detecting solely viable Salmonella cells. However, even if a positive result is obtained by RT-qPCR it does not eliminate the need for the BAM method to obtain a physical isolate for regulatory purposes. RT-qPCR is an inexpensive assay that will reduce time and resources expended in the laboratory, because only positive samples (indicative of viable cells) will be processed after the enrichment step (13). We have tested this RNA-based assay with several matrices, and it has been reported that the invA mRNA-based assay detected live Salmonella cells in pork (41). Nonetheless, we recommend an assessment of this technique for use with any new food matrix. Collaborative studies should be undertaken to assess the interlaboratory reproducibility of this much-needed RNA-based assay if it is to be used extensively for detection of viable Salmonella spp. on produce.
Supplementary Material
ACKNOWLEDGMENT
This project was supported by the FDA Foods Program Intramural Funds.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Altekruse S. F., Cohen M. L., Swerdlow D. L. 1997. Emerging foodborne diseases. Emerg. Infect. Dis. 3:285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrews W. A., Hammack T. S. December 2007, posting date. Salmonella. In Jackson G. J., et al. (ed.), Bacteriological analytical manual online. Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, Gaithersburg, MD: http://www.cfsan.fda.gov/∼ebam/bam-5.html [Google Scholar]
- 3. Anonymous. 2010. State-specific trends in fruit and vegetable consumption among adults — United States, 2000–2009. MMWR Morb. Mortal. Wkly. Rep. 59:1125–1130 [PubMed] [Google Scholar]
- 4. Bohaychuk V. M., Gensler G. E., McFall M. E., King R. K., Renter D. G. 2007. A real-time PCR assay for the detection of Salmonella in a wide variety of food and food-animal matrices. J. Food Prot. 70:1080–1087 [DOI] [PubMed] [Google Scholar]
- 5. Bureau of the Census, U.S. Department of Commerce 1996. Per capita utilization of selected commercially produced fresh fruits and vegetables: 1970 to 1994, p. 148 Statistical Abstract of the United States. U.S. Government Printing Office, Washington, DC [Google Scholar]
- 6. Bustin S. A., et al. 2010. MIQE precis: practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol. Biol. 11:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bustin S. A., et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 8. Deer D. M., Lampel K. A., Gonzalez-Escalona N. 2010. A versatile internal control for use as DNA in real-time PCR and as RNA in real-time reverse transcription PCR assays. Lett. Appl. Microbiol. 50:366–372 [DOI] [PubMed] [Google Scholar]
- 9. Drahovska H., et al. 2001. Detection of Salmonella by PCR targeted to fimC gene. Biologia 56:611–616 [Google Scholar]
- 10. Feder I., et al. 2001. Comparison of cultivation and PCR-hybridization for detection of Salmonella in porcine fecal and water samples. J. Clin. Microbiol. 39:2477–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fey A., et al. 2004. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl. Environ. Microbiol. 70:3618–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frahm E., Obst U. 2003. Application of the fluorogenic probe technique (TaqMan PCR) to the detection of Enterococcus spp. and Escherichia coli in water samples. J. Microbiol. Methods 52:123–131 [DOI] [PubMed] [Google Scholar]
- 13. Gonzalez-Escalona N., et al. 2009. Detection of live Salmonella sp. cells in produce by a TaqMan-based quantitative reverse transcriptase real-time PCR targeting invA mRNA. Appl. Environ. Microbiol. 75:3714–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammack T. S., Valentin-Bon I. E., Jacobson A. P., Andrews W. H. 2004. Relative effectiveness of the Bacteriological Analytical Manual method for the recovery of Salmonella from whole cantaloupes and cantaloupe rinses with selected preenrichment media and rapid methods. J. Food. Prot. 67:870–877 [DOI] [PubMed] [Google Scholar]
- 15. Hara-Kudo Y., Yoshino M., Kojima T., Ikedo M. 2005. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253:155–161 [DOI] [PubMed] [Google Scholar]
- 16. Hartman L. J., Coyne S. R., Norwood D. A. 2005. Development of a novel internal positive control for Taqman based assays. Mol. Cell. Probes 19:51–59 [DOI] [PubMed] [Google Scholar]
- 17. Hoorfar J., et al. 2004. Diagnostic PCR: making internal amplification control mandatory. Lett. Appl. Microbiol. 38:79–80 [DOI] [PubMed] [Google Scholar]
- 18. Jacobsen C. S., Holben W. E. 2007. Quantification of mRNA in Salmonella sp seeded soil and chicken manure using magnetic capture hybridization RT-PCR. J. Microbiol. Methods 69:315–321 [DOI] [PubMed] [Google Scholar]
- 19. Jalal H., et al. 2006. Development and validation of a rotor-gene real-time PCR assay for detection, identification, and quantification of Chlamydia trachomatis in a single reaction. J. Clin. Microbiol. 44:206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones T. F., Gerber D. E. 2001. Perceived etiology of foodborne illness among public health personnel. Emerg. Infect. Dis. 7:904–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klontz K. C., Klontz J. C., Mody R. K., Hoekstra R. M. 2010. Analysis of tomato and jalapeno and Serrano pepper imports into the United States from Mexico before and during a national outbreak of Salmonella serotype Saintpaul infections in 2008. J. Food Prot. 73:1967–1974 [DOI] [PubMed] [Google Scholar]
- 22. Krascsenicsova K., Piknova L., Kaclikova E., Kuchta T. 2008. Detection of Salmonella enterica in food using two-step enrichment and real-time PCR. Lett. Appl. Microbiol. 46:483–487 [DOI] [PubMed] [Google Scholar]
- 23. Li J., et al. 1995. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc. Natl. Acad. Sci. U. S. A. 92:7252–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X., et al. 2009. A loop-mediated isothermal amplification method targets the phoP gene for the detection of Salmonella in food samples. Int. J. Food Microbiol. 133:252–258 [DOI] [PubMed] [Google Scholar]
- 25. Longo M. C., Berninger M. S., Hartley J. L. 1990. Use of uracil DNA glycosylase to control carry-over contamination in PCRs. Gene 93:125–128 [DOI] [PubMed] [Google Scholar]
- 26. Lu Y. X., et al. 2009. Specific detection of viable Salmonella cells by an ethidium monoazide-loop mediated isothermal amplification (EMA-LAMP) method. J. Health Sci. 55:820–824 [Google Scholar]
- 27. Malorny B., Bunge C., Helmuth R. 2007. A real-time PCR for the detection of Salmonella Enteritidis in poultry meat and consumption eggs. J. Microbiol. Methods 70:245–251 [DOI] [PubMed] [Google Scholar]
- 28. Malorny B., et al. 2004. Diagnostic real-time PCR for detection of Salmonella in food. Appl. Environ. Microbiol. 70:7046–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mody R. K., et al. 2011. National outbreak of Salmonella serotype saintpaul infections: importance of Texas restaurant investigations in implicating jalapeno peppers. PLoS One 6:e16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mori Y., Nagamine K., Tomita N., Notomi T. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150–154 [DOI] [PubMed] [Google Scholar]
- 31. Nde C. W., Fakhr M. K., Doetkott C., Logue C. M. 2008. An evaluation of conventional culture, invA PCR, and the real-time PCR iQ-Check kit as detection tools for Salmonella in naturally contaminated premarket and retail turkey. J. Food Prot. 71:386–391 [DOI] [PubMed] [Google Scholar]
- 32. Notomi T., et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Novinscak A., Surette C., Filion M. 2007. Quantification of Salmonella spp. in composted biosolids using a TaqMan qPCR assay. J. Microbiol. Methods 70:119–126 [DOI] [PubMed] [Google Scholar]
- 34. Ohtsuka K., Yanagawa K., Takatori K., Hara-Kudo Y. 2005. Detection of Salmonella enterica in naturally contaminated liquid eggs by loop-mediated isothermal amplification, and characterization of Salmonella isolates. Appl. Environ. Microbiol. 71:6730–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okamura M., et al. 2008. Loop-mediated isothermal amplification for the rapid, sensitive, and specific detection of the O9 group of Salmonella in chickens. Vet. Microbiol. 132:197–204 [DOI] [PubMed] [Google Scholar]
- 36. Okamura M., et al. 2009. Rapid, sensitive, and specific detection of the O4 group of Salmonella enterica by loop-mediated isothermal amplification. Avian Dis. 53:216–221 [DOI] [PubMed] [Google Scholar]
- 37. Rodriguez-Lazaro D., Hernandez M., Esteve T., Hoorfar J., Pla M. 2003. A rapid and direct real time PCR-based method for identification of Salmonella spp. J. Microbiol. Methods 54:381–390 [DOI] [PubMed] [Google Scholar]
- 38. Rump L. V., Asamoah B., Gonzalez-Escalona N. 2010. Comparison of commercial RNA extraction kits for preparation of DNA-free total RNA from Salmonella cells. BMC Res. Notes 3:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scallan E., et al. 2011. Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Techathuvanan C., Draughon F. A., D'Souza D. H. 2010. Loop-mediated isothermal amplification (LAMP) for the rapid and sensitive detection of Salmonella Typhimurium from pork. J. Food Sci. 75:M165–M172 [DOI] [PubMed] [Google Scholar]
- 41. Techathuvanan C., Draughon F. A., D'Souza D. H. 2010. Real-time reverse transcriptase PCR for the rapid and sensitive detection of Salmonella typhimurium from pork. J. Food Prot. 73:507–514 [DOI] [PubMed] [Google Scholar]
- 42. Tirado C., Schmidt K. 2001. WHO surveillance programme for control of foodborne infections and intoxications: preliminary results and trends across greater Europe. J. Infect. 43:80–84 [DOI] [PubMed] [Google Scholar]
- 43. Valasek M. A., Repa J. J. 2005. The power of real-time PCR. Adv. Physiol. Educ. 29:151–159 [DOI] [PubMed] [Google Scholar]
- 44. Wang L., Shi L., Alam M. J., Geng Y., Li L. 2008. Specific and rapid detection of foodborne Salmonella by loop-mediated isothermal amplification method. Food Res. Int. 41:69–74 [Google Scholar]
- 45. Way J. S., et al. 1993. Specific detection of Salmonella spp. by multiplex PCR. Appl. Environ. Microbiol. 59:1473–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wolffs P. F., Glencross K., Thibaudeau R., Griffiths M. W. 2006. Direct quantitation and detection of salmonellae in biological samples without enrichment, using two-step filtration and real-time PCR. Appl. Environ. Microbiol. 72:3896–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang J. L., et al. 2010. Simple and rapid detection of Salmonella serovar Enteritidis under field conditions by loop-mediated isothermal amplification. J. Appl. Microbiol. 109:1715–1723 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.