Abstract
“Candidatus Liberibacter asiaticus” is a psyllid-transmitted, phloem-limited alphaproteobacterium and the most prevalent species of “Ca. Liberibacter” associated with a devastating worldwide citrus disease known as huanglongbing (HLB). Two related and hypervariable genes (hyvI and hyvII) were identified in the prophage regions of the Psy62 “Ca. Liberibacter asiaticus” genome. Sequence analyses of the hyvI and hyvII genes in 35 “Ca. Liberibacter asiaticus” DNA isolates collected globally revealed that the hyvI gene contains up to 12 nearly identical tandem repeats (NITRs, 132 bp) and 4 partial repeats, while hyvII contains up to 2 NITRs and 4 partial repeats and shares homology with hyvI. Frequent deletions or insertions of these repeats within the hyvI and hyvII genes were observed, none of which disrupted the open reading frames. Sequence conservation within the individual repeats but an extensive variation in repeat numbers, rearrangement, and the sequences flanking the repeat region indicate the diversity and plasticity of “Ca. Liberibacter asiaticus” bacterial populations in the world. These differences were found not only in samples of distinct geographical origins but also in samples from a single origin and even from a single “Ca. Liberibacter asiaticus”-infected sample. This is the first evidence of different “Ca. Liberibacter asiaticus” populations coexisting in a single HLB-affected sample. The Florida “Ca. Liberibacter asiaticus” isolates contain both hyvI and hyvII, while all other global “Ca. Liberibacter asiaticus” isolates contain either one or the other. Interclade assignments of the putative HyvI and HyvII proteins from Florida isolates with other global isolates in phylogenetic trees imply multiple “Ca. Liberibacter asiaticus” populations in the world and a multisource introduction of the “Ca. Liberibacter asiaticus” bacterium into Florida.
INTRODUCTION
Citrus huanglongbing (HLB), also known as citrus greening, is one of the most destructive diseases of citrus. Occurring in most of the global citrus-producing regions, HLB affects all citrus cultivars by causing rapid decline and shortening the life span of infected trees (6, 18, 22). From the first documentation in the early 20th century in China (55) to the recent finding in Sao Paulo (44) and Florida (8), this century-old disease has been a major problem for citrus production, especially in the top-producing countries: China, Brazil, and the United States (19). The causal agents of the disease are believed to be three species of Alphaproteobacteria in the genus of “Candidatus Liberibacter,” namely, “Ca. Liberibacter asiaticus,” “Ca. Liberibacter americanus,” and “Ca. Liberibacter africanus.” These fastidious bacteria reside in the phloem of the plant hosts (25) and are vectored by the citrus psyllids Diaphorina citri and/or Trioza erytreae (6, 22). Of the three bacterial species, “Ca. Liberibacter asiaticus” is the most prevalent and draws the most research attention worldwide.
As an obligate plant pathogen and insect symbiont, “Ca. Liberibacter asiaticus” has a significantly reduced genome with 1.23 Mb (16). However, “Ca. Liberibacter asiaticus” bacteria have evolved into two distinct ecological niches, plant and insect. This small-genome bacterium not only multiplies in most if not all citrus species, citrus hybrids in the genus of Citrus, and many other members of the Rutaceae family as well as in periwinkle of the Apocynaceae family (22) and tomato in the Solanaceae family (15) but also multiplies in psyllids, reaching 1010 bacteria per individual insect (16). Disease symptoms vary with bacterial titers in HLB-affected citrus and periwinkle plants both in the field and in the greenhouse (57). These differences were noted within the same variety of citrus and periwinkle plants, suggesting that the genetic variations of the pathogen may play important roles in these host-microbe interactions (57).
Annotation of the “Ca. Liberibacter asiaticus” Psy62 genome did not reveal any known transposon or insertion sequence (IS) elements (16). The potential contributions of prophage and phage-related sequences to the genetic diversity of “Ca. Liberibacter asiaticus” were then investigated. Prophages can constitute as much as 10 to 20% of a bacterium's genome and are major contributors to genetic variability, including virulence factors and pathogenicity islands between individuals within species (10). Consequently, prophage DNA can play an important role in the evolution of bacterial pathogenicity (7). In silico analyses revealed multiple regions of the “Ca. Liberibacter asiaticus” Psy62 genome contained prophage-related sequences, and two of them were identified as prophages in psyllids and as temperate phage in plants, which were similar to those described in 2011 by Zhang et al. (54). In these prophage regions, two candidate genes were identified and tentatively named hyvI and hyvII.
Little was known about the genetic diversity of the HLB bacterial pathogens since the bacteria were not cultivable in vitro and limited sequence data were available before the recent publication of the complete sequence of the “Ca. Liberibacter asiaticus” genome (16). Prior to the availability of the “Ca. Liberibacter asiaticus” genome, diversity studies were restricted to 16S/23S rRNA genes, the omp gene region, or the rplKAJL-rpoBC operon sequence (3, 12, 17, 26, 38, 42, 49, 57). The genetic variation in these housekeeping genes was limited, and most consisted of single-nucleotide polymorphisms (SNPs). However, since the publication of the “Ca. Liberibacter asiaticus” genome sequences, a number of loci including simple repeat sequences (SSRs) and prophage genes have been demonstrated to be the better markers for differentiating the “Ca. Liberibacter asiaticus” isolates from different geographic origins, including Southeast Asia (47), China versus Florida (11), Japan, Taiwan, Indonesia (28), and two citrus-growing provinces in China (33).
Here we present characteristics and phylogenetics of two closely related prophage genes (hyvI and hyvII) cloned from “Ca. Liberibacter asiaticus” isolates in different hosts and geographic origins. Hypervariations in these two genes among “Ca. Liberibacter asiaticus” isolates or within a single isolate imply a potentially important mechanism of adaptation available to “Ca. Liberibacter asiaticus,” although the functions of these genes remain to be determined.
MATERIALS AND METHODS
Bacterial strains, plasmid, and media.
TOPO TA vector pCR2.1 (Invitrogen, Carlsbad, CA) was used for cloning PCR products containing the hyvI and hyvII genes. Escherichia coli Top10 cells (Invitrogen) were used to maintain the TOPO cloning plasmid. Individual clones of E. coli were grown in Luria-Bertani (LB) medium supplemented with 50 mg/ml ampicillin or 50 mg/ml kanamycin at 37°C (40).
“Ca. Liberibacter asiaticus”-infected samples and DNA extraction.
A total of 266 DNA extracts from “Ca. Liberibacter asiaticus”-infected hosts (112 citrus, 36 periwinkle, 20 dodder, and 98 psyllid samples) from Florida were used to characterize the hyvI and hyvII genes. Representative samples with cloning and sequencing analysis are listed below (see Table 2). DNA samples from citrus and psyllids were prepared either by our collaborators or in our laboratory. “Ca. Liberibacter asiaticus”-infected citrus samples were collected from citrus groves in different countries and across the state of Florida, including from the United States Horticulture Research Laboratory farm (Ft. Pierce, FL). Some “Ca. Liberibacter asiaticus”-infected source plants were propagated by grafting and maintained in the USHRL insect-proof greenhouse (Ft. Pierce, FL). The “Ca. Liberibacter asiaticus”-infected periwinkle plants were obtained by dodder (Cuscuta campestri) transmission from “Ca. Liberibacter asiaticus”-infected citrus, and the repeated propagation of “Ca. Liberibacter asiaticus”-infected periwinkles was obtained either by dodder transmission (56) or by graft transmission from “Ca. Liberibacter asiaticus”-infected periwinkle to healthy periwinkle (53). The “Ca. Liberibacter asiaticus”-infected psyllid DNA used for the identification of hyvI and hyvII genes was the same as that for Psy62 genome sequencing (16). All the other psyllids (Diaphorina citri) collected from “Ca. Liberibacter asiaticus”-infected citrus plants were subjected to DNA extraction or preserved in 70% ethanol for later extraction.
Table 2.
hyvI and hyvII genes in isolates of “Ca. Liberibacter asiaticus” from infected plant or psyllid DNA samples of various geographic origins
| Name of “Ca. Liberibacter asiaticus” isolate | Source | Yr of collection | Location | CT value of 16S rRNA gene- based real-time PCR |
hyvI genea |
hyvII geneb |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cloning ID | Accession no. | No. of full/partial repeatsc | Cloning ID | Accession no. | No. of full/partial repeats | |||||
| Isolates from Florida | ||||||||||
| FL-Psy62(MDA) | Diaphorina citri | 2007 | Picos Farm, Ft. Pierce | 19.44 | pLJ108.1 | YP_003084345.1 | 12/4 | HQ263703 | 0/1 | |
| FL-Psy12 | Diaphorina citri | 2007 | Picos Farm, Ft. Pierce | 19.71 | pLJ266.2 | HQ263675 | 2/1 | pLJ391.1 | HQ263704 | 0/1 |
| FL-Psy32 | Diaphorina citri | 2007 | Picos Farm, Ft. Pierce | 19.41 | pLJ267.3 | HQ263676 | 11/3 | pLJ394.1 | HQ263705 | 0/1 |
| FL-14879 | Citrus sp. | 2008 | Hardee | 28.92 | pLJ148.9 | HQ263677 | 1/6 | NC | NC | |
| FL-15704 | Citrus aurantiifolia | 2008 | Osceola | 21.26 | pLJ150.9 | HQ263678 | 10/4 | NC | NC | |
| pLJ150.3 | HQ263679 | 0/1 | ||||||||
| FL-R8T1 | Citrus maxima | 2007 | Picos Farm, Ft. Pierce | 27.54 | pLJ144.2 | HQ263680 | 6/5 | NC | NC | |
| FL-CG3MV | Citrus sinensis | 2008 | Greenhouse, Ft. Pierce | 23.95 | pLJ153.1 | HQ263681 | 1/1 | NC | NC | |
| FL-CG8-S | Citrus limon | 2008 | Greenhouse, Ft. Pierce | 21.36 | pLJ169.2 | HQ263682 | 12/4 | pLJ399 | HQ263706 | 0/1 |
| FL-CG8-R | Poncirus trifoliata | 2008 | Greenhouse, Ft. Pierce | 18.85 | pLJ171.6 | HQ263683 | 5/3 | pLJ400 | HQ263707 | 0/1 |
| FL-P1-B1 | Catharanthus roseus | 2008 | Greenhouse, Ft. Pierce | 19.05 | pLJ158.14 | HQ263684 | 12/4 | NC | NC | |
| FL-P1-B2 | Catharanthus roseus | 2009 | Greenhouse, Ft. Pierce | 19.12 | pLJ163.1 | HQ263685 | 2/1 | pLJ233 | HQ263708 | 0/1 |
| FL-PP11-B1 | Catharanthus roseus | 2009 | Greenhouse, Ft. Pierce | 18.94 | pLJ168.9 | HQ263686 | 10/3 | pLJ401.1 | HQ263709 | 2/4 |
| pLJ401.3 | HQ263710 | 0/1 | ||||||||
| FL-PP11-B2 | Catharanthus roseus | 2009 | Greenhouse, Ft. Pierce | 19.23 | NC | NC | pLJ392 | HQ263711 | 0/1 | |
| FL-PG15 | Catharanthus roseus | 2008 | Greenhouse, Ft. Pierce | 18.86 | pLJ157.1 | HQ263687 | 12/4 | NC | NC | |
| pLJ157.3 | HQ263688 | 0/1 | ||||||||
| FL-Dod-citrus | Cuscuta campestri | 2008 | Greenhouse, Ft. Pierce | 20.86 | NC | NC | NC | NC | ||
| FL-Dod-periwinkle | Cuscuta campestri | 2008 | Greenhouse, Ft. Pierce | 18.37 | NC | NC | pLJ395.3 | HQ263712 | 0/1 | |
| FL-Dod-tobacco | Cuscuta campestri | 2007 | Greenhouse, Ft. Pierce | 17.87 | NC | NC | NC | NC | ||
| FL-Dod-potato | Cuscuta campestri | 2009 | Greenhouse, Ft. Pierce | 20.53 | pLJ161.5 | HQ26389 | 12/4 | pLJ396.2 | HQ263713 | 2/4 |
| pLJ396.14 | HQ263714 | 0/1 | ||||||||
| Isolates from China | ||||||||||
| CHA-Cit4 | Citrus reticulata | 2009 | Fujian | 25.56 | pLJ341.1 | HQ263690 | 4/4 | − | − | |
| CHA-Cit5 | Citrus sp. | 2009 | Fujian | 25.49 | − | − | pLJ393.1 | HQ263715 | 1/0 | |
| CHA-Cit18 | Citrus reticulata | 2009 | Fujian | 26.65 | pLJ316.1 | HQ263691 | 7/2 | − | − | |
| CHA-Psy7 | Diaphorina citri | 2009 | Fujian | 22.36 | − | − | pLJ225 | NC | 1/0 | |
| CHA-Psy42 | Diaphorina citri | 2009 | Fujian | 24.24 | pLJ222.1 | HQ263692 | 7/2 | − | − | |
| TW-Psy17 | Diaphorina citri | 2009 | Taiwan | 27.07 | pLJ402.1 | HQ263701 | 7/2 | − | − | |
| TW-Psy27 | Diaphorina citri | 2009 | Taiwan | 25.87 | pLJ405.2 | HQ263702 | 10/2 | − | − | |
| Isolates from Thailand | ||||||||||
| THA-Cit2 | Citrus sp. | 2009 | Kamphaeng Phet | 26.02 | − | − | pLJ232.1 d | HQ263716 | 3/3 | |
| pLJ232.2 d | HQ263717 | 1/13 | ||||||||
| THA-TF | Poncirus trifoliata | 2010 | Chanthaburi | 25.76 | − | − | pLJ397.1 | HQ263718 | 1/0 | |
| THA-KP2.3 | Citrus reticulata | 2010 | Chanthaburi | 24.16 | pLJ342.1 | HQ263693 | 7/2 | − | − | |
| THA-Psy25 | Diaphorina citri | 2010 | Kamphaeng Phet | 28.44 | pLJ343.1 | HQ263694 | 5/2 | − | − | |
| THA-Psy22 | Diaphorina citri | 2010 | Kamphaeng Phet | 26.30 | − | − | pLJ398.4 | HQ263719 | 1/0 | |
| Isolates from Philippines | ||||||||||
| PHA-Psy5 | Diaphorina citri | 2009 | Unknown | 18.67 | pLJ313.4 | HQ263695 | 7/2 | − | − | |
| pLJ313.5 | HQ263696 | 5/2 | ||||||||
| pLJ313.6 | HQ263697 | 1/2 | ||||||||
| Isolates from India | ||||||||||
| IND-citrus | Citrus sp. | 2009 | Unknown | 22.01 | − | − | − | − | ||
| IND-Psy1 | Diaphorina citri | 2009 | Unknown | 26.63 | pLJ315.1 | HQ263698 | 1/1 | − | − | |
| IND-Psy2 | Diaphorina citri | 2009 | Unknown | 24.66 | pLJ314.1 | HQ263699 | 4/1 | pLJ286.1 | 0/0 | |
| Isolate from Brazil | ||||||||||
| Bra-Cit1 | Citrus sp. | 2007 | São Paulo | 23.96 | pLJ236.1 | HQ263700 | 7/2 | − | − | |
The hyvI gene was cloned from the LJ729/LJ730 PCR product.
The hyvII gene was cloned from the LJ812/LJ1089 PCR product.
The number of full and partial repeats in hyvI and hyvII genes with the dominant size in each clone library.
Cloning from LJ776/730 PCR product: NC, PCR positive but not cloned; −, PCR negative by all hyvI and hyvII primers.
Total DNA was extracted from dodder haustoria or midribs of citrus and periwinkle as described in 2006 by Irey et al. (24) with some modifications. Briefly, 0.1 g dodder or periwinkle tissue or 0.2 g citrus tissue was cut and transferred to an autoclaved 2-ml screw-cap tube containing 1 silicone-carbide sharp particle (4 mm in diameter) and 3 chrome-steel beads (2.3 mm in diameter) in 800 μl of extraction buffer (100 mM Tris base, 50 mM EDTA, 500 mM NaCl, 2.5% polyvinylpyrrolidone, and 10 mM β-mercaptoethanol); homogenized by a Fast Prep-24 homogenizer (MP Bio., Solon, OH) at speed 4.0 for 60 s; and incubated at 65°C for 30 min after addition of 1.5% SDS (vol/vol). After adding 1/3 volume of 5 M potassium acetate, the tube was incubated on ice for 20 min and centrifuged at 14,000 × g at 4°C for 5 min to remove plant debris. The supernatant was centrifuged for another 10 min at the same speed, and 800 μl of supernatant was transferred to a new 1.5-ml tube containing 2/3 volume of cold isopropanol. The resulting DNA pellet was washed once with 70% ethanol and then resuspended in 100 μl of sterile water. DNA from individual psyllids was extracted as described previously (16).
Real-time PCR.
TaqMan real-time PCR amplifications were performed in a 7500 real-time PCR system (Applied Biosystems, Foster City, CA), using primers HLBasf and HLBr and probe HLBp targeting 16S rRNA genes of “Ca. Liberibacter asiaticus” as listed in Table 1 (31). PCR mixtures with a total volume of 15 μl contained 7.5 μl of TaqMan PCR master mix (Applied Biosystems), 250 nM each primer, 150 nM probe, and 100 ng of template DNA. Primer LJ900fr and probe LJ900p (Table 1) targeting the tandem repeats of hyvI and hyvII genes were employed in the same real-time system, following the protocol described elsewhere (J. K. Morgan et al., submitted for publication). Real-time PCR data were analyzed with Applied Biosystems 7500 system SDS version 1.2 software.
Table 1.
Primers and probes used in this study
| Name | Sequence (5′–3′) | Target gene or flanking region | Source or reference | |
|---|---|---|---|---|
| Conventional PCR primers | ||||
| LJ729 | TTGCGACTAAAGACAACGAG | 5′ flanking region of hyvI gene | This study | |
| LJ730 | TTGCTAGTCTTATCGGCTTATC | 3′ flanking region of hyvI and hyvII genes | This study | |
| LJ788 | GCCGTTTTCTGAAGGATAAGC | hyvI gene | This study | |
| LJ776 | TGAAGTTAAATATCCTGATGGCAAC | hyvII gene | This study | |
| LJ812 | CCACGGAATACATCAAAGCTC | 5′ flanking region of hyvII gene | This study | |
| LJ1089 | TTAGTCATCAAAATTAATAAC | hyvII gene | This study | |
| Real-time PCR primers and probes | ||||
| LJ900f | GCCGTTTTAACACAAAAGATGAATATC | hyvI and hyvII | This study | |
| LJ900r | ATAAATCAATTTGTTCTAGTTTACGAC | hyvI and hyvII | This study | |
| LJ900pa | ACATCTTTCGTTTGAGTAGCTAGATCATTGA | hyvI and hyvII | This study | |
| HLBasfb | TCGAGCGCGTATGCGAATACG | 16S rRNA genes | 31 | |
| HLBr | GCGTTATCCCGTAGAAAAAGGTAG | 16S rRNA genes | 31 | |
| HLBpa | AGACGGGTGAGTAACGCG | 16S rRNA genes | 31 |
6-Carboxyfluorescein (FAM) at 5′ end; Iowa Black FQ at 3′ end.
Additional “G” (boldface underlined) was added in the HLBas primer sequence (31) based on 16S rRNA gene sequence in the “Ca. Liberibacter asiaticus” Psy62 genome, and this modified primer was named HLBasf.
Conventional PCR.
All the primers used in this study are listed in Table 1. Primers targeting hyvI and hyvII or their flanking regions were designed using primer analysis software Oligo 7.23 (Molecular Biology Insights, Inc., Cascade, CO). LJ729/LJ730 were used to amplify a 3,513-bp fragment including the hyvI full gene and its flanking region; LJ729/LJ788 were designed for the conserved but partial hyvI region of all “Ca. Liberibacter asiaticus” isolates; LJ776/LJ730 or LJ812/LJ1089 were used to amplify hyvII gene and its flanking region. To reduce the error rate in sequencing the hyvI and hyvII genes, high-fidelity platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) was used. One unit of the polymerase was added to a 20-μl reaction mixture containing 1× high-fidelity PCR buffer, 0.2 mM each deoxynucleoside triphosphate (dNTP), 2.0 mM MgSO4, 250 nM forward/reverse primer, and 1.5 μl template DNA. The PCR cycles were started with denaturation at 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 54°C for 30 s, 68°C for 3.5 min, and a final extension at 68°C for 10 min. For regular PCR testing, 10 μl of 2× buffer D (Epicentre Biotechnologies, Madison, WI), 250 nM forward/reverse primer, 1.25 U Taq DNA polymerase (New England BioLabs Inc., Ipswich, MA), and 1 to 2 μl template DNA were added to a 20-μl reaction mixture. The PCR cycles were started with 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 50 to 54°C for 30 s, 72°C for 2.0 to 3.5 min for different primer sets, and a final extension at 72°C for 10 min.
Cloning, sequencing, and sequence assembly.
PCR products containing the hyvI or hyvII gene from different isolates were ligated into TOPO TA vector pCR2.1 and transferred to TOPO 10 chemical competent cells following the manufacturer's protocol. Plasmid DNA was isolated from E. coli cultures by using the QIAprep Spin Miniprep kit (Qiagen). DNA sequencing was performed in the U.S. Horticulture Research Laboratory Core Genomics Facility using BigDye Terminator version 3.1 and the 3730xl DNA analyzer (Applied Biosystems). Sequences were assembled by ContigExpress of Vector NTI (Invitrogen) and analyzed by Align X in Vector NTI.
Bioinformatics and phylogenetic analysis.
Tandem repeats finder version 4.04 (4) was used to find repeats in hyvI and hyvII sequences. Bacterial gene predictions were carried out using FGENESB software (SoftBerry Inc., Mount Kisco, NY). BLASTP of WU-BLAST v 2.0 in TIGR CMR server (http://blast.jcvi.org/cmr-blast/) or PSI-BLAST in NCBI (2) was used to predict the potential function of hyvI and hyvII genes.
Phylogenetic relationships for protein sequences were inferred using parsimony and maximum likelihood (ML) methods. For all representative proteins, a multiple sequence alignment was produced in Mesquite version 2.73 using ClustalW version 2.0.12 with default settings (34, 46), followed by manual adjustments. These data were analyzed in ProtTest version 2.4 with the Akaike Information Criterion (AIC), resulting in a best-fit substitution model JTT+G+F (1, 13, 21, 27) for amino acid replacement for both HyvI and HyvII proteins. Using maximum parsimony, a heuristic search with random stepwise addition and tree bisection-reconnection (TBR) was implemented in PAUP version 4.0b10 (43). Support was assessed using neighbor-joining (NJ) bootstrapping (1,000 replicates). ML inference was obtained using RAxML v7.2.6 (41) on the CIPRES teragrid portal with default settings and JTT, followed by 800 (1,000 for HyvII) bootstrap pseudoreplicates (37). These trees were then used to construct a majority rule consensus tree in PAUP (43).
Nucleotide sequence accession numbers.
The nucleotide sequences of hyvI and hyvII genes and their flanking regions of the global isolates from various geographic origins have been deposited in GenBank under accession numbers YP_003084345.1 and HQ263675 to 263719, as listed in Table 2.
RESULTS
Identification of hyvI and hyvII genes in the “Ca. Liberibacter asiaticus” Psy62 genome.
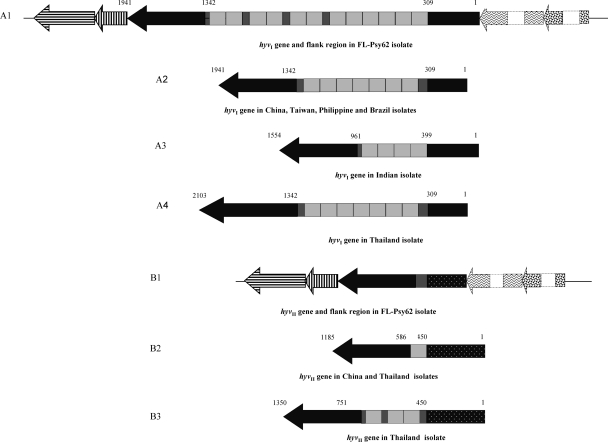
While closing gaps for the “Ca. Liberibacter asiaticus” Psy2 genome, we found two prophage regions containing a large number of phage-related genes (16, 54). Based on cloning and sequencing of a 3,513-bp DNA fragment (cloning ID pLJ108) from one of the prophage regions, one gene containing multiple nearly identical tandem repeats (NITRs) was identified and named as hyvI. The hyvI gene is 2,760 bp long and putatively encodes a 919-amino-acid (aa) acidic protein (pI = 4.54) with a molecular mass of 103.5 kDa. Using Tandem Repeats Finder (4) and manual arrangements, the intragenic tandem repeat region was identified in the hyvI gene. This region includes 12 full NITRs and 4 partial tandem repeats. Each full repeat is of 132 bp, with three partial repeats of 48 bp sitting between full repeats 6 and 7, 8 and 9, and 10 and 11. There is an additional 33-bp partial repeat at the 3′ end of the entire tandem repeat region (Fig. 1A1). The partial repeat sequences are nearly identical to the first 48 or 33 bp of the 132-bp full repeat. The similarity among 12 full repeats within the hyvI gene was 93 to 100% at the nucleic acid level (see Fig. S1A in the supplemental material) and 82 to 100% at the putative protein level (see Fig. S1B in the supplemental material).
Fig. 1.
General features of hyvI and hyvII genes (scaled) in the genome of “Candidatus Liberibacter asiaticus” from various geographic isolates. The black arrow boxes in A1 to A4 and B1 to B3 represent the open reading frames of hyvI and hyvII genes with different numbers of full or partial repeat units in the middle. Each light grey box in the middle of the gene represents a 132-bp full tandem repeat unit, and each deep grey box stands for partial repeat unit. The numbers above the hyvI and hyvII genes indicate the gene size and the start and end positions of repeat regions in the hyvI and hyvII genes. A1 and B1 are hyvI and hyvII genes with two flanking genes on both 5′ and 3′ ends in the FL-Psy62 genome. Dark horizotal- and vertical-arrow boxes in A1 and B1 represent two hypothetical genes neighboring the 3′ end of the hyvI (A1) and hyvII (B1) genes; zigzag and large-confetti arrow boxes with dashed outline boxes represent two other hypothetical genes neighboring the 5′ ends of the hyvI and hyvII genes (not to scale). The spotted box or arrow in B1 to B3 represents the sequence variations (similarity of less than 80%) compared to the sequence in same region within the hyvI gene or the flanking genes.
Based on the hyvI gene sequence, the hyvII gene was identified from another prophage region (“Ca. Liberibacter asiaticus” Psy62-FP2, 38,551 bp long, GenBank accession number: JF773396) of the “Ca. Liberibacter asiaticus” Psy62 genome. The hyvII gene is 1,026 bp long and putatively encodes a 341-aa acidic protein (pI = 5.1) with a molecular mass of 38.9 kDa. In the “Ca. Liberibacter asiaticus” Psy62 genome, hyvII contained only one partial repeat unit and shared 92% identity with hyvI on downstream sequence outside 3′ end of the repeat unit (Fig. 1B1).
hyvI and hyvII localized to homologous gene clusters within each prophage region (Fig. 1A1 and B1), and all the genes in these two clusters encoded hypothetical proteins of unknown function. Two genes from corresponding loci flanking 5′ or 3′ of the hyvI and hyvII genes were either identical or closely related at the protein level. A BLAST search of the protein database from 723 prokaryote genomes and the protein database in NCBI yielded little information on the HyvI and HyvII proteins. In the HyvI repeat region, 586 aa shared 25% identity to the leucine-rich repeat protein of Colwellia psychrerythraea 34H (GenBank accession number: AAZ26055), and 560 aa exhibited 22% identity with cell wall-associated biofilm protein of Staphylococcus epidermidis (ZP_06614153). The N-terminal (133-aa) and C-terminal (195-aa) sequences outside of the repeat region did not share a significant homology with any other protein (E-value better than the threshold) of known function in the databases.
Variations of hyvI and hyvII genes among Florida “Ca. Liberibacter asiaticus” isolates from different hosts.
A total of 265 DNA isolates were selected from different “Ca. Liberibacter asiaticus”-infected host plants and vector psyllids in Florida to evaluate the diversity of hyvI and hyvII genes. The isolates, confirmed “Ca. Liberibacter asiaticus” positive with threshold cycle (CT) values of between 17 and 29 by 16S rRNA gene-based real-time PCR (Table 2), were equivalent to “Ca. Liberibacter asiaticus” populations ranging between 2.95 × 108 and 1.08 × 105 bacterial cells/μl DNA extracts based on the grand universal regression equation Y = 13.82 − 0.2866X, where X is the mean CT value and Y is the log concentration of the target DNA copies (32). All the Florida “Ca. Liberibacter asiaticus”-positive samples tested positive for both hyvI and hyvII genes by conventional PCR using hyvI- and hyvII-specific primer sets LJ729/LJ788 and LJ812/LJ1089, respectively. All negative control DNA extracted from healthy plant and psyllid samples tested negative for “Ca. Liberibacter asiaticus” by conventional PCR.
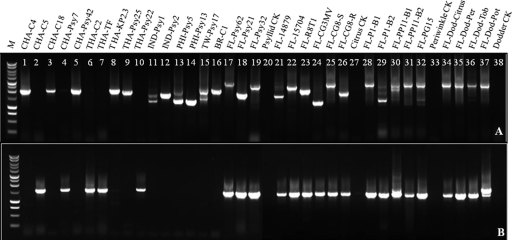
According to the hyvI gene sequence from the Psy62 genome (GenBank accession number CP001677), LJ729/LJ788 should amplify a 2,867-bp product. However, different sizes and/or multiple sizes of PCR products were observed in most of the Florida “Ca. Liberibacter asiaticus”-infected DNA samples. The hyvI amplicons showed more variation than those of hyvII (Fig. 2A and B). In addition, variation of the hyvI amplicons was greater in citrus than in periwinkle, dodder, or psyllid (Fig. 2A). Furthermore, the differences in the hyvI amplicons were observed not only among different host plants or vector psyllids but also between citrus scions and rootstocks (Fig. 2A, lanes 25 and 26) and among different periwinkle branches with the same HLB inoculum (Fig. 2A, lanes 30 and 31). Most of the samples yielded multiple weak bands in addition to the dominant band for the hyvI region (Fig. 2A). Eighteen hyvI and nine hyvII amplicons were cloned and sequenced from the “Ca. Liberibacter asiaticus” isolates of different sources. Sequence analyses confirming the presence of different sizes of amplicons belonging to hyvI or hyvII, and the clone(s) reflecting one to three dominant sizes in each library, are listed in Table 2. Further sequence analyses of these amplicons revealed that the amplicon size differences were due to changes in the repeat (full or partial) numbers and the arrangement of individual units in each hyvI gene.
Fig. 2.
PCR amplification of the hyvI and hyvII gene target regions in isolates of “Candidatus Liberibacter asiaticus” from Florida and other global origins, using primer sets LJ788/LJ729 for hyvI (A) and LJ812/LJ1089 for hyvII (B). Lane M: 1-kb DNA ladder (Promega); lanes 1 to 16, DNA from “Ca. Liberibacter asiaticus”-infected plants or psyllids of global origins outside the United States; lanes 17 to 38, DNA from “Ca. Liberibacter asiaticus”-infected different hosts in Florida; lanes 17 to 19, DNA from “Ca. Liberibacter asiaticus”-positive psyllid from field; lane 20, DNA from “Ca. Liberibacter asiaticus”-free psyllid control from greenhouse; lanes 20 to 23, DNA from “Ca. Liberibacter asiaticus”-positive citrus from field; lanes 24 to 26, DNA from “Ca. Liberibacter asiaticus”-positive citrus from greenhouse, CG8-S/CG8-R leaf samples collected from scion/rootstock of grafted citrus CG8; lane 27, “Ca. Liberibacter asiaticus”-free citrus control from greenhouse; lanes 28 to 32, “Ca. Liberibacter asiaticus”-positive periwinkle from greening, B1 and B2 from branch 1 and branch 2 in dodder-transmitted periwinkle plants P1 (lanes 28 and 29) and PP11 (lanes 30 and 31), PG from grafted-transmitted periwinkle plant; lane 33, “Ca. Liberibacter asiaticus”-free periwinkle control; lanes 34 to 37, “Ca. Liberibacter asiaticus”-positive dodder collected from citrus, periwinkle, tobacco, and potato; lane 38, “Ca. Liberibacter asiaticus”-free dodder control.
The number of full repeats varied from 1 (pLJ153) to 12 (pLJ108), and the arrangement of full and partial repeats in each hyvI gene was also highly variable (Table 2). However, all repeats were found to be in frame when the gene was translated to protein by FGENESB software. Therefore, deletion or insertion of a full or partial repeat unit did not disrupt their open reading frames in any hyvI or hyvII clone.
In comparison to hyvI amplification, hyvII PCR products were more uniform (Fig. 2B). All PCR products from HLB-affected citrus showed one single band. More than one amplicon was observed only in a periwinkle DNA sample (Fig. 2B, lane 30) and a dodder DNA sample (Fig. 2B, lane 37). The difference in hyvII amplicon sizes was also due to variation in the number of repeat units. The largest amplicon of hyvII was cloned from periwinkle FL-PP11-B1 and contained 2 full repeats and 4 partial repeats. Interestingly, periwinkle DNA sample FL-PP11-B2, isolated from a different branch of the same periwinkle plant as FL-PP11-B1, only had one additional amplicon containing one partial repeat. Similarly, multiple amplicons were observed only in one out of four dodder samples used to transmit “Ca. Liberibacter asiaticus” from HLB-affected citrus to the experimental host, potato.
Diversity of hyvI or hyvII genes in “Ca. Liberibacter asiaticus” isolates of other global origins.
“Ca. Liberibacter asiaticus” isolates were kindly provided by our collaborators from HLB-affected citrus plants and psyllids collected in China, Thailand, Philippines, India, and Brazil (Table 2). All these isolates were confirmed “Ca. Liberibacter asiaticus” positive with CT values between 18 and 29 using 16S rRNA gene-based real-time PCR (Table 2), indicating “Ca. Liberibacter asiaticus” populations between 1.53 × 1011 and 1.08 × 108 cells/gram of fresh plant tissue. Unlike the Floridian isolates evaluated, only one gene, either hyvI or hyvII, was detected in these international “Ca. Liberibacter asiaticus” isolates (Fig. 2A and B, lanes 1 through 16) regardless of citrus or psyllid origins (Table 2). The numbers of “Ca. Liberibacter asiaticus” DNA isolates from “Ca. Liberibacter asiaticus”-infected citrus and psyllid samples of global origin are listed in Table S1 in the supplemental material. DNA fragments of the hyvI gene were amplified by the primer set LJ729/LJ730 or LJ729/LJ788 from isolates of “Ca. Liberibacter asiaticus”-infected citrus or psyllids in China, Taiwan, Thailand, India, Philippines, and Brazil, while hyvII fragments were obtained by the primer set LJ776/LJ730 or LJ812/LJ1089 only from isolates collected in China and Thailand (Table 2; also see Table S1 in the supplemental material). However, using the same primer sets, neither the hyvI nor the hyvII gene was amplified from eight Indian citrus DNA samples and six Philippine psyllid DNA samples, although all these DNA samples were from HLB-symptomatic tissue and tested “Ca. Liberibacter asiaticus” positive with CT values from 17.7 to 27.8 (Table 3) by 16S rRNA gene-based real-time PCR. To further evaluate the presence or absence of the hyvI and hyvII genes in these samples, a newly developed real-time PCR assay with hyvI repeat-based primers and probe LJ900fpr was performed, and this confirmed the presence of the NITR in both the Indian citrus samples and the Philippine psyllid samples (Table 3).
Table 3.
Results of 16S rRNA gene- and hyvI- and hyvII-based TaqMan real-time PCR for DNA extracts from HLB-infected Indian citrus and Philippine psyllid samples testing negative by hyvI- and hyvII-based conventional PCR
| Name of DNA extract | Source | Real-time PCR CT value by: |
|
|---|---|---|---|
| 16S rRNA gene-based HLBasfpr | hyvI- and hyvII-based LJ900fpr | ||
| IND-C1-1 | Citrus | 27.80 | 33.35 |
| IND-C1-11 | Citrus | 23.52 | 27.30 |
| IND-C1-15 | Citrus | 22.98 | 32.66 |
| INDC1-23 | Citrus | 23.31 | 31.12 |
| IND-C2-15 | Citrus | 22.01 | 33.38 |
| IND-C2-16 | Citrus | 24.66 | 31.10 |
| IND-C2-17 | Citrus | 23.05 | 32.27 |
| IND-C2-18 | Citrus | 23.24 | 32.86 |
| PHA-Psy4 | Psyllid | 17.70 | 17.15 |
| PHA-Psy7 | Psyllid | 23.18 | 23.17 |
| PHA-Psy11 | Psyllid | 17.74 | 16.56 |
| PHA-Psy14 | Psyllid | 19.30 | 18.18 |
| PHA-Psy16 | Psyllid | 27.07 | 26.74 |
| PHA-Psy17 | Psyllid | 19.83 | 17.86 |
Sixteen clone libraries were made from the PCR amplicons of international “Ca. Liberibacter asiaticus” DNA by primers targeting the hyvI and hyvII regions. Sequencing of the hyvI and hyvII genes in international “Ca. Liberibacter asiaticus” isolates showed various NITR numbers that were similar to those from the Florida isolates (Fig. 1A2 to A4, B2, and B3; Table 2). The dominant form of hyvI clones from China, Philippines, Taiwan, and Brazil were of the same size and contained seven NITRs and two partial repeats (Fig. 1A2; Table 2). There were three additional sizes of the hyvI gene: a Taiwanese psyllid sample containing 10 and 2 full and partial repeats, respectively, and two Philippine psyllid samples containing 5 and 2 (i) and 1 and 2 (ii) full and partial repeats, respectively. The hyvI upstream and downstream sequences flanking the repeat region shared 93% to 100% similarity among all isolates (see Table S2 in the supplemental material). The hyvI genes from Indian and Thailand were different in terms of both numbers of NITR and downstream sequences flanking the repeat region (Fig. 1A3 and A4; see Table S2 in the supplemental material). The hyvII genes obtained from the Florida, China, and Thailand isolates shared 91% to 100% sequence similarity outside of the repeat region (see Table S3 in the supplemental material). The hyvII gene sequences amplified by LJ776/730 and LJ812/1089 were identical among the Florida and Chinese isolates. However, two variants of the hyvII gene were amplified from a Thai citrus DNA (THA-C2) by two sets of primers targeting the hyvII gene. The primer set LJ812/LJ1089 amplified a sequence identical to the hyvII sequence from China (Fig. 1B2; Fig. 2, lane 6), while LJ776/LJ730 amplified two products with 3 and 3 (i) and 1 and 11 (ii) full and partial repeats, respectively (Table 2). The sequence flanking the repeat region was more closely related to the same region in hyvII from Florida.
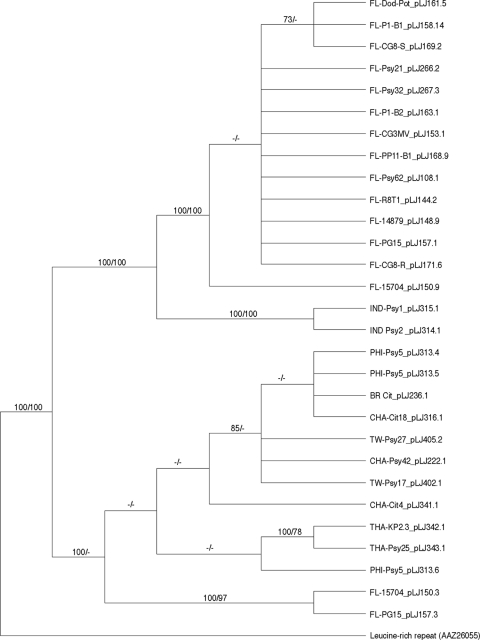
Phylogenetic analysis of hyvI and hyvII protein sequences.
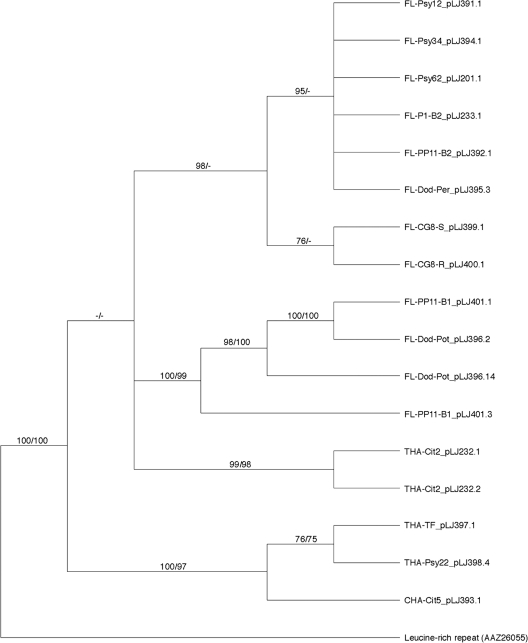
Based on the variable nucleotide sequences from “Ca. Liberibacter asiaticus” isolates of global origins, amino acid sequences deduced from these hyvI and hyvII gene sequences were aligned and integrated into phylogenetic analyses using parsimony and maximum likelihood methods. The leucine-rich repeat protein homolog from Colwellia psychrerythraea was employed as an outgroup for both Hyv proteins. The results of these phylogenetic analyses for HyvI and HyvII demonstrated that both genes bifurcate into clades on the basis of sampling locations (Fig. 3 and 4). HyvI phylogenetic inferences, containing 30 taxa and 948 informative characters, resulted in a tree with two well-supported clades that reflect the geographic origins of the sample sets (Fig. 3). The samples from Indian psyllids form a well-supported sister clade (100% bootstrap) to the sample set for most of the Florida isolates. The Florida samples were not well resolved by either method and are illustrated in the parsimonious tree as an unresolved polytomy. Two samples from Thailand formed a well-supported sister clade to a psyllid isolate from Philippines, although without bootstrap support.
Fig. 3.
Majority rule consensus tree based on maximum parsimony analyses of 29 amino acid sequences representing hyvI gene from global isolates of “Candidatus Liberibacter asiaticus.” The leucine-rich repeat protein sequence from Colwellia psychrerythraea is used as the outgroup. Branch support values represent maximum-parsimony bootstrap (≥70%) before the slash and RAxML (≥70%) bootstrap values after the slash. Each branch label reflects the identification listed in Table 2 and indicates the geographic origin of the sample.
Fig. 4.
Majority rule consensus tree based on maximum parsimony analyses of 17-amino-acid sequences representing the hyvII gene from global isolates of “Candidatus Liberibacter asiaticus.” The leucine-rich repeat protein sequence from Colwellia psychrerythraea is used as the outgroup. Branch support values represent maximum-parsimony bootstrap (≥70%) before the slash and RAxML (≥70%) bootstrap values after the slash. Each branch label reflects the identification listed in Table 2 and indicates the geographic origin of the sample.
Although the hyvII gene did not display as much size variability as the hyvI gene, these sequences were also different in “Ca. Liberibacter asiaticus” isolates based on geographic origins (Fig. 4). HyvII phylogenetic inferences, containing 18 taxa and 584 informative characters, resulted in a tree with well-supported clades whose segregation was based on geographic site of origin. One citrus sample from China and two psyllid samples from Thailand were grouped into one well-supported sister clade to those of the Florida samples. The Florida “Ca. Liberibacter asiaticus” isolates clustered into two clades, although without support at deep internodes. Only the clade consisting of periwinkle and dodder samples demonstrated strong branch support (100% bootstrap).
DISCUSSION
The prophage/phage accounts for approximately 1/15 of the significantly reduced “Ca. Liberibacter asiaticus” genome (16, 54). Characterization of the variations of these prophages/phages in the HLB bacteria may provide insight into their evolution and adaptation to host plants and insects. Through genomic approaches, two genes (hyvI and hyvII) were identified in two prophage regions of the “Ca. Liberibacter asiaticus” Psy62 genome. Although these two genes are closely related and share a homologous repetitive sequence unit, the numbers of repeats and sequences flanking the repeat region are significantly different. These differences were found not only in samples of distinct geographical origins but also from a single origin and even from a single “Ca. Liberibacter asiaticus”-infected sample, as indicated by the hyvI gene in citrus FL-15704 and periwinkle FL-PG15. The variation of repeat numbers and sequences flanking the repeat region of hyvI in the same isolate indicates coinfection by different “Ca. Liberibacter asiaticus” populations in a single HLB-affected sample. Therefore, HLB disease may be caused by coinfection of either different species of Ca. Liberibacter (45) or different stains/isolates of a Ca. Liberibacter species. Further investigation of population dynamics in these coinfected samples may reveal their potential roles in HLB disease development and epidemiology. Hypervariation of sequences, especially in the repeat number, were observed in both the hyvI and hyvII genes of the “Ca. Liberibacter asiaticus” isolates from Florida, Brazil, China, Thailand, India, and Philippines. The repeat numbers of these two genes varied from 1 partial to 12 full and 4 partials, and no apparent correlations were found between repeat number and bacterial titers, disease symptoms, or different hosts, reflecting the plasticity of the bacterial genome (Table 2 and Fig. 2). Potential mechanisms for deletion or insertion of the tandem repeats include slipped-strand mispairing, unequal crossover, rolling-circle or circle excision, and reinsertion occurring during bacterial replication (30, 39). This size variation caused by different numbers of tandem repeats provides the opportunity for functional diversity and alterations in phenotypes (20, 29, 48). Although the functions of hyvI and hyvII genes are unknown, BLASTP analysis using position-specific iterative basic local alignment search tool (PSI-BLAST) indicated that the repeat region of HyvI protein shares homology with a leucine-rich repeat protein, cell wall-associated biofilm protein, and a cell surface protein. These results suggest that the variations in the intragenic tandem repeats of hyvI gene may be associated with cell surface diversity (36). It is worthy to note that the structure of tandem repeats present in hyvI and hyvII is similar to those in pthA, the pathogenicity gene (transcription activator-like effector) of the citrus canker bacterium (Xanthomonas citri pv. citri) (5, 51). pthA contains 17.5 copies of a nearly identical tandem repeat unit that encodes 34 amino acids and is responsible for pathogenicity and host specificity (50). It undergoes intragenic recombination resulting in rapid adaptations to new hosts (9, 52), while the expression of pthA within plant cells elicits division, enlargement, and cell death, mimicking canker (14). Genes that contain coding tandem repeats often encode proteins with diverse functions, and the variable numbers of tandem repeats affect the pathogenicity or antigenicity in several human/animal pathogens, such as Treponema denticola and Streptococcus agalactiae (23, 35, 36). Experiments to reveal the functions of the hyvI and hyvII genes are under way.
It is interesting to note that all the Florida isolates in this study contained both the hyvI and hyvII genes, while the isolates from other geographic origins only contained one or the other. The presence or absence of hyvI and/or hyvII genes varied with the origins of the “Ca. Liberibacter asiaticus”-infected samples. Nevertheless, at least one of them was always associated with the presence of a “Ca. Liberibacter asiaticus” bacterium. Results from real-time PCR using the hyvI and hyvII repeat-based primers and probe indicate the presence of NITR in eight Indian citrus and six Philippine psyllid DNA samples that previously tested negative for hyvI and hyvII genes by conventional PCR using hyvI and hyvII gene-based primers. This result implies that more distinct variations of hyvI and hyvII might exist in these isolates. Recent findings revealed two circular phage genomes, SC1 and SC2, which are present in the Florida “Ca. Liberibacter asiaticus” bacterial genome (54). The forms of the phage/prophage genomes, circular, linear, or integrated into the “Ca. Liberibacter asiaticus” chromosome, varied with their different hosts (54). Since hyvI and hyvII were localized in two different prophage regions of the “Ca. Liberibacter asiaticus” genome or the two circular phage genomes, SC1 and SC2, it is hypothesized that the “Ca. Liberibacter asiaticus” isolates from the other six countries or regions may contain only one of the two prophage/phages or other unknown prophage/phages with sequences divergent from those of SC1 and SC2. This hypothesis was supported by PCR analyses of different loci, which indicated the presence of only one prophage/phage in these international samples (data not shown).
Phylogenetic placements were inferred for hyvI and hyvII by using two distinct methods. The most parsimonious, yielding the least number of steps on the trees, found several well-supported clades with clear demarcations based on the geographic origin of the sample. Maximum likelihood, utilizing a predicted model of evolution for both Hyv proteins, supported the resulting clades based on sample locations. Interestingly, different hosts—insect, citrus, dodder, or periwinkle—did not influence phylogenetic placements. The clustering of the Florida samples based on the full/partial repeat numbers, syntenic arrangement, or sequences flanking repeat region provides strong evidence of multiple introductions of HLB disease into Florida. Based on the phylogenetic tree established on the basis of the hyvI gene variations, an early introduction of the disease agent (containing only the hyvI gene) may have occurred in India, and the introduction into the majority of the Florida “Ca. Liberibacter asiaticus” isolates occurred later. More-recent introductions of the HLB disease into Florida may be evident, as two of the Florida “Ca. Liberibacter asiaticus” isolates were placed with good support in the HyvI clade with samples from Brazil, Philippines, Thailand, and China, including Taiwan (Fig. 3). In addition, the full hyvI genes from these international “Ca. Liberibacter asiaticus” isolates were identical, indicating a high possibility of the same source of “Ca. Liberibacter asiaticus” introduction into these four regions. The HyvII phylogenetic tree suggests that another major introduction of the HLB disease may have occurred from Thailand to Florida.
The high copy numbers of NITR (132 bp each) in the hyvI and hyvII genes provided excellent targets for the development of more sensitive diagnostic methods. This is especially important for the HLB bacterial pathogens because they are fastidious and often exist at extremely low titers in host plants and insect vectors. Protocols targeting this repeat unit were developed using SYBR green 1 (LJ900fr) and TaqMan (LJ900fpr) methods. Compared to the 16S rRNA gene-based real-time PCR method, an increase in Ca. Liberibacter detection likelihood at a range of 100- to 2,000-fold was demonstrated. This highly sensitive method consequently reduces false-negative results frequently associated with other detection methods (Morgan et al., submitted). This method is especially useful in seed transmission studies where “Ca. Liberibacter asiaticus” populations are usually at extremely low titer in the seedlings (data not shown). The conservation and variations observed in the sequences flanking the repeats in the hyvI and hyvII genes (Fig. 1; also see Tables S2 and S3 in the supplemental material) provide an excellent resolution of genetic diversity, which is useful for identifying the origins of individual isolates and developing the phylogenetics of “Ca. Liberibacter asiaticus” around the world.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Jerry Mozoruk and Nicola Faraci for their sequencing assistance, J. Kent Morgan for the help with real-time PCR analysis, Christina T. Latza for technical support, and Lesley Benyon and Melissa Doud for her critical review of the manuscript.
Funding for this work was provided by the Florida Citrus Advanced Technology Program awards 162 and 310.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Abascal F., Zardoya R., Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bastianel C., Garnier-Semancik M., Renaudin J., Bové J. M., Eveillard S. 2005. Diversity of “Candidatus Liberibacter asiaticus,” based on the omp gene sequence. Appl. Environ. Microbiol. 71:6473–6478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boch J., et al. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512 [DOI] [PubMed] [Google Scholar]
- 6. Bové J. M. 2006. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88:7–37 [Google Scholar]
- 7. Boyd E. F., Brüssow H. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521–529 [DOI] [PubMed] [Google Scholar]
- 8. Bruce D. S., et al. 2005. Detection and identification of citrus Huanglongbing (greening) in Florida, abstr. H-11, p. 59 Proceedings of the Second International Citrus Canker and Huanglongbing Research Workshop Orlando, FL [Google Scholar]
- 9. Brunings A. M., Gabriel D. W. 2003. Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4:141–157 [DOI] [PubMed] [Google Scholar]
- 10. Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277–300 [DOI] [PubMed] [Google Scholar]
- 11. Chen J., et al. 2010. Guangdong and Florida populations of ‘Candidatus Liberibacter asiaticus’ distinguished by a genomic locus with short tandem repeats. Phytopathology 100:567–572 [DOI] [PubMed] [Google Scholar]
- 12. Ding F., et al. 2009. Phylogenetic analysis of the citrus Huanglongbing (HLB) bacterium based on the sequences of 16S rDNA and 16S/23S rDNA intergenic regions among isolates in China. Eur. J. Plant Pathol. 124:495–503 [Google Scholar]
- 13. Drummond A., Strimmer K. 2001. PAL: an object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics 17:662–663 [DOI] [PubMed] [Google Scholar]
- 14. Duan Y. P., Castaneda A., Zhao G., Erdos G., Gabriel D. W. 1999. Expression of a single, host-specific, bacterial pathogenicity gene in plant cells elicits division, enlargement and cell death. Mol. Plant Microbe Interact. 12:556–560 [Google Scholar]
- 15. Duan Y. P., Gottwald T., Zhou L. J., Gabriel D. W. 2008. First report of dodder transmission of ‘Candidatus Liberibacter asiaticus’ to tomato (Lycopersicon esculentum). Plant Dis. 92:831. [DOI] [PubMed] [Google Scholar]
- 16. Duan Y. P., et al. 2009. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 22:1011–1020 [DOI] [PubMed] [Google Scholar]
- 17. Furuya N., et al. 2010. Sequence homogeneity of the ψserA-trmU-tufB-secE-nusG-rplKAJL-rpoB gene cluster and the flanking regions of ‘Candidatus Liberibacter asiaticus’ isolates around Okinawa Main Island in Japan. J. Gen. Plant Pathol. 76:122–131 [Google Scholar]
- 18. Gottwald T. R., Da Graca J. V., Bassanezi R. B. 2007. Citrus huanglongbing: the pathogen and its impact. Plant Health Progr. http://www.plantmanagementnetwork.org/pub/php/review/2007/huanglongbing/
- 19. Gottwald T. R. 2010. Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48:119–139 [DOI] [PubMed] [Google Scholar]
- 20. Gravekamp C., Rosner B., Madoff L. C. 1998. Deletion of repeats in the alpha C protein enhances the pathogenicity of group B streptococci in immune mice. Infect. Immun. 66:4347–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 22. Halbert S. E., Manjunath K. L. 2004. Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: a literature review and assessment of risk in Florida. Fla. Entomol. 87:330–353 [Google Scholar]
- 23. Ikegami A., Honma K., Sharma A., Kuramitsu H. K. 2004. Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect. Immun. 72:4619–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Irey M. S., Gast T., Gottwald T. 2006. Comparison of visual assessment and PCR assay testing to estimate the incidence of the huanglongbin pathogen in commercial Florida citrus. Proc. Fla. State Hortic. Soc. 42:17–21 [Google Scholar]
- 25. Jagoueix S., Bové J. M., Garnier M. 1994. The phloem-limited bacterium of greening disease of citrus is a member of the subdivision of the proteobacteria. Int. J. Syst. Bacteriol. 44:379–386 [DOI] [PubMed] [Google Scholar]
- 26. Jagoueix S., Bové J. M., Garnier M. 1997. Comparison of the 16S/23S ribosomal intergenic regions of “Candidatus Liberobacter asiaticum” and “Candidatus Liberobacter africanum,” the two species associated with citrus huanglongbing (greening) disease. Int. J. Syst. Bacteriol. 47:224–227 [DOI] [PubMed] [Google Scholar]
- 27. Jones D. T., Taylor W. R., Thornton J. M. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. (Camb.) 8:275–282 [DOI] [PubMed] [Google Scholar]
- 28. Katoh H., et al. 2011. Differentiation of “Candidatus Liberibacter asiaticus” isolates by variable number of tandem repeat analysis. Appl. Environ. Microbiol. 77:1910–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levdansky E., et al. 2007. Coding tandem repeats generate diversity in Aspergillus fumigatus genes. Eukaryot. Cell 6:1380–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levinson G., Gutman G. A. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203–221 [DOI] [PubMed] [Google Scholar]
- 31. Li W., Hartung J. S., Levy L. 2006. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 66:104–115 [DOI] [PubMed] [Google Scholar]
- 32. Li W., Li D., Twieg E., Hartung J. S., Levy L. 2008. Optimized quantification of unculturable Candidatus Liberibacter spp. in host plants using real-time PCR. Plant Dis. 92:854–861 [DOI] [PubMed] [Google Scholar]
- 33. Liu R., et al. 2011. Analysis of a prophage gene frequency revealed population variation of ‘Candidatus Liberibacter asiaticus’ from two citrus-growing provinces in China. Plant Dis. 95:431–435 [DOI] [PubMed] [Google Scholar]
- 34. Maddison W. P., Maddison D. R. 2010. Mesquite: a modular system for evolutionary analysis. Version 2.73. http://mesquiteproject.org
- 35. Madoff L. C., Michel J. L., Gong E. W., Kling D. E., Kasper D. L. 1996. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc. Natl. Acad. Sci. U. S. A. 93:4131–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Michel J. L., et al. 1992. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc. Natl. Acad. Sci. U. S. A. 89:10060–10064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller M. A., Pfeiffer W., Schwartz T. 2010. Creating the CIPRES gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop, New Orleans, LA [Google Scholar]
- 38. Planet P., Jagoueix S., Bové J. M., Garnier M. 1995. Detection and characterization of the African citrus greening Liberibacter by amplification, cloning and sequencing of the rplKAJL-rpoBC operon. Curr. Microbiol. 30:137–141 [DOI] [PubMed] [Google Scholar]
- 39. Romero D., Palacios R. 1997. Gene amplification and genomic plasticity in prokaryotes. Annu. Rev. Genet. 31:91–111 [DOI] [PubMed] [Google Scholar]
- 40. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 41. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 42. Subandiyah S., et al. 2000. Comparison of 16S rDNA and 16S/23S intergenic region sequences among citrus greening organisms in Asia. Plant Dis. 84:15–18 [DOI] [PubMed] [Google Scholar]
- 43. Swofford D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4b10. Sinauer Associates, Sunderland, MA [Google Scholar]
- 44. Teixeira D. C., et al. 2005. First report of a huanglongbing-like disease of citrus in Sao Paulo State, Brazil, and association of a new liberibacter species, ‘Candidatus Liberibacter americanus’, with the disease. Plant Dis. 89:107. [DOI] [PubMed] [Google Scholar]
- 45. Teixeira D. C., et al. 2005. Citrus huanglongbing in São Paulo, Brazil: PCR detection of the ‘Candidatus’ Liberibacter species associated with the disease, Mol. Cell. Probes 19:173–179 [DOI] [PubMed] [Google Scholar]
- 46. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tomimura K., et al. 2009. Evaluation of genetic diversity among ‘Candidatus Liberibacter asiaticus’ isolates collected in southeast Asia. Phytopathology 99:1062–1069 [DOI] [PubMed] [Google Scholar]
- 48. Verstrepen K. J., Jansen A., Lewitter F., Fink G. R. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37:986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Villechanoux S., Garnier M., Laigret F., Renaudin J., Bové J. M. 1993. The genome of the non-cultured, bacterial-like organism associated with citrus greening disease contains the nusG-rplKAJLrpoBC gene cluster and the gene for a bacteriophage type DNA polymerase. Curr. Microbiol. 26:161–166 [DOI] [PubMed] [Google Scholar]
- 50. Yang Y., De Feyter R., Gabriel D. W. 1994. Host-specific symptoms and increased release of Xanthomonas citri and X. campestris pv. malvacearum from leaves are determined by the 102-bp tandem repeats of pthA and avrb6, respectively. Mol. Plant Microbe Interact. 7:345–355 [Google Scholar]
- 51. Yang Y., Gabriel D. W. 1995. Xanthomonas avirulence/pathogenicity gene family encodes functional plant nuclear targeting signals. Mol. Plant Microbe Interact. 8:627–631 [DOI] [PubMed] [Google Scholar]
- 52. Yang Y., Gabriel D. W. 1995. Intragenic recombination of a single plant pathogen gene provides a mechanism for the evolution of new host specificities. J. Bacteriol. 177:4963–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang M. Q., et al. 2010. Screening molecules for control of citrus huanglongbing using an optimized regeneration system for ‘Candidatus Liberibacter asiaticus’ infected periwinkle (Catharanthus roseus) cuttings. Phytopathology 100:239–245 [DOI] [PubMed] [Google Scholar]
- 54. Zhang S. J., et al. 2011. Ca. Liberibacter asiaticus carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant. Mol. Plant Microbe Interact. 24:458–468 [DOI] [PubMed] [Google Scholar]
- 55. Zhao X. 1981. Citrus yellow shoot disease (Huanglongbing) - a review. Proc. Int. Soc. Citric. 1:466–469 [Google Scholar]
- 56. Zhou L. J., Gabriel D. W., Duan Y. P., Halbert S. E., Dixon W. N. 2007. First report of dodder transmission of Huanglongbing from naturally infected Murraya paniculata to citrus. Plant Dis. 91:227. [DOI] [PubMed] [Google Scholar]
- 57. Zhou L. J., Duan Y. P. 2008. Genetic diversity of citrus huanglongbing bacterium 'Candidatus Liberibacter asiaticus,' p. 172–175 Proceedings of the International Research Conference on Huanglongbing, Orlando, FL [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.