Abstract
Protein-encoding and 16S rRNA genes of Pasteuria penetrans populations from a wide range of geographic locations were examined. Most interpopulation single nucleotide polymorphisms (SNPs) were detected in the 16S rRNA gene. However, in order to fully resolve all populations, these were supplemented with SNPs from protein-encoding genes in a multilocus SNP typing approach. Examination of individual 16S rRNA gene sequences revealed the occurrence of “cryptic” SNPs which were not present in the consensus sequences of any P. penetrans population. Additionally, hierarchical cluster analysis separated P. penetrans 16S rRNA gene clones into four groups, and one of which contained sequences from the most highly passaged population, demonstrating that it is possible to manipulate the population structure of this fastidious bacterium. The other groups were made from representatives of the other populations in various proportions. Comparison of sequences among three Pasteuria species, namely, P. penetrans, P. hartismeri, and P. ramosa, showed that the protein-encoding genes provided greater discrimination than the 16S rRNA gene. From these findings, we have developed a toolbox for the discrimination of Pasteuria at both the inter- and intraspecies levels. We also provide a model to monitor genetic variation in other obligate hyperparasites and difficult-to-culture microorganisms.
INTRODUCTION
The genus Pasteuria, a group of endospore-producing Gram-positive bacteria, belongs to the Bacillus-Clostridium clade (9). Of those characterized, all are parasites of free-living and plant parasitic nematodes, with the exception of Pasteuria ramosa (24), a parasite of the water flea (Cladocera, Daphnia spp.).
Pasteuria penetrans is an obligate parasite of plant parasitic nematodes belonging to Meloidogyne spp., which are responsible worldwide for more than $100 billion worth of annual crop damage (30). Populations, defined here as collections of endospores obtained from field isolates of particular host female nematodes in distinct geographic locations, have been isolated from Meloidogyne spp. from around the globe, but there is very little understanding of the genetic variation between and within populations. Previous studies have focused on the position of P. penetrans in the bacterial kingdom (9, 31, 34) and the use of consensus 16S rRNA gene sequences (6, 7, 18) to differentiate between species and populations of Pasteuria (5, 16, 33). However, this information has not been extensively supported with sequence data from protein-encoding genes, which potentially provide an additional source of discrimination, with the exception of one study examining partial coding sequences of some genes in P. ramosa populations (32), as well as a recent study of genes encoding collagen-like proteins in this species (22a).
Geographically distinct populations of P. penetrans display various degrees of host specificity (11) through mechanisms that are not fully understood. It has been shown that cuticle heterogeneity, as exhibited by endospore attachment, is not linked in any simple way to the phylogeny of the nematode (12). Intraspecific functional variation in spore attachment to the infective second-stage juvenile cuticle has been observed, and it has been suggested that a Velcro-like mechanism operates, involving carbohydrate recognition domains on the surface of the cuticle and collagen-like fibers on the surface of the endospore (11). Although no simple relationship has been observed between endospore attachment and the phylogeny of Meloidogyne spp. (12), a greater understanding of the genetic variation between populations of Pasteuria isolated from root-knot nematodes would provide researchers with an enhanced knowledge of the bacterial “strains” with which they are working and expedite the classification of newly discovered strains. Currently, application of this organism for control of nematodes has not produced robust and repeatable levels of control on a field scale (10), and the development of a suite of diagnostic tools for discrimination of Pasteuria at the inter- and intrapopulation level will undoubtedly enhance our understanding of inoculant diversity and their use in biological control systems either by direct application or through soil amendments. The primary aim of this study was to develop molecular diagnostic tools for P. penetrans and to examine the relationship and variation between representative protein-encoding and 16S rRNA genes, which we have good reason to suspect are single- or low-copy-number genes in difficult-to-culture bacteria such as Pasteuria (19, 20, 22). They are distributed throughout the genomes of closely related Bacillus spp. (27) and so should provide a clear understanding of protein-encoding gene variation on a global scale in P. penetrans. We also examine equivalent genes in two other related species of Pasteuria to improve the classification of this family. Work on the Pasteuria group of bacteria, which has considerable biotechnological potential despite difficulty in obtaining and maintaining culture and extracting sufficient genetic material for analysis, can be seen as a model for the study of other fastidious microorganisms.
MATERIALS AND METHODS
Pasteuria populations.
Pasteuria endospores from the Rothamsted collection used in this study were EL48, PP1, PP3, PPE, RES147, RES148, Thies, and Pasteuria hartismeri, and all of which were originally isolated from Meloidogyne spp. from around the globe (13, 34). Table 1 lists the original host and geographic location of each population. Populations are defined as collections of endospores obtained from field isolates of particular host female nematodes in distinct geographic locations.
Table 1.
Pasteuria populations and species used in this study
| Populationa | Host | Geographic location (reference) |
|---|---|---|
| EL48 | Meloidogyne spp. | USA (this report) |
| RES147 | Meloidogynejavanica/ Meloidogyneincognita | Papua New Guinea (11) |
| PPE | Meloidogyne spp. | Queensland, Australia (11) |
| PP1 | M. javanica | California, USA (38) |
| PP3 | M. javanica/M. incognita | South Africa (37) |
| P. hartismeri | M. ardenensis | UK (7) |
| P. ramosa | Daphnia magna | Russia |
| RES148 | M. incognita | USA (this report) |
| Thies | Meloidogyne spp. | Thies, Senegal (38) |
All populations listed are P. penetrans with the exception of P. hartismeri and P. ramosa.
DNA release from endospores using microLYSIS-Plus.
Approximately 106 endospores were pelleted and resuspended in microLYSIS-Plus buffer (Microzone Ltd., Haywords Heath, W. Sussex, United Kingdom), processed, and subsequently treated with microCLEAN (Microzone Ltd.) to obtain template DNA as previously described (22).
Multiple-strand DNA amplification using whole-genome amplification (WGA).
The illustra GenomiPhi V2 DNA amplification kit (GE Healthcare) was used with Pasteuria genomic DNA template as described previously (22).
PCR.
PCR was performed on WGA Pasteuria DNA with the primers listed in Table S1 in the supplemental material. Only one set of primers designed for each gene fragment were required for PCR amplification, with the exception of the two genes whose amplification is described below. For gyrB-targeted PCRs, a first round of PCR was performed using the universal primers Up1 long and Up2r long. Next, nested PCRs were performed on these products using the Up1 and Pasteuria-specific gyrB rev primers as well as Up2r and Pasteuria-specific primer gyrB for. For 16S rRNA gene-targeted PCRs, two separate PCRs were performed using the primers 39F and 1166R as well as the primers PsppF4 and PsppR5. PCR conditions were used as previously described (22).
Cloning and sequencing of PCR products.
PCR amplification products were ligated into the pSC-A-amp/kan cloning vector (Stratagene, La Jolla, CA), and ligation products were then transformed into StrataClone recombinant cells (Stratagene, La Jolla, CA) as previously described (22). A proportion of colonies arising from the transformation were screened by colony PCR, with T3 and T7 primers flanking the insertion site. Clones were cultured in Luria-Bertani medium overnight with ampicillin selection, and plasmid DNA was then extracted using the GeneJet plasmid miniprep kit (Fermentas). Sequencing of plasmid inserts was performed by Eurofins MWG/Operon (Ebersberg, Germany) using either the T3 or T7 primer. Sequences retrieved were subjected to BLASTn analysis using default settings (4).
Sequence analysis and multiple sequence alignments.
Cloned PCR product DNA sequences were used to create “in silico” molecules, contigs, and alignments in the Vector NTI suite (Invitrogen, Carlsbad, California). In silico translation of nucleic acid sequences was done using the ExPASy translate tool (http://www.expasy.ch/tools/dna.html).
Hierarchical cluster analysis.
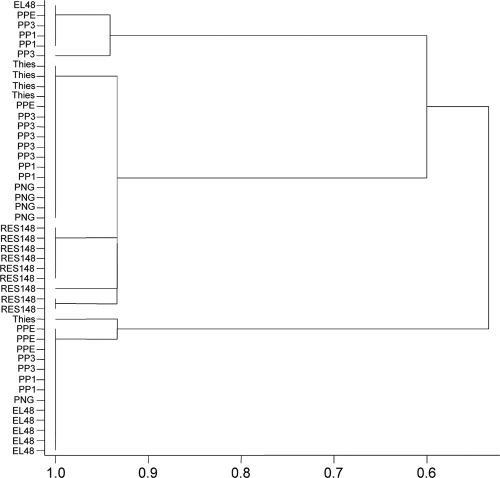
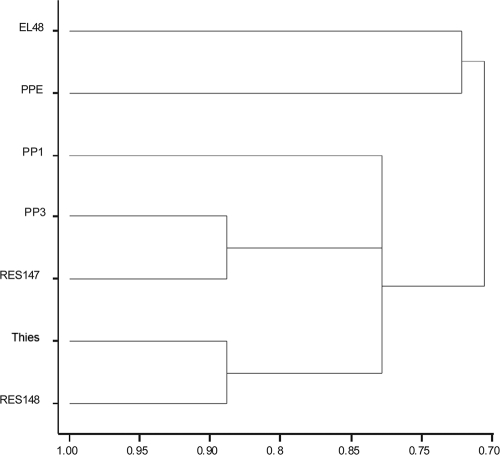
The GenStat statistical system (12th edition of GenStat 2010, Lawes Agricultural Trust [Rothamsted Research], VSN International Ltd., Hemel Hempstead, United Kingdom) was used to perform cluster analysis (14) based on the 16S rRNA gene single nucleotide polymorphism (SNP) as well as the multiple locus sequence typing (MLST)-SNP “barcode” data sets (see Tables S2 and S4, respectively, in the supplemental material). It was necessary to convert the SNP data, i.e., the A, C, G, and T codes, to the following respective numerical identifiers 1, 2, 3, and 4 in order for the similarity matrix to be constructed, using the simple matching measure of similarity. The similarity matrix was then used in the cluster analysis, incorporating the single-link (nearest-neighbor) method of clustering. This allowed for construction of the dendrograms in Fig. 1 and 2.
Fig. 1.
Dendrogram from hierarchical cluster analysis of 16S rRNA gene SNP data for all individuals from the seven P. penetrans populations. The x axis shows the similarity measure. The single-link (nearest-neighbor) clustering method was applied to the similarity matrix (formed using simple matching) among the 45 clones.
Fig. 2.
Dendrogram from hierarchical cluster analysis of the MLST-SNP barcode data for each of the seven P. penetrans populations (see Table S4 in the supplemental material), using a similarity matrix formed by simple matching and the single-link (nearest-neighbor) method of clustering. The x axis shows the similarity measure.
RESULTS
Range and depth of genes sequenced.
The genetic variability within seven populations of P. penetrans and their relationship to P. hartismeri and P. ramosa were investigated by sequence analysis of sections of the 13 protein-encoding and 16S rRNA genes amplified using the primers listed in Table S1 in the supplemental material for P. penetrans and P. hartismeri, whereas P. ramosa sequences were retrieved from GenBank. Initially, individual molecules from two separate PCR products from the WGA template for each gene from each P. penetrans population were cloned and sequenced to account for Taq amplification error. The sequences for each gene from all populations tested were then aligned, those genes that contained SNPs were further investigated, and SNPs were confirmed by cloning additional individual molecules from the relevant PCR products. In some cases, individual molecules from a third PCR product were also cloned and sequenced. Table S3 in the supplemental material shows the total number of clones analyzed for each gene in individual species/populations, which range from 2- to 5-fold coverage of sequences in genes for which no SNPs were detected and 3- to 23-fold coverage in genes with SNPs. The total number of clones sequenced for each respective gene when pooled from all populations tested ranged from 20 to 31 clone sequences for genes without SNPs and 26 to 64 clone sequences for genes with SNPs. In total, 95 new sequences were obtained from this study and deposited in the National Center for Biotechnology (NCBI) database, and their accession numbers can be found in Table S3.
Intrapopulation diversity analysis of P. penetrans.
The individual sequence traces from each population for each gene were examined, and this allowed a more subtle examination of potential differences that occur within a population, as well as the detection of parallel SNPs that occur in the minority across populations. It was found that in protein-encoding genes, there were very few examples of additional SNPs occurring in more than one clone that were not represented in the consensus sequence for that population. Exceptions were a single synonymous SNP in populations RES147, RES148, and PPE for the spoIIAA-spoIIAB genes. Overall, it seems that P. penetrans populations are clonal but nevertheless harbor intrapopulation variation, albeit at a relatively low frequency, in protein-encoding genes. These result in only synonymous substitutions in the genes examined to date.
When pooling all 45 contigs of the non-protein-encoding 16S rRNA gene, several SNPs were found in a minority of members of some P. penetrans populations at the same position (Table 2). Indeed, the total number of SNPs rose from 12 to 17 because of “cryptic” SNPs occurring at locations 92, 193, 197, 236, and 773 in the 16S rRNA gene that do not appear in the consensus sequence of any population.
Table 2.
SNP analysis of P. penetrans populations for the 16S rRNA gene sequence
| Population | SNP population analysisa |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 72 (V1) (A>C) | 92 (V1) (—>C) | 93 (V1) (T>—) | 100 (V1) (G>C) | 104 (V1) (—, G, A) | 151 (T>C) | 193 (V2) (C>T) | 194 (V2) (C>T) | 195 (V2) (C>T) | 197 (V2) (C>T) | 210 (V2) (T>A) | 214 (V2) (G>A) | 215 (V2) (G>A) | 236 (C>T) | 276 (A>G) | 417 (V3) (G>A) | 773 (C>T) | |
| PP1 | 0/6 (A) | 2/6 (—) | 2/6 (T) | 2/6 (G) | 2,2,2 | 2/6 (T) | 2/6 (C) | 2/6 (C) | 4/6 (T) | 2/6 (C) | 2/6 (T) | 4/6 (A) | 2/6 (G) | 2/6 (C) | 2/6 (A) | 2/6 (G) | 0/6 (C) |
| RES147 | 0/5 (A) | 0/5 (—) | 1/5 (T) | 1/5 (G) | 4,1,0 (—) | 1/5 (T) | 0/5 (C) | 1/5 (C) | 1/5 (C) | 0/5 (C) | 1/5 (T) | 1/5 (G) | 1/5 (G) | 0/5 (C) | 4/5 (G) | 4/5 (A) | 0/5 (C) |
| EL48 | 0/6 (A) | 1/6 (—) | 5/6 (—) | 5/6 (C) | 0,5,1 (G) | 5/6 (C) | 1/6 (C) | 5/6 (T) | 6/6 (T) | 1/6 (C) | 5/6 (A) | 6/6 (A) | 5/6 (A) | 1/6 (C) | 0/6 (A) | 0/6 (G) | 0/6 (C) |
| RES148 | 9/9 (C) | 0/9 (—) | 0/9 (T) | 0/9 (G) | 9,0,0 (—) | 0/9 (T) | 0/9 (C) | 0/9 (C) | 0/9 (C) | 0/9 (C) | 0/9 (T) | 0/9 (G) | 0/9 (G) | 0/9 (C) | 9/9 (G) | 8/9 (A) | 2/9 (C) |
| PPE | 0/5 (A) | 1/5 (—) | 3/5 (—) | 3/5 (C) | 1,3,1 (G) | 3/5 (C) | 1/5 (C) | 3/5 (T) | 4/5 (T) | 1/5 (C) | 3/5 (A) | 4/5 (A) | 3/5 (A) | 1/5 (C) | 1/5 (A) | 1/5 (G) | 0/5 (C) |
| PP3 | 0/9 (A) | 2/9 (—) | 2/9 (T) | 2/9 (G) | 5,2,2 (—) | 2/9 (T) | 2/9 (C) | 2/9 (C) | 4/9 (C) | 1/9 (C) | 2/9 (T) | 4/9 (G) | 2/9 (G) | 2/9 (C) | 5/9 (G) | 5/9 (A) | 0/9 (C) |
| Thies | 0/5 (A) | 0/5 (—) | 1/5 (T) | 1/5 (G) | 4,1,0 (—) | 1/5 (T) | 0/5 (C) | 1/5 (C) | 1/5 (C) | 0/5 (C) | 1/5 (T) | 1/5 (G) | 1/5 (G) | 1/5 (C) | 4/5 (G) | 4/5 (A) | 0/5 (C) |
The numbers at the top of the columns refer to the SNP base pair location in the 16S rRNA gene, and the V code refers to the variable region, if any, in which this SNP is located. The proportion of clones from each population showing an SNP is shown as a fraction of the total number of individual sequenced clones for that population. Cryptic SNP locations are given in boldface. —, SNP deletion.
Further analysis of the SNPs in the 45 individual 16S rRNA gene sequences by cluster analysis revealed that they fall into four main groups (Fig. 1). One of the groups was comprised exclusively of sequences derived from the highly passaged population RES148, the other three groups contained mixtures of the other populations.
Table 2 and Table S2 in the supplemental material show the distribution of the SNPs throughout the 16S rRNA gene sequences from individual clones for each Pasteuria population. The majority of SNPs occurring among P. penetrans populations in the 16S rRNA gene are located within the first 215 bp from the start of the approximately 1,500-bp gene. These correspond mainly to hypervariable regions V1 and V2. There are also lone SNPs located in V3 and V4 and two further SNPs which are not located in any of the nine characterized variable regions (26) (Table 2). Overall, the amount of intrapopulation variation is low, though there are clear differences in the 16S rRNA gene.
Interpopulation diversity analysis of P. penetrans.
The consensus sequences of the 13 gene fragments from each of the seven P. penetrans populations were compared. The differences between gene sequences for all P. penetrans populations are minor, with three or fewer SNPs identified (with the exception of the 16S rRNA gene) (see Table 4). When examining the seven P. penetrans populations, it was found that there was a total of 19 SNPs among all P. penetrans population consensus sequences. The 16S rRNA gene exhibited the highest number (12 SNPs) of polymorphic sites located at positions 72, 93, 100, 104, 151, 194, 195, 210, 214, 215, 276, and 417. This was followed by the gyrB gene (3 SNPs), and gene fragments with one polymorphic site identified were yvjB, hemN, dnaE, and pheT. The remaining seven protein-encoding gene fragments did not display any polymorphic sites. Translation of the seven SNPs from the protein-encoding genes resulted in synonymous substitutions.
Table 4.
SNPs identified in P. penetrans housekeeping genes
| Gene | Fragment size (bp) | No. of SNPsa | % SNPs |
|---|---|---|---|
| ATP synthase B subunit (atpF) | 245 | 0 | 0 |
| Cytochrome C oxidase subunit C (ctaC) | 267 | 0 | 0 |
| Carboxy-terminal processing protease (ctpA) | 367 | 0 | 0 |
| DNA polymerase III alpha subunit (dnaE) | 285 | 1 | 0.35 |
| Beta-ketoacyl-acyl-carrier protein synthase II (fabF) | 490 | 0 | 0 |
| DNA gyrase subunit B (gyrB) | 1,183 | 3 | 0.25 |
| Coproporphyrinogen III oxidase (hemN) | 233 | 1 | 0.43 |
| Phenylalanyl-tRNA synthetase alpha subunit (pheT) | 473 | 1 | 0.2 |
| Sigma factor E (sigE) | 307 | 0 | 0 |
| Sporulation protein/anti-sigma regulatory factor (spoIIAA/AB) | 250 | 0 (1) | 0.4 |
| Hypothetical protein (yacL) | 330 | 0 | 0 |
| Carboxy-terminal processing protease (yvjB) | 202 | 1 | 0.5 |
| 16S ribosomal RNA (16S rRNA) | 1,430 | 12 (5) | 0.84 |
Values in parentheses represent the additional number of cryptic SNPs found for that gene.
The 16S rRNA gene alone was unable to distinguish between all P. penetrans populations. However, when used in conjunction with other SNP-containing protein-encoding genes, a “barcode” was constructed, as was a cluster analysis of these data, and was able to discriminate between all P. penetrans populations examined (Fig. 2; see also Table S4 in the supplemental material).
Interspecies diversity analysis of selected Pasteuria genes.
The occurrence and significance of interspecies gene polymorphisms were also examined between P. penetrans and two other species of Pasteuria, namely, Pasteuria hartismeri, a parasite of the ash parasitic nematode Meloidogyne ardenensis (7), and the Daphnia endoparasite P. ramosa (24). The results from these analyses are summarized in Table S5a to c in the supplemental material. It was found that selected protein-encoding genes give much clearer discrimination between Pasteuria species than the 16S rRNA gene. For example, when comparing P. penetrans with P. hartismeri, the protein-encoding genes displayed, on average, 16% SNP frequency compared with 3% for the 16S rRNA gene. The opposite is found when discriminating P. penetrans populations, as the 16S rRNA gene was found to be the best discriminator (see Table 4), although the frequency of SNPs in this gene was lower than that between Pasteuria species.
When considering the location of SNPs within the 16S rRNA gene between all combinations of Pasteuria tested, it was found that intraspecies variation was concentrated at the 5′ end of the gene, whereas SNPs were more evenly distributed throughout the gene between different Pasteuria species (Tables 2 and 3).
Table 3.
SNP analysis of the distribution of Pasteuria 16S rRNA gene sequences between all combinations of Pasteuria tested
| Combination | SNP distribution analysisa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | Other | |
| P. penetrans vs P. penetrans | 5 (29.4) | 7 (41.2) | 1 (5.9) | 0 | 0 | 0 | 0 | 0 | 0 | 4 (23.5): 1 between V2 and V3; 2 between V2 and V3; 1 between V4 and V5 |
| P. penetrans vs P. hartismeri | 5 (10.9) | 9 (19.6) | 1 (2.2) | 5 (10.9) | 0 | 6 (13) | 1 (2.2) | 6 (13) | 2 (4.3) | 11 (23.9): 3 between V1 and V2; 6 between V2 and V3; 2 between V3 and V4 |
| P. penetrans vs P. ramosa | 10 (7.6) | 12 (9.2) | 7 (5.3) | 13 (9.9) | 1 (0.8) | 26 (19.8) | 4 (3.1) | 7 (5.3) | 8 (6.1) | 43 (32.8): 8 between V1 and V2; 20 between V2 and V3; 7 between V3 and V4; 3 between V4 and V5; 2 between V5 and V6; 3 between V7 and V8 |
| P. ramosa vs P. hartismeri | 9 (7.4) | 9 (7.4) | 6 (5) | 11 (9.1) | 1 (0.8) | 23 (19) | 4 (3.3) | 10 (8.3) | 6 (5) | 42 (34.7): 8 between V1 and V2; 19 between V2 and V3; 7 between V3 and V4; 3 between V4 and V5; 2 between V5 and V6; 3 between V7 and V8 |
The V codes refer to the variable regions of this gene, and the numbers of SNPs in each region are recorded along with their percent contribution (in parentheses) to the total number of SNPs between each combination of Pasteuria.
Although the analyses performed here do not include all of the protein-encoding genes examined for P. penetrans populations, those examined seemingly have great potential to discriminate between Pasteuria species. As expected, the percentages of SNPs and associated amino acid changes were greater between the nematode-parasitizing Pasteuria (P. penetrans and P. hartismeri) and the parasite of Daphnia, P. ramosa. For example, even though the 256-bp sigE fragments compared in P. penetrans and P. hartismeri contained 32 SNPs, all of these were synonymous. This was not the case for the equivalent genes in P. ramosa and nematode-parasitizing Pasteuria, where many amino acid changes are predicted by the sequence polymorphisms (see Table S5a to c in the supplemental material).
DISCUSSION
This work shows that P. penetrans populations from geographically distinct locations, including South Africa, Australia, and the United States, show a high level of homogeneity in most genes tested. This can largely be explained, as they are obligate hyperparasites of root-knot nematodes, which are obligate parasites of many crop plants. In evolutionary terms, genetic variation both in root-knot nematodes and consequently in P. penetrans is constrained by the host plant. As such, the lack of genetic diversity observed in P. penetrans is likely to be indicative of human agricultural practice and the transferal and cultivation of crops around the globe with their associated parasites (35, 36). This lack of diversity is in accordance with the Mycobacterium tuberculosis species complex, a fastidious group which have static genomes sharing 99.9% sequence identity (15), as well as other monomorphic bacteria such as Salmonella enterica serovar Typhi and Bacillus anthracis (23).
To date, this is the largest study examining the genetic diversity of P. penetrans populations, which encompasses a variety of genes encoding proteins and the 16S rRNA. Our results show that a number of SNPs were identified in these genes (Table 4) and that the 16S rRNA gene gave the best discrimination between populations of this organism. This is likely to be due to the tight constraints on protein-encoding genes of an obligate organism to maintain correct folding and presentation of protein active sites. This is less marked in the 16S rRNA gene sequences where the scaffold for ribosomal assembly and interaction with mRNA is interspersed with well-defined variable regions that form helical secondary structures and are shown to have base substitutions in closely related organisms (40). It was somewhat surprising that the protein-encoding gene sequences between P. penetrans populations did not display a higher degree of synonymous substitution, but perhaps their relatively recent distribution through agricultural cultivation on a global scale has meant that there has not yet been sufficient time for divergence to occur.
When examining the genetic diversity between Pasteuria species, it was found that the protein-encoding genes were better relative discriminators than the 16S rRNA gene (see Table S5a to c in the supplemental material). This is presumably because these species have adapted to different ecological niches over a sufficient evolutionary time period, which has led to a large number of changes between the protein-encoding genes, the majority of which are synonymous, although a proportion result in amino acid changes, which presumably reflect differences in protein activity. The poor discrimination observed between 16S rRNA gene sequences could be due to the fact that the 16S rRNA gene is not protein encoding, and so variation is restricted to particular regions of the gene, whereas protein-encoding genes are able to vary throughout their sequence due to the wobble phenomenon. Indeed, work on Pseudomonas intragenic phylogenies have shown that the 16S rRNA gene does not discriminate sufficiently at the interspecies level (40). In addition, it has been suggested that a multiple locus sequence typing (MLST) approach of highly expressed core genes, such as those studied in this work, is a viable phylogenetic approach (17, 21). The rationale is that these genes form part of the core genome, are highly conserved, exhibit a high degree of codon bias, and evolve more slowly than typical accessory protein-encoding genes but more rapidly than the 16S rRNA gene. Our results suggest that this approach would be suited for interspecific and intraspecific Pasteuria diagnostics. This approach is particularly well suited for fastidious obligate organisms such as Pasteuria, for which obtaining pure genomic DNA is problematic (22), and it has been used for the typing of obligate intracellular bacteria belonging to the Chlamydiales (28) and Wolbachia (41) spp.
The degree of variability within populations of P. penetrans was also examined. It was shown that the protein-encoding genes showed only a very small amount of variation, but it was revealed that the 16S rRNA gene displayed more variability and that cryptic SNPs exist within populations in this gene (Table 2). The 16S rRNA gene sequence is known to display an alternating pattern of conserved and variable regions that reflect the functional importance of the conserved regions (26). We found that P. penetrans populations vary from each other mainly in regions V1 and V2, whereas Pasteuria interspecies differences are not constrained to these regions and are more evenly distributed throughout the gene (Table 3). The biological significance of variation within P. penetrans being focused in these regions is unclear, though it has also been shown that intraspecies variation of Streptococcus equi subsp. zooepidemicus is focused in V2 (1), indicating that the 5′ end of the 16S rRNA gene is under less selection pressure than the other variable regions in terms of maintaining the rRNA secondary and tertiary structures. It has been shown that other fastidious organisms like Rickettsia, Mycobacteria, and Wolbachia possess relatively few and often single copies of the 16S rRNA gene (20), and so, populations of Pasteuria could be composed of a mixture of “biotypes.” In this study, cluster analysis revealed that individual 16S rRNA gene sequences from seven P. penetrans populations separated into four groups. With the exception of the highly passaged population RES148, which showed very little intrapopulation variation and formed its own group, sequences derived from the other populations were fairly evenly represented in the other three groupings (Fig. 1). This shows that 16S rRNA gene sequences in these populations contain a degree of genetic variability and that their consensus sequences are dictated by the proportion of each sequence type in a population. As such, care needs to be taken when typing fastidious clonal organisms such as Pasteuria when based on this gene. Additionally, the observation that the highly passaged population RES148 shows less genetic variation than the other P. penetrans populations tested indicates that the composition of Pasteuria population structures can be manipulated and suggests that there is very little intragenomic heterogeneity in individual P. penetrans cells and/or that the copy number of this gene is very low. Indeed, Mauchline et al. (22) previously provided evidence that the 16S rRNA gene copy number in P. penetrans is low.
The identification of SNPs in this paper facilitates the future use of high-throughput analyses of Pasteuria populations with techniques such as MID-tagged PCR product pyrosequencing. Such techniques will enhance the quality control of Pasteuria inoculants produced for biological control applications, give better resolution of biodiversity, and provide the wherewithal to monitor population dynamics of Pasteuria in field situations. This will enable us to answer key biological questions regarding Pasteuria epidemiology, which until now has not been possible. For example, is infection of a single nematode achieved by a single spore, or do many spores coinfect hosts? If the latter is true, then a mixed-population inoculant could increase nematode control by Pasteuria.
As a concluding remark, this paper has highlighted that for discrimination of Pasteuria, both the 16S rRNA gene and protein-encoding genes should be studied. For intraspecies comparisons, the variable regions of the 16S rRNA gene provide sufficient resolution to “type” some populations. However, the SNPs identified in P. penetrans protein-encoding genes provide greater coverage of the core and dispensable genome than the 16S rRNA gene alone and allow resolution of all populations tested in this study. In addition, other genes, such as rpoB, have been reported to be useful taxonomic markers for phylogenetic analyses and identification of closely related bacterial isolates (2). Furthermore, groEL and sodA have been usefully deployed for analysis of the closely related Bacillus cereus group (8). For interspecies phylogenies, protein-encoding genes show more diversity than the 16S rRNA gene and are well suited to a MLST approach. Indeed, in organisms more amenable to culture, they have been shown to compare favorably with the DNA-DNA reassociation values (21, 25, 29). The development of a panel of tools based on selected protein-encoding genes that evolve mainly by synonymous substitution in a MLST assay is therefore likely to provide an accurate interpretation of intra- and interspecies Pasteuria phylogeny. Finally, the approaches described here provide a model for phylogenetic analyses and for the development of diagnostics in other obligate hyperparasites and difficult-to-culture microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council and the British Council through the United Kingdom-India Education and Research Initiative (UKIERI) titled “Multitrophic Interactions in the Rhizosphere.”
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Abdulmawjood A., Lämmler C. H. 2000. Determination of intraspecies variations of the V2 region of the 16S rRNA gene of Streptococcus equi subsp. zooepidemicus. Res. Vet. Sci. 68: 33–39 [DOI] [PubMed] [Google Scholar]
- 2. Adékambi T., Drancourt M., Raoult D. 2009. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 17: 37–45 [DOI] [PubMed] [Google Scholar]
- 3. Reference deleted.
- 4. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- 5. Anderson J. M., et al. 1999. Phylogenetic analysis of Pasteuria penetrans by 16S rRNA gene cloning and sequencing. J. Nematol. 31: 319–325 [PMC free article] [PubMed] [Google Scholar]
- 6. Atibalentja N., Noel G. R., Domier L. L. 2000. Phylogenetic position of the North American isolate of Pasteuria that parasitizes the soybean cyst nematode, Heterodera glycines, as inferred from 16S rDNA sequence analysis. Int. J. Syst. Evol. Microbiol. 50: 605–613 [DOI] [PubMed] [Google Scholar]
- 7. Bishop A. H., Gowen S. R., Pembroke B., Trotter J. R. 2007. Morphological and molecular characteristics of a new species of Pasteuria parasitic on Meloidogyne ardenensis. J. Invertebr. Pathol. 96: 28–33 [DOI] [PubMed] [Google Scholar]
- 8. Chang Y. H., Shangkuan Y. H., Lin H. C., Liu H. W. 2003. PCR assay of the groEL gene for detection and differentiation of Bacillus cereus group cells. Appl. Environ. Microbiol. 69: 4502–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charles L., et al. 2005. Phylogenetic analysis of Pasteuria penetrans by use of multiple genetic loci. J. Bacteriol. 187: 5700–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dabiré K. B., et al. 2005. Influence of irrigation on the distribution and control of the nematode Meloidogyne javanica by the biocontrol bacterium Pasteuria penetrans in the field. Biol. Fert. Soils 41: 205–211 [Google Scholar]
- 11. Davies K. G. 2009. Understanding the interaction between an obligate hyperparasitic bacterium, Pasteuria penetrans and its obligate plant-parasitic nematode host, Meloidogyne spp. Adv. Parasitol. 68: 211–245 [DOI] [PubMed] [Google Scholar]
- 12. Davies K. G., et al. 2001. Cuticle heterogeneity as exhibited by Pasteuria spore attachment is not linked to the phylogeny of parthenogenetic root-knot nematodes (Meloidogyne spp.). Parasitology 122: 111–120 [DOI] [PubMed] [Google Scholar]
- 13. Davies K. G., Redden M. 1997. Diversity and partial characterization of putativevirulence determinants in Pasteuria penetrans, the hyperparasitic bacterium of root knot nematodes (Meloidogyne spp.). J. Appl. Microbiol. 83: 227–235 [DOI] [PubMed] [Google Scholar]
- 14. Digby P. G. N., Kempton R. A. 1987. Multivariate analysis of ecological communities. Chapman and Hall, London, United Kingdom [Google Scholar]
- 15. Dos Vultos T., et al. 2008. Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS One 3: e1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duan Y. P., Castro H. F., Hewlett T. E., White J. H., Ogram A. V. 2003. Detection and characterization of Pasteuria 16S rRNA gene sequences from nematodes and soils. Int. J. Syst. Evol. Microbiol. 53: 105–112 [DOI] [PubMed] [Google Scholar]
- 17. Feil E. J., Li B. C., Aanensen D. M., Hanage W. P., Spratt B. G. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186: 1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giblin-Davis R. M., et al. 2003. ‘Candidatus Pasteuria usgae’ sp. nov., an obligate endoparasite of the phytoparasitic nematode Belonolaimus longicaudatus. Int. J. Syst. Evol. Microbiol. 53: 197–200 [DOI] [PubMed] [Google Scholar]
- 19. Klappenbach J. A., Saxman P. R., Cole J. R., Schmidt T. M. 2001. rrndb: the rRNA Operon Copy Number Database. Nucleic Acids Res. 29: 181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee Z. M., Bussema C., III, Schmidt T. M. 2009. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 37: D489–D493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maiden M. C., et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95: 3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mauchline T. H., et al. 2010. A method for release and multiple strand amplification of small quantities of DNA from endospores of the fastidious bacterium Pasteuria penetrans. Lett. Appl. Microbiol. 50: 515–521 [DOI] [PubMed] [Google Scholar]
- 22a. McElroy K., Mouton L., Du Pasquier L., Qi W., Ebert D. 2011. Characterisation of a large family of polymorphic collagen-like proteins in the endospore-forming bacterium Pasteuria ramosa. Res. Microbiol. 162: 701–714 [DOI] [PubMed] [Google Scholar]
- 23. Medini D., et al. 2008. Microbiology in the post-genomic era. Nat. Rev. Microbiol. 6: 419–430 [DOI] [PubMed] [Google Scholar]
- 24. Metchnikoff M. E. 1888. Pasteuria ramosa un representant des bacteries a division longitudinale. Ann. Inst. Pasteur 2: 165–170 [Google Scholar]
- 25. Mulet M., Lalucat J., García-Valdés E. 2010. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12: 1513–1530 [DOI] [PubMed] [Google Scholar]
- 26. Neefs J.-M., Van De Peer Y., Hendriks L., De Wachter R. 1990. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 18: 2237–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nong G., Chow V., Schmidt L. M., Dickson D. W., Preston J. F. 2007. Multiple-strand displacement and identification of single nucleotide polymorphisms as markers of genotypic variation of Pasteuria penetrans biotypes infecting root-knot nematodes. FEMS Microbiol. Ecol. 61: 327–336 [DOI] [PubMed] [Google Scholar]
- 28. Pannekoek Y., et al. 2008. Multilocus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol. 28: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richter M., Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106: 19126–19131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sasser J. N., Freckman D. W. 1987. A world perspective on nematology: the role of the society. In Veech J. A., Dickson D. W. (ed.), Vistas on nematology. Society of Nematologists, Hyattsville, MD [Google Scholar]
- 31. Schmidt L. M., Preston J. F., Nong G., Dickson D. W., Aldrich H. C. 2004. Detection of Pasteuria penetrans infection in Meloidogyne arenaria race 1 in planta by PCR. FEMS Microbiol. Ecol. 48: 457–464 [DOI] [PubMed] [Google Scholar]
- 32. Schmidt L. M., Mouton L., Nong G., Ebert D., Preston J. F. 2008. Genetic and immunological comparison of the cladoceran parasite Pasteuria ramosa with the nematode parasite Pasteuria penetrans. Appl. Environ. Microbiol. 74: 259–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sturhan D. 1988. New host and geographical records of nematode-parasitic bacteria of the Pasteuria penetrans group. Nematologica 34: 350–356 [Google Scholar]
- 34. Trotter J. R., Bishop A. H. 2003. Phylogenetic analysis and confirmation of the endospore-forming nature of Pasteuria penetrans based on the spo0A gene. FEMS Microbiol. Lett. 225: 249–256 [DOI] [PubMed] [Google Scholar]
- 35. Trudgill D. L. 1995. Origins of root knot nematodes (Meleoidogyne spp., Nematoda) in relation to their control. Phytoparasitica 23: 191–194 [Google Scholar]
- 36. Trudgill D. L., Blok V. C. 2001. Apomictic, polyphagous root-knot nematdoes: exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 39: 53–77 [DOI] [PubMed] [Google Scholar]
- 37. Tzortzakakis E. A., Channer A. G. D. R., Gowen S. R., Ahmed R. 1997. Studies on the potential use of Pasteuria penetrans as a biocontrol agent of root-knot nematodes (Meloidogyne spp.). Plant Pathol. 46: 44–55 [Google Scholar]
- 38. Wishart J., Blok V., Philips M. S., Davies K. G. 2004. Pasteuria penetrans and P. nishizawae attachment to Meloidogyne chitwoodi, M. fallax and M. hapla. Nematology 6: 507–510 [Google Scholar]
- 39. Reference deleted.
- 40. Yamamoto S., et al. 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146: 2385–2394 [DOI] [PubMed] [Google Scholar]
- 41. Yun Y., et al. 2010. Wolbachia strains typing in different geographic population spider, Hylyphantes graminicola (Linyphiidae). Curr. Microbiol. doi:10.1007/s00284-010-9686-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.