Abstract
The gut microbiota is strongly associated with the well-being of the host. Its composition is affected by environmental factors, such as food and maternal inoculation, while the relative impact of the host's genetics have been recently uncovered. Here, we studied the effect of the host genetic background on the composition of intestinal bacteria in a murine model, focusing on lactic acid bacteria (LAB) as an important group that includes many probiotic strains. Based on 16S rRNA gene genotyping, variation was observed in fecal LAB populations of BALB/c and C57BL/6J mouse lines. Lactobacillus johnsonii, a potentially probiotic bacterium, appeared at significantly higher levels in C57BL/6J versus BALB/c mouse feces. In the BALB/c gut, the L. johnsonii level decreased rapidly after oral administration, suggesting that some selective force does not allow its persistence at higher levels. The genetic inheritance of L. johnsonii levels was further tested in reciprocal crosses between the two mouse lines. The resultant F1 offspring presented similar L. johnsonii levels, confirming that mouse genetics plays a major role in determining these levels compared to the smaller maternal effect. Our findings suggest that mouse genetics has a major effect on the composition of the LAB population in general and on the persistence of L. johnsonii in the gut in particular. Concentrating on a narrow spectrum of culturable LAB enables the isolation and characterization of such potentially probiotic bacterial strains, which might be specifically oriented to the genetic background of the host as part of a personalized-medicine approach.
INTRODUCTION
The human digestive tract is a habitat for bacteria, which are 10-fold more abundant than the total number of human cells. Colonization of the gastrointestinal tract begins at birth, when the newborn is first exposed to the maternal microbiota, and continues thereafter via prolonged exposure to maternal and other environmental bacteria (13, 33). Eventually, the adult gut microbiota will consist of hundreds to thousands of bacterial species. The composition of the intestinal microbiota is known to differ among individual hosts (6, 12, 14, 29) as a result of variations in environmental factors, such as food consumption, exposure to maternal bacteria during infancy, antibiotic intake, or other stochastic factors (8, 11, 16, 23, 28, 33, 36, 45). However, the importance of the host's genetics in determining the intestinal microbiota has also been demonstrated in animal models, such as mice (3, 8, 15, 24, 48, 53), as well as in humans (27, 50, 55). The composition of the gut microbiota may be affected by genetically inherited components, such as the immune system major histocompatibility complex (MHC) (43), as well as by intestinal surfaces, including the intestinal mucus, gut epithelial cells, and extracellular matrix. These intestinal surfaces are a target for bacterial adhesion, implying that variation in the surface-presented components may result in differential bacterial adhesion (17, 41, 46). Along with the impact of host genetics on the composition of the gut microbiota, genetic biodiversity among bacterial strains may influence bacterial adhesion in particular and persistence in the gut in general (2, 10), contributing to the complexity of host-bacterium interactions.
The intestinal microbiota is highly important for maintaining a healthy organism. Recent studies have shown a tight connection between the gut microbiota and different health disorders, such as inflammatory bowel diseases, atopic diseases, obesity, metabolic syndrome, intestinal cancers, and diabetes (1, 19, 32, 39, 44, 52, 53). One important group of gut colonizers is the lactic acid bacteria (LAB), a heterogeneous group of Gram-positive rods and cocci that belong to the phylum Firmicutes. There are indications of an association between oral administration of different LAB strains and improvement of gut health disorders, such as pouchitis, ulcerative colitis, infectious diarrhea, antibiotic-associated diarrhea, traveler's diarrhea, necrotizing enterocolitis, atopic eczema, and Helicobacter pylori infections (20–22, 26, 31, 54). Lactobacillus is the largest genus in the LAB, composed of over 100 species and containing several probiotic strains (i.e., live microorganisms, which when administered in adequate amounts, confer health benefits on the host [ftp://ftp.fao.org/es/esn/food/wgreport2.pdf]), which are well characterized. The genus Lactobacillus is highly divergent but can nevertheless be divided into seven or eight groups (5). One of them is the acidophilus group, which contains, among others, the bacterium Lactobacillus johnsonii. This species includes commensal, as well as probiotic, strains, such as L. johnsonii NCC533. The genome sequence of this strain reveals genes responsible for interactions with the host, as well as the absence of genes responsible for several important biosynthetic pathways, suggesting strong dependence on the host or on other microbes (38). Such dependence might partially explain the variation in the abundance of L. johnsonii in the guts of different animal hosts (K. Buhnik-Rosenblau, V. Matsko-Efimov, M. Jung, H. Shin, Y. Danin-Poleg, and Y. Kashi, submitted for publication).
In a parallel study, we tested the variation in fecal LAB populations in animal hosts belonging to different taxonomic groups by using terminal restriction fragment length polymorphism (TRFLP) of 16S rRNA genes. These bacterial populations were found to vary with the host taxonomy (Buhnik-Rosenblau et al., submitted). Here, we concentrate on the host's subspecies level by comparing the intestinal LAB populations of two genetically diverse mouse lines, C57BL/6J and BALB/c, held under the same environmental conditions (food, cages, etc.). The intestinal LAB populations were analyzed by TRFLP, followed by precise strain typing for L. johnsonii using variable number tandem repeat (VNTR) and multilocus sequence typing (MLST). Our results suggest that mouse genetics plays a major role in determining L. johnsonii levels in particular and the composition of the LAB population in general.
MATERIALS AND METHODS
Mouse lines.
C57BL/6J, BALB/c, and reciprocal F1 hybrid mice, all aged 5 to 7 weeks, were obtained from Harlan Laboratories Ltd. (Jerusalem, Israel). For the TRFLP analysis (see below), C57BL/6J and BALB/c mice from self-breeding stocks were used, as well. For generating F1 hybrids, BALB/c females were mated with C57BL/6J males to obtain F1 offspring designated ♀BALB/c × ♂C57BL/6J, and C57BL/6J females were mated with BALB/c males to obtain the reciprocal ♀C57BL/6J × ♂BALB/c F1 offspring. The male parents were removed from the cages after mating to prevent contact with their offspring. Five males and five females were analyzed from each reciprocal cross. All mice (inbred lines and F1 mice) were housed under specific-pathogen-free conditions in individually ventilated cages with five mice per cage. Females and males were housed in separate cages. Autoclaved food (Teklad Certified Global 18% Protein Rodent Diet; Harlan Laboratories Ltd.) and sterilized deionized water were provided ad libitum. The cages were held in a room with a 12-h/12-h light/dark cycle (lights on at 7:00 am) at 24 ± 1°C and 60 to 70% humidity and with centrally controlled ventilation with HEPA filters (75 air changes/hour). All experiments employing mice were approved by the animal care and use committee of the Technion, Israel Institute of Technology. Feces samples were collected from each individual mouse separately. Equal weights of feces collected from five individuals of the same mouse line were pooled for further analysis.
Isolation of fecal bacteria.
Feces samples were suspended in 0.1 M sodium phosphate buffer, pH 7, to a final concentration of 10% (wt/vol) by vigorous vortexing, followed by centrifugation at 1,500 × g at 4°C for 5 min. The supernatant containing the bacterial suspension was transferred to a clean tube, and 100 μl of bacterial suspension was spread on Difco m-Enterococcus agar plates (BD, Sparks, MD) in four dilutions (nondiluted sample, 1:10, 1:100, and 1:1,000); these were grown under anaerobic conditions at 37°C for 48 h. For the TRFLP analysis (see below), bacteria were grown under aerobic conditions, as well, to compare the bacterial populations obtained from each of the growth conditions. Cells from a loopful of undiluted fecal-bacterial populations grown on m-Enterococcus agar plates were suspended in 70% ethanol (1 ml) by vigorous vortexing, 33 μl of sodium acetate (3 M; pH 5.2) was added, and the samples were incubated at −80°C for 20 min, followed by centrifugation at 12,000 × g for 15 min. The supernatant was removed, and the pellet was dissolved in 30 μl 0.1× Tris-EDTA (TE). The crude DNA was diluted 10-fold in double-distilled H2O (ddH2O) and stored at −20°C.
TRFLP of fecal-bacterial populations.
The 16S rRNA genes of fecal-bacterial populations were amplified in a total volume of 50 μl using 27F-FAM and 1492R primers (40) and 10 μl of the crude extract of 10-fold-diluted bacterial DNA at an annealing temperature of 60°C. The PCR products were purified by ethanol precipitation and dissolved in ddH2O. The purified PCR product (1,000 ng) was digested with 20 U of MspI restriction enzyme (New England BioLabs, Ipswich, MA) in a total volume of 20 μl for 2.25 h at 37°C, followed by 20 min at 65°C. The digested DNA (50 ng) was loaded into a 3130 genetic analyzer together with 9 μl of formamide and 0.5 μl of Genescan 1200 Liz size standard (lot 0709012; Applied Biosystems, Foster City, CA) for size determination. The results were analyzed with GeneMapper 4.0 software (Applied Biosystems).
L. johnsonii identification and colony hybridization.
An L. johnsonii-specific probe was chosen by in silico screening of the L. johnsonii NCC 533 genome. Three genes not belonging to any cluster of orthologous genes (at http://www.ncbi.nlm.nih.gov/sutils/cogtik.cgi?gi=382&cog=−) were screened for uniqueness against all bacterial sequences using the Blastn algorithm (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). Gene LJ0244 (locus tag) was chosen as an L. johnsonii-specific locus. The probe was prepared by PCR amplification of L. johnsonii DNA isolated from C57BL/6J mice using LJ0244-specific primers (probLJ_F, 3′-CAAAAACCAACTTTCTATGTG-5′, and probLJ_R, 3′-TTAATAGTGGCTACCCATA-5′; annealing temperature, 51°C).
Colony hybridization was performed in two dilutions of fecal-bacterial population grown on m-Enterococcus agar plates that were selected based on the number of bacterial colonies, ranging from dozens to hundreds. The colonies were transferred to an Amersham Hybond N+ membrane (GE Healthcare) according to the manufacturer's instructions with minor modifications. Briefly, the membranes were placed on the plates and left for overnight incubation. Then, the membranes were denatured for 3 min and neutralized for 3 min twice, followed by agitation in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 2 min or until the membranes were cleaned of cell debris. DNA was fixed to the membranes by UV radiation (0.15 J/cm2). Hybridization was further performed using the Amersham AlkPhos Direct Labeling and Detection system with CDP-Star (GE Healthcare, Buckinghamshire, United Kingdom) according to the manufacturer's instructions. Briefly, the membranes were agitated in hybridization buffer (1 ml buffer per 30-cm2 membrane) at 55°C for 15 min and then reacted with alkaline phosphatase-labeled L. johnsonii-specific probe (LJ0244; 4.2 ng labeled probe per 1-cm2 membrane) overnight at 55°C. The membranes were washed twice with primary wash buffer (1 ml per 1-cm2 membrane) at 55°C and then washed twice with secondary wash buffer (1 ml per 1-cm2 membrane) at room temperature, followed by a 3-min exposure to CDP-Star substrate to produce a chemiluminescent reaction (1 ml substrate per 180-cm2 membrane). The membranes were exposed to Kodak Biomax Light Film for 4 h, and L. johnsonii colonies were enumerated.
The specificity of L. johnsonii identification using the LJ0244 colony hybridization probe was confirmed by 16S rRNA gene sequencing and by PCR amplification using L. johnsonii-specific primers targeted to L. johnsonii 23S rRNA genes (LJ1_F, 3′-GTGAGAGCCCCGTAC-5′, and LJ1_R, 3′-CTACCACGCATATAATATA-5′; annealing temperature, 51°C). Species-specific primers were designed based on the 23S rRNA gene sequences of lactobacilli available in the NCBI database. The forward primer was designed so that the last nucleotide at the 3′ end was unique to L. johnsonii. The reverse primer was designed based on an L. johnsonii-specific probe (37). Species-specific PCR amplification was performed directly on the colonies of the suspected L. johnsonii isolates.
PCR.
Each PCR mixture contained 0.2 mM deoxynucleoside triphosphates, 0.4 μM forward and reverse primers, 0.02 U of Taq polymerase (SuperNova; JMR Holding, Kent, England) per μl, 1× reaction buffer (containing 1.5 mM MgCl2), and 5 μl of 1:10 diluted crude DNA or, alternatively, spiked cells from a colony in a total volume of 25 μl. The reactions were carried out in a Veriti 96-Well Thermal Cycler (Applied Biosystems) as follows: 95°C for 3 min; 30 cycles of 30 s at 95°C, 30 s at the annealing temperature, and 90 s at 72°C; 10 min at 72°C; and cooling to 12°C. PCR amplification products were verified by gel (1.2%) electrophoresis and observed by UV fluorescence.
Administration of L. johnsonii to mice.
A feeding trial was performed following the method of Denou et al. (9), with minor modifications. Briefly, all mouse groups received a fresh supply of erythromycin daily (catalog no. E0774; Sigma-Aldrich, Rehovot, Israel) at 10 μg/ml through the drinking water for 3 days to decrease the endogenous gut microbiota. Then, a fresh culture of L. johnsonii isolated from C57BL/6J mouse feces (termed L. johnsonii 16) was administered to the mice by intragastric gavage at 109 CFU in 200 μl. This was performed once a day for 3 days for each BALB/c and C57BL/6J experimental group. Feces samples from all mouse groups were collected; the first sampling was performed right before the first antibiotic administration (day −2), and 11 additional samplings were conducted daily, starting just before the first feeding. Bedding was renewed daily in all cages until day 1 to prevent exposure to fecal bacteria during the antibiotic treatment or possible exposure to remnants of antibiotics on the first feeding day.
In preliminary experiments conducted to test the resistance of fecal LAB populations to erythromycin, the MIC was determined for a pure culture of L. johnsonii 16 and for fecal bacteria of either C57BL/6J or BALB/c mice in MRS broth (De Man Rogosa and Sharp; Oxoid, Hampshire, United Kingdom) for growth of total LAB. The observed MICs were 2 μg/ml, 4 μg/ml, and 1 μg/ml, respectively. Cultures incubated for 20 h with erythromycin concentrations above the MICs did not show regrowth of any bacteria on MRS agar, indicating bacterial death rather than growth inhibition. In vivo resistance of intestinal LAB populations to erythromycin was tested at erythromycin concentrations of 5 and 10 μg/ml, both resulting in similar decreases in the tested fecal-bacterial population.
Growth kinetics.
Six lyophilized isolates from C57BL/6J and BALB/c mice were subcultured twice in 5 ml of MRS broth and grown aerobically at 37°C for 20 h. For the kinetics experiments, cultures were diluted 1:100 in fresh MRS broth and placed in a 96-well plate (200 μl from each culture in 8 replications). Experiments were conducted in a Bio-Tek Synergy HT Multi-Detection Microplate Reader (MTX Lab Systems, Inc., Vienna, VA) at 37°C for 24 h under slow agitation, with a reading every 15 min.
Data analysis.
For the TRFLP analysis, reproducible peaks with an average peak area larger than 1% of the summed area of all peaks were designated major peaks. Quantitative analysis of L. johnsonii by colony hybridization was performed in duplicate: the values for L. johnsonii levels in all experiments are expressed as average values.
L. johnsonii levels, L. johnsonii direct counts, and total counts for the two mouse lines and genders were compared by two-way analysis of variance (ANOVA), followed by means separation by Tukey's honestly significant difference (HSD) test using the statistical software JMP 8 (2008 version; SAS Institute Inc., Cary, NC). Significantly different groups (α = 0.05), as determined by a Tukey test, are indicated. For the feeding trial, ANOVA was used to compare L. johnsonii fractions, direct counts, and total bacterial counts among the experimental and control groups of the two mouse lines during all experiments and on days 4 through 10. ANOVA was carried out for analysis of L. johnsonii levels in F1 hybrid mice and parental lines, as the gender parameter was nonsignificant by two-way ANOVA.
Nucleotide sequence accession numbers.
Nucleotide sequence data determined in the present study have been submitted to the GenBank database under the following accession numbers: 16S rRNA gene sequences of Enterococcus hirae (2 sequences), Lactobacillus intestinalis, L. johnsonii (L. johnsonii 16), and Enterococcus faecalis, JF923641, JF923642, JF923643, JF923644, and JF923645, respectively; E. hirae rpoA, JF923647; L. johnsonii-specific probe (LJ_0244), JF923646; and L. johnsonii 16 (similar to eight clones from C57BL/6J and BALB/c mice) at the three “conserved hypothetical” genes, JN012106, JN012145, and JN012184.
RESULTS
Profile of fecal LAB populations and L. johnsonii levels in C57BL/6J and BALB/c mice.
We tested the variation in the gut microbiota between C57BL/6J and BALB/c mouse lines, concentrating on the fecal-bacterial subpopulations that grow on m-Enterococcus agar. This agar was chosen as a highly selective medium that enables the growth of a limited number of bacterial species. Experiments were applied to a total of 50 C57BL/6J mice and 33 BALB/c mice over a 2-year period.
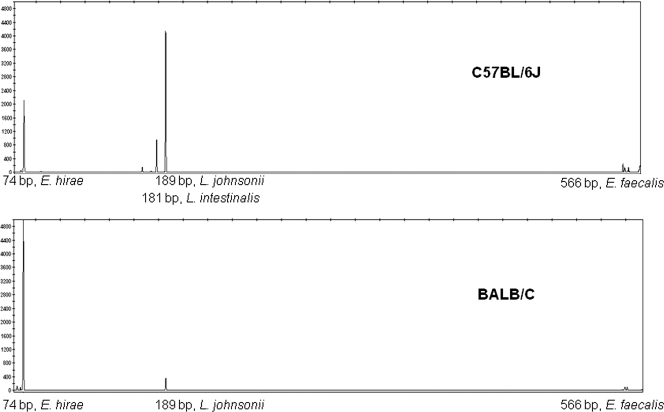
Fecal samples from the two mouse lines were collected from five individual 5- to 7-week-old mice and pooled to minimize the variation between individuals. TRFLP analysis of the bacterial population revealed different patterns for C57BL/6J and BALB/c mouse lines, based on two biological replications. These results were highly reproducible regardless of the mouse origin: mice obtained from Harlan Laboratories Ltd. and self-bred mice presented similar patterns. Generally, the TRFLP patterns consisted of three major peaks: 74 bp, 189 bp, and 566 bp (Fig. 1). Minor peaks were excluded from the analysis in order to focus on the main bacterial species composing the bacterial population isolated from each line. To identify the bacterial species corresponding to each of the peaks, representative colonies were analyzed for their 16S terminal restriction DNA fragments (TRFs), followed by 16S rRNA gene sequencing. The 566-bp TRFLP peak was found to represent E. faecalis, the 189-bp peak represented L. johnsonii, and the 74-bp peak represented bacterial species belonging to the Enterococcus faecium cluster. Precise species identification based on sequencing of the housekeeping gene rpoA (34) revealed that the isolated species was E. hirae. The TRFLP patterns suggested that L. johnsonii was consistently present in bacterial populations originating from C57BL/6J mice, whereas it was nearly absent in BALB/c mice. Higher levels of E. hirae were present in BALB/c mice than in C57BL/6J mice. E. faecalis was generally present at low levels in both mouse lines (Fig. 1). Males and females within each line showed similar patterns with slight nonsignificant differences. The TRFLP patterns of bacterial populations grown under both aerobic and anaerobic conditions were similar, with the exception of a 181-bp peak, representing L. intestinalis, which appeared in C57BL/6J mice only after anaerobic incubation. The most significant variation between mouse lines was in L. johnsonii, which was highly abundant in C57BL/6J relative to BALB/c mice, and we therefore chose to focus on this species in further experiments.
Fig. 1.
TRFLP patterns of fecal LAB populations obtained from C57BL/6J and BALB/c mice. LAB were grown on m-Enterococcus agar under anaerobic conditions. DNA fragments were observed using fluorescent 6-carboxyfluorescein (FAM)-labeled primer for 16S PCR amplification, followed by digestion with MspI restriction enzyme and fragment size analysis using an ABI 3130 genetic analyzer. The sizes of specific fragments are given.
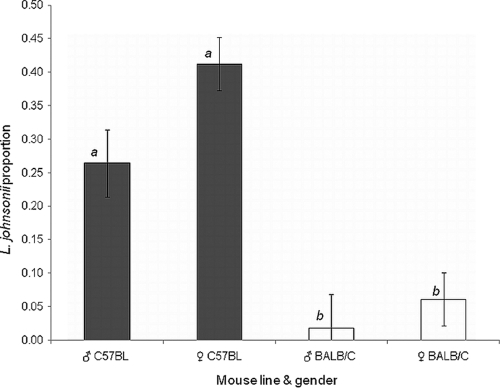
Quantitative estimation of the relative abundance of L. johnsonii in fecal samples from the two mouse lines, C57BL/6J and BALB/c, was performed by hybridization of L. johnsonii-specific probe (corresponding to the LJ0244 gene) to colonies grown on m-Enterococcus agar. The results were based on three biological replicates in which females and males were sampled separately. Hybridizations were performed in duplicate for each fecal sample. C57BL/6J mice were found to contain significantly higher levels of L. johnsonii in their feces than BALB/c mice (Fig. 2), analyzed either as direct L. johnsonii counts or as their fractions of the total counts (F = 6.079, P = 0.0025, and F = 17.867, P < 0.0001, respectively; two-way ANOVA of gender and line). Both direct colony counts of L. johnsonii and their fraction of the total bacterial counts were 10-fold higher in C57BL/6J mice (1.7 × 107 ± 0.4 × 107 CFU/g feces and 0.36 ± 0.04, respectively) than in BALB/c mice (7.6 × 105 ± 3.5 × 105 CFU/g feces and 0.04 ± 0.02, for L. johnsonii counts and their fraction, respectively). In contrast, the total bacterial counts were highly variable, with nonsignificant differences between mouse lines and genders (F = 2.002, P = 0.1365). Thus, we chose to further refer to the parameter of L. johnsonii proportions, normalizing the variable total counts. Slight differences were observed between genders (F = 4.25, P = 0.0487), with the females always presenting a higher fraction of L. johnsonii than the males; nevertheless, the mouse line had the major effect (F = 41.36, P < 0.0001). Comparison of all means by Tukey HSD analysis further confirmed that the significant differences in L. johnsonii levels are indeed between the two mouse lines (Fig. 2) (α = 0.05).
Fig. 2.
Levels of L. johnsonii in fecal samples of mouse lines C57BL/6J (black bars) and BALB/c (white bars). Males (♂) and females (♀) were tested separately (five mice per group). The levels are expressed as proportions of L. johnsonii colonies out of the total number of colonies grown on m-Enterococcus agar plates and are given as average values of three and five independent biological replicates for males and females, respectively. L. johnsonii was enumerated using colony hybridization with an L. johnsonii-specific probe. Bars labeled with different letters are significantly different at an α value of <0.05 by two-way ANOVA followed by a Tukey HSD test. The error bars indicate standard errors.
Strain typing of L. johnsonii isolates from C57BL/6J and BALB/c mice.
Since host-bacterium interactions are highly dependent on the bacterial strain, we tested whether both mouse lines carry the same L. johnsonii clone by strain genotyping. Four representative L. johnsonii colonies were isolated from each mouse line and identified as L. johnsonii using species-specific PCR amplification of 23S rRNA genes. Further confirmation was achieved by sequencing of the 16S rRNA genes (1,534 bp) and of 258 bp of the LJ0244 gene. Precise typing was achieved using multilocus sequence typing (MLST) at three conserved hypothetical genes and VNTR (or simple sequence repeat [SSR]) analysis at 11 genomic loci. Both MLST and VNTR analysis were found to be useful for fine discrimination among L. johnsonii from various hosts, as described elsewhere (Buhnik-Rosenblau et al., submitted).
Two strategies were used to characterize the genetic variation among L. johnsonii isolates: sizing for the 10 VNTR (SSR) loci using gel or capillary electrophoresis and sequencing for one VNTR locus with a motif size of 1 bp and for three conserved hypothetical genes. Variation analysis of the eight isolates from C57BL/6J and BALB/c mice at the 11 VNTR loci and the three conserved hypothetical genes revealed identical results, i.e., all isolates presented the same alleles or identical sequences in each of the tested genomic loci. This genotyping identity was in contrast to the high variation found among 47 isolates from diverse origins, showing 2 to 10 alleles in the tested VNTR loci and 10 to 24 sequence types at each conserved hypothetical gene (Buhnik-Rosenblau et al., submitted). An additional 21 L. johnsonii isolates from both mouse lines were tested for polymorphism at the two most polymorphic VNTR loci, giving identical alleles for all isolates at both loci (data not shown). These typing results supported the similarity of L. johnsonii clones isolated from the two mouse lines. In addition, the growth rates of isolates from C57BL/6J and BALB/c mice were further compared in 8 replications. Similar growth rates were observed for isolates from the two mouse lines, presenting an average growth rate of 0.79 ± 0.04 h−1 and 0.78 ± 0.05 h−1 for isolates originating in C57BL/6J and BALB/c mice, respectively (ANOVA, F = 0.7084, P = 0.404). These results further supported the similarity of L. johnsonii clones isolated from the two mouse lines.
L. johnsonii gut persistence in a feeding experiment.
To test whether variation in the gut persistence of L. johnsonii is responsible for the observed difference in L. johnsonii abundance in the fecal samples from the two mouse lines, we performed a force-feeding experiment with L. johnsonii.
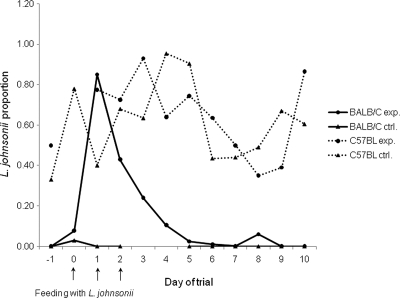
The feeding trial included one experimental and one control group for each of the mouse lines. All mouse groups received erythromycin prior to L. johnsonii administration to the experimental groups following the method of Denou et al. (9). Antibiotics were used prior to the feeding to decrease the normal mouse gut microbiota, especially the endogenous LAB population that might compete with L. johnsonii for the same niches. In a preliminary test, the fecal culturable LAB populations from the two mouse lines and the endogenous L. johnsonii (L. johnsonii 16) were found to be susceptible to erythromycin by MIC determination in vitro. Twelve feces samplings were conducted during the feeding experiment. Quantification of L. johnsonii levels in each sample was performed in duplicate by colony hybridization (see above). The results showed that 10 μg/ml erythromycin given to mice via the drinking water generally decreased the LAB populations grown on m-Enterococcus agar in half orders of magnitude to ∼105 CFU/g feces (see Table S1 in the supplemental material). The same results were obtained in the preliminary experiment run with different individual mice. L. johnsonii direct counts, as well as their proportions of the total counts in C57BL/6J mice during the feeding trial, were high and were similar in both control and experimental groups (Fig. 3) (1.5 × 106 ± 0.4 × 106 and 1 × 106 ± 0.2 × 106 CFU/g feces and proportions of 0.63 ± 0.05 and 0.65 ± 0.05 for control and experimental groups, respectively). In contrast, BALB/c mice presented wide differences between control and experimental groups: L. johnsonii direct counts and their proportion of the control BALB/c mice remained low during the entire trial (< 104 CFU/g feces and 0.002 ± 0.002, respectively) (see Table S1 in the supplemental material), while the BALB/c experimental group presented a sharp increase in L. johnsonii levels immediately after the first feeding (Fig. 3, day 1) (1.9× 106 CFU/g feces of L. johnsonii and a proportion of 0.85), which rapidly decreased to the basic low level (Fig. 3). Further analysis of L. johnsonii fractions following the feeding (on days 4 to 10) using ANOVA revealed significant differences only between mouse lines and not between experimental and control groups (F = 56.0, P < 0.0001). In contrast, the total counts showed nonsignificant differences among all mouse groups. These results further indicated that variation in the persistence of L. johnsonii between mouse lines might have a genetic basis.
Fig. 3.
Fecal levels of L. johnsonii after feeding C57BL/6J and BALB/c mice with L. johnsonii isolated from C57BL/6J mice. Both experimental (exp.) and control (ctrl.) groups (five mice per group) received antibiotic treatment for 3 days prior to the feeding. Experimental groups were fed L. johnsonii on days 0, 1, and 2 of the trial (indicated by arrows). L. johnsonii levels are expressed as proportions of L. johnsonii colonies out of the total number of colonies grown on m-Enterococcus agar plates. Levels are given as average values of two technical replicates. L. johnsonii was enumerated using colony hybridization with an L. johnsonii-specific probe.
L. johnsonii levels in F1 offspring of reciprocal C57BL/6J × BALB/c crosses.
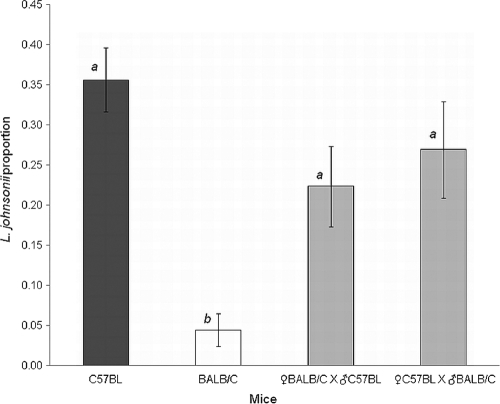
To confirm that L. johnsonii levels are mainly determined by host genetic factors rather than exposure to the maternal microbiota, we tested C57BL/6J × BALB/c reciprocal crosses. Basically, the reciprocal hybrids are genetically similar (except for maternally inherited elements), whereas during and following birth, the F1 mice are exposed to either C57BL/6J or BALB/c maternal microbiota. The levels of L. johnsonii in fecal samples of the F1 hybrids were quantified in quadruplicate by colony hybridization (see above), and the average proportion of L. johnsonii was calculated. The values obtained were compared with L. johnsonii levels of the parental lines C57BL/6J and BALB/c. ANOVA was performed to test the difference in L. johnsonii levels among the genetic groups, as nonsignificant differences were observed between genders (two-way ANOVA, F = 3.63, P = 0.064). The results showed significant differences among BALB/c, C57BL/6J, and F1 hybrids of the reciprocal crosses (F = 13.8513, P < 0.0001). Comparison of all means by Tukey HSD analysis showed significantly lower L. johnsonii proportions in the parental BALB/c line than in the two F1 hybrids and the parental C57BL/6J line (Fig. 4) (α = 0.05). The proportions of L. johnsonii were similar between hybrids of the cross ♀C57BL/6J × ♂BALB/c and the reciprocal cross ♀BALB/c × ♂C57BL/6J, presenting an average of 0.27 ± 0.06 and 0.22 ± 0.05, respectively (Fig. 4). Slightly higher L. johnsonii proportions were observed for the parental line C57BL/6J (0.36 ± 0.04). These results indicated that mouse genetics is the major parameter determining the levels of L. johnsonii, whereas the maternal effect is relatively low. Comparison of L. johnsonii proportions between the BALB/c parental line and the offspring of a ♀BALB/c × ♂C57BL/6J cross, which differ only in their paternal genetics, showed highly significant differences of almost 1 order of magnitude (0.04 ± 0.02 and 0.22 ± 0.05, respectively). Analysis of L. johnsonii direct counts revealed equivalent results. C57BL/6J presented significantly higher L. johnsonii counts (F = 12.1841, P < 0.0001), and the two F1 hybrids of the reciprocal crosses presented similar low levels (1.0 × 106 ± 0.6× 106 and 0.6 × 106 ± 0.2 × 106 CFU/g feces for the cross ♀C57BL/6J × ♂BALB/c and the reciprocal cross ♀BALB/c × ♂C57BL/6J, respectively) resembling the BALB/c L. johnsonii counts (see above). All the above data suggest that mouse genetics plays a major role in determining the fecal levels of L. johnsonii.
Fig. 4.
L. johnsonii levels in fecal samples from F1 offspring (gray bars) and the parental mouse lines C57BL/6J (black bars) and BALB/c (white bars), with five mice per group. F1 offspring resulted from two separate reciprocal crosses: ♀BALB/c × ♂C57BL and ♂BALB/c × ♀C57BL. Levels are expressed as proportions of L. johnsonii colonies out of the total number of colonies grown on m-Enterococcus agar plates and are given as average values of four independent biological replicates. L. johnsonii was enumerated using colony hybridization with an L. johnsonii-specific probe. Bars labeled with different letters are significantly different at an α value of <0.05 by ANOVA, followed by a Tukey HSD test. The error bars indicate standard errors.
DISCUSSION
An increasing number of studies are indicating a role for host genetics in determining the gut microbiota composition. Here, we show a major effect of a mouse's genetics on its intestinal-bacterial population, focusing on a narrow spectrum: LAB at the species and strain level. Mouse feces were assigned to represent the murine gut microbiota. Both mucosa and feces are commonly used to study intestinal-microbiota profiles, presenting distinct bacterial populations (14, 57). While the mucosa mainly contains mucosa-associated bacteria, feces were previously postulated to represent a combination of shed mucosal bacteria and a separate nonadherent luminal population (14). Fecal communities were previously shown to be host specific (3, 14) and stable (56), and their use further allows monitoring of the gut microbiota of the same individual over time. Here, we chose m-Enterococcus agar as a highly selective medium that enables the growth of a limited number of bacterial species. Indeed, this narrow spectrum allowed us to specifically isolate and identify four bacterial species that display variation in the fecal-bacterial population.
C57BL/6J and BALB/c mice showed highly reproducible variation in their fecal LAB populations (Fig. 1), as analyzed by TRFLP. The same variation was observed between these inbred lines regardless of mouse origin (commercial source or self-breeding), indicating that the composition of the gut LAB population is genetically controlled. Similarly, a previous study observed variation in the cellular fatty acid profiles of the intestinal-bacterial population between these two mouse lines, indicating that mouse genetics has an influence on the gastrointestinal microbiota (48a). Four bacterial species were found to dominate mouse feces in the bacterial spectrum tested here. E. faecalis was present in both mouse lines, whereas levels of L. johnsonii, L. intestinalis, and E. hirae were found to differ between the two mouse lines. Both L. johnsonii and L. intestinalis dominated in C57BL/6J versus BALB/c mice. Quantitative analysis of L. johnsonii in mouse feces by colony hybridization using an L. johnsonii-specific probe demonstrated that the levels of L. johnsonii in BALB/c mice are an order of magnitude lower than in C57BL/6J mice (Fig. 2), further confirming the significant differences observed in L. johnsonii levels between inbred mouse lines with diverse genetic backgrounds. In contrast to C57BL/6J mice, which presented high levels of both L. johnsonii and L. intestinalis, BALB/c mice presented high levels of E. hirae, indicating that these taxonomically related bacterial species might compete for the same niche or have similar metabolic functions.
The observed variation in the compositions of intestinal-bacterial species from genetically diverse hosts has been previously observed in humans (11, 12), where it was claimed that the combination of bacterial species and strains is unique to each individual, calling for precise identification of the isolated bacterial strains. Strain identification is highly important, as different strains belonging to the same bacterial species might vary in their abilities to persist in the gut of a specific host. Denou et al. (10) previously showed that different L. johnsonii strains vary significantly in their gut residence times after oral feeding of mice having a single genetic background. Indications of strain-dependent persistence have also been found in other bacterial species (2, 35). In Lactobacillus reuteri, phylogenic analysis of strains revealed host-specific clusters. Moreover, in a feeding trial, administering a mixture of L. reuteri strains from various origins to germ-free mice resulted in better persistence of the mouse and rat isolates, suggesting host-specific ecological fitness up to the strain level (35). Strain characterization was conducted on L. johnsonii, a potentially probiotic bacterium (38) that was found here to populate the gut of C57BL/6J mice at higher levels than in BALB/c mice. Molecular typing of L. johnsonii was performed by MLST and VNTR analysis, widely used methods for strain typing and for epidemiological studies in many bacterial species (4, 7, 47, 49). Both VNTR and MLST of conserved hypothetical genes did not discriminate between strains isolated from the two mouse lines, in contrast to the high discrimination that we had achieved among other L. johnsonii strains isolated from various hosts in general and mice in particular (Buhnik-Rosenblau et al., submitted). The identical genotyping results obtained at 14 genomic loci, including VNTR, known as highly polymorphic genetic markers as a result of their relatively high mutation rate (51), indicates that the isolates from the two mouse lines are similar and cannot be distinguished. In addition, strains from both mouse lines presented similar growth rates. All the above data suggest that the two mouse lines carry the same L. johnsonii strain. Nevertheless, only a comparison of the complete bacterial genomic sequences can give unequivocal proof of their similarity. Our results indicate that the same L. johnsonii strain might coevolve with the two mouse lines, which in turn suggests that its differential persistence in the guts of C57BL/6J and BALB/c mice is a result of the genetic variation between these mouse lines rather than genetic variation in L. johnsonii. In order to test L. johnsonii's ability to persist in the guts of BALB/c and C57BL/6J mice, the two mouse lines were force fed the same L. johnsonii strain, in addition to their conventional diet. Mice received antibiotic treatment prior to the feeding to give L. johnsonii a better chance of integrating into the gut bacterial population. The results demonstrated that fecal levels of L. johnsonii in C57BL/6J mice are not influenced by feeding, as L. johnsonii levels in both experimental and control groups were high and generally similar. However, in BALB/c mice after the feeding, L. johnsonii levels in the experimental group immediately increased and rapidly decreased to the initial low level observed in the control group throughout the trial (Fig. 3). This suggests that there is some selective force preventing L. johnsonii's persistence at higher levels in the guts of BALB/c mice. In contrast, the high levels of L. johnsonii in C57BL/6J mice indicate that it can persist in that host's genetic background. Taken together, the results suggest that the variation in persistence ability of L. johnsonii between the two mouse lines has a genetic basis. Nevertheless, the following environmental factors may also influence L. johnsonii persistence.
The maternal microbiota is an additional factor that may differentially influence L. johnsonii levels, as each BALB/c or C57BL/6J mouse is exposed to a different maternal microbiota at an early stage of its life. The gut microbiota is known to be dynamic in the early stages of life, as the young mammal is exposed to the maternal microbiota starting at birth (36), and to become relatively stable at later stages (30, 56). To test the effects of the maternal microbiota on the bacterial populations of the two mouse lines, they were subjected to reciprocal crosses. The F1 offspring of the two crosses had the same genetics but differed in their maternally inherited genetic elements, as well as in exposure to different maternal microbiotas. The resultant reciprocal F1 groups presented similar levels of L. johnsonii (Fig. 4), suggesting that L. johnsonii levels are governed mainly by mouse genetics rather than by the maternal microbiota or by other maternal effects. Both reciprocal F1 groups showed high proportions of L. johnsonii, like those in the C57BL/6J mice and much higher than those in the BALB/c mice, suggesting that in the F1 mice, L. johnsonii is allowed to persist, as in C57BL/6J mice. Furthermore, comparing mice with the same maternal parent (BALB/c) but different genomic contents revealed significantly higher L. johnsonii levels in F1 offspring of the C57BL/6J male parent than in those of the BALB/c male parent (i.e., BALB/c mice). The results clearly demonstrated that changing the paternal genetics results in a dramatic change in L. johnsonii levels, further confirming that the mouse's genetic background is the major parameter in determining fecal levels of L. johnsonii, whereas the maternal effect is relatively small. The use of F1 reciprocal crosses provided clear proof of the strong impact of the mouse genetic background on L. johnsonii levels in BALB/c and C57BL/6J inbred lines. The genetically inherited levels of L. johnsonii observed here might be a result of different kinds of interactions between L. johnsonii and the host. These may include direct interplay with the host, like adhesion of L. johnsonii to the mouse gut, as well as indirect host-bacterium interplay, such as interaction of L. johnsonii with other intestinal bacteria. Ongoing genome sequencing of L. johnsonii 16 conducted in our laboratory revealed the presence of genes responsible for increased gut persistence, such as the gene LJ1680, which was previously found to affect the gut persistence of L. johnsonii NCC 533 in a murine model (10). Comparison of this gene between the above-mentioned genomes revealed 75% amino acid identity (in a region of 1,020 amino acids). The presence of such genes in L. johnsonii 16 may support direct interaction with the mouse gut; however, the interaction is dependent on the host genetics. The findings presented here are further supported by numerous studies indicating the influence of the host's genetics on its gut microbiota, conducted with animal models and humans (8, 24, 27, 42, 48, 50, 53, 55). Eighteen host quantitative trait loci were identified in an advanced intercross mouse line originating from a cross between C57BL/6J and an ICR mouse line-derived outbred line (HR) showing linkage with the relative abundance of specific microbial taxa. Among these, the L. johnsonii-Lactobacillus gasseri group was found to segregate with two genomic loci on murine chromosomes 7 and 14 (3). That study and others made use of novel DNA-sequencing techniques that provide a broad view of the gut microbiome (18, 39). However, concentrating on the narrow spectrum of LAB enables the isolation and characterization of potentially probiotic bacterial strains. The culture-dependent approach used here enabled the isolation of a potentially probiotic strain belonging to L. johnsonii, a species that is already used in industry. Furthermore, such an approach is expected to provide insight into the highly specific host-bacterium interactions, down to the subspecies level of both host and bacteria. Indeed, our results present a highly specific interaction between the mouse genetic background and a specific L. johnsonii strain.
In conclusion, our findings suggest that mouse genetics has a major effect on determining the composition of LAB populations in general and the gut persistence of L. johnsonii, a potentially probiotic bacterial species, in particular. Environmental factors had relatively small or undetectable effects on the levels of the tested bacteria. Finding LAB strains with specificity for host genetics in a murine model, as demonstrated here, could lead to the discovery of genes in both the bacterium and its host that may be involved in bacterial persistence in the gut. Such a discovery is expected to have a major impact, especially with respect to health-promoting bacteria (i.e., probiotics), and could lead to future development of probiotic products specifically oriented to the consumer's genetics as part of the personalized-medicine approach.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Bajzer M., Seeley R. J. 2006. Physiology: obesity and gut flora. Nature 444: 1009–1010 [DOI] [PubMed] [Google Scholar]
- 2. Barbas A. S., et al. 2009. Altering and assessing persistence of genetically modified E. coli MG1655 in the large bowel. Exp. Biol. Med. 234: 1174–1185 [DOI] [PubMed] [Google Scholar]
- 3. Benson A. K., et al. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U. S. A. 107: 18933–18938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Broza Y. Y., et al. 2009. Epidemiologic study of Vibrio vulnificus infections by using variable number tandem repeats. Emerg. Infect. Dis. 15: 1282–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Claesson M. J., van Sinderen D., O'Toole P. W. 2007. The genus Lactobacillus: a genomic basis for understanding its diversity. FEMS Microbiol. Lett. 269: 22–28 [DOI] [PubMed] [Google Scholar]
- 6. Costello E. K., et al. 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Danin-Poleg Y., et al. 2007. Vibrio cholerae strain typing and phylogeny study based on simple sequence repeats. J. Clin. Microbiol. 45: 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deloris Alexander A., et al. 2006. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm. Genome 17: 1093–1104 [DOI] [PubMed] [Google Scholar]
- 9. Denou E., et al. 2007. Gene expression of commensal Lactobacillus johnsonii strain NCC533 during in vitro growth and in the murine gut. J. Bacteriol. 189: 8109–8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Denou E., et al. 2008. Identification of genes associated with the long-gut-persistence phenotype of the Probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190: 3161–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dethlefsen L., Eckburg P. B., Bik E. M., Relman D. A. 2006. Assembly of the human intestinal microbiota. Trends Ecol. Evol. 21: 517–523 [DOI] [PubMed] [Google Scholar]
- 12. Dethlefsen L., McFall-Ngai M., Relman D. A. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ducluzeau R. 1983. Implantation and development of the gut flora in the newborn animal. Ann. Rech. Vet. 14: 354–359 [PubMed] [Google Scholar]
- 14. Eckburg P. B., et al. 2005. Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Esworthy R. S., Smith D. D., Chu F. F. 2010. A strong impact of genetic background on gut microflora in mice. Int. J. Inflam. 2010: 986046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farano S., Chierici R., Guerrini P., Vigi V. 2003. intestinal microflora in early infancy: composition and development. Acta Paediatr. 91: 48–55 [DOI] [PubMed] [Google Scholar]
- 17. Firon N., Duksin D., Sharon N. 1985. Mannose-specific adherence of Escherichia coli to Bhk cells that differ in their glycosylation patterns. FEMS Microbiol. Lett. 27: 161–165 [Google Scholar]
- 18. Frank D. N., Pace N. R. 2008. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 24: 4–10 [DOI] [PubMed] [Google Scholar]
- 19. Garrett W. S., Gordon J. I., Glimcher L. H. 2010. Homeostasis and inflammation in the intestine. Cell 140: 859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guandalini S. 2010. Update on the role of probiotics in the therapy of pediatric inflammatory bowel disease. Exp. Rev. Clin. Immunol. 6: 47–54 [DOI] [PubMed] [Google Scholar]
- 21. Haller D., et al. 2010. Guidance for substantiating the evidence for beneficial effects of probiotics: probiotics in chronic inflammatory bowel disease and the functional disorder irritable bowel syndrome. J. Nutr. 140: 690S–697S [DOI] [PubMed] [Google Scholar]
- 22. Holubar S. D., Cima R. R., Sandborn W. J., Pardi D. S. 2010. Treatment and prevention of pouchitis after ileal pouch-anal anastomosis for chronic ulcerative colitis. Cochrane Database Syst. Rev. 6:CD001176 [DOI] [PubMed] [Google Scholar]
- 23. Jakobsson H. E., et al. 2010. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 5: e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang H. Q., Bos N. A., Cebra J. J. 2001. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect. Immun. 69: 3611–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reference deleted.
- 26. Kalliomaki M., et al. 2010. Guidance for substantiating the evidence for beneficial effects of probiotics: prevention and management of allergic diseases by probiotics. J. Nutr. 140: 713S–721S [DOI] [PubMed] [Google Scholar]
- 27. Khachatryan Z. A., et al. 2008. PLoS One 3: e3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ley R. E., et al. 2008. Evolution of mammals and their gut microbes. Science 320: 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ley R. E., Lozupone C. A., Hamady M., Knight R., Gordon J. I. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6: 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023 [DOI] [PubMed] [Google Scholar]
- 31. Lionetti E., et al. 2010. Role of probiotics in pediatric patients with Helicobacter pylori infection: a comprehensive review of the literature. Helicobacter 15: 79–87 [DOI] [PubMed] [Google Scholar]
- 32. Mai V., Draganov P. V. 2009. Recent advances and remaining gaps in our knowledge of associations between gut microbiota and human health. World J. Gastroenterol. 15: 81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mshvildadze M., Neu J., Mai V. 2008. Intestinal microbiota development in the premature neonate: establishment of a lasting commensal relationship? Nutr. Rev. 66: 658–663 [DOI] [PubMed] [Google Scholar]
- 34. Naser S. M., et al. 2005. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 151: 2141–2150 [DOI] [PubMed] [Google Scholar]
- 35. Oh P. L., et al. 2010. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 4: 377–387 [DOI] [PubMed] [Google Scholar]
- 36. Palmer C., Bik E. M., DiGiulio D. B., Relman D. A., Brown P. O. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5: 1556–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park S. H., Itoh K. 2005. Species-specific oligonucleotide probes for the detection and identification of Lactobacillus isolated from mouse faeces. J. Appl. Microbiol. 99: 51–57 [DOI] [PubMed] [Google Scholar]
- 38. Pridmore R. D., et al. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. U. S. A. 101: 2512–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qin J., et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakamoto M., Hayashi H., Benno Y. 2003. Terminal restriction fragment length polymorphism analysis for human fecal microbiota and its application for analysis of complex bifidobacterial communities. Microbiol. Immunol. 47: 133–142 [DOI] [PubMed] [Google Scholar]
- 41. Sharon N. 1987. Bacterial lectins, cell-cell recognition and infectious disease. FEBS Lett. 217: 145–157 [DOI] [PubMed] [Google Scholar]
- 42. Spor A., Koren O., Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9: 279–290 [DOI] [PubMed] [Google Scholar]
- 43. Toivanen P., Vaahtovuo J., Eerola E. 2001. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect. Immun. 69: 2372–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Turnbaugh P. J., et al. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031 [DOI] [PubMed] [Google Scholar]
- 45. Turnbaugh P. J., et al. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1: 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uchida H., et al. 2006. Lactobacilli binding human A-antigen expressed in intestinal mucosa. Res. Microbiol. 157: 659–665 [DOI] [PubMed] [Google Scholar]
- 47. Urwin R., Maiden M. C. J. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11: 479–487 [DOI] [PubMed] [Google Scholar]
- 48. Vaahtovuo J., Toivanen P., Eerola E. 2003. Bacterial composition of murine fecal microflora is indigenous and genetically guided. FEMS Microbiol. Ecol. 44: 131–136 [DOI] [PubMed] [Google Scholar]
- 48a. Vaahtovuo J., Eerola E., Toivanen P. 2005. Comparison of cellular fatty acid profiles of the microbiota in different gut regions of BALB/c and C57BL/6J mice. Antonie Van Leeuwenhoek 88: 67–74 [DOI] [PubMed] [Google Scholar]
- 49. Van der Mee-Marquet N., et al. 2009. Variable-number tandem repeat analysis and multilocus sequence typing data confirm the epidemiological changes observed with Staphylococcus aureus strains isolated from bloodstream infections. J. Clin. Microbiol. 47: 2863–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van de Merwe J. P., Stegeman J. H., Hazenberg M. P. 1983. The resident faecal flora is determined by genetic characteristics of the host. Implications for Crohn's disease? Antonie Van Leeuwenhoek 49: 119–124 [DOI] [PubMed] [Google Scholar]
- 51. Van Ert M. N., et al. 2007. Global genetic population structure of Bacillus anthracis. PLoS One 2: e461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vijay-Kumar M., et al. 2010. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science 328: 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wen L., et al. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455: 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wolvers D., et al. 2010. Guidance for substantiating the evidence for beneficial effects of probiotics: prevention and management of infections by probiotics. J. Nutr. 140: 698S–712S [DOI] [PubMed] [Google Scholar]
- 55. Zoetendal E. G., Akkermans A. D. L., Akkermans-van Vliet W. M., de Visser J. A. G. M., de Vos W. M. 2001. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb. Ecol. Health Dis. 13: 129–134 [Google Scholar]
- 56. Zoetendal E. G., Akkermans A. D. L., de Vos W. M. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64: 3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zoetendal E. G., et al. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68: 3401–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.