Abstract
α-l-Rhamnosidases play an important role in the hydrolysis of glycosylated aroma compounds (especially terpenes) from wine. Although several authors have demonstrated the enological importance of fungal rhamnosidases, the information on bacterial enzymes in this context is still limited. In order to fill this important gap, two putative rhamnosidase genes (ram and ram2) from Pediococcus acidilactici DSM 20284 were heterologously expressed, and the respective gene products were characterized. In combination with a bacterial β-glucosidase, both enzymes released the monoterpenes linalool and cis-linalool oxide from a muscat wine extract under ideal conditions. Additionally, Ram could release significant amounts of geraniol and citronellol/nerol. Nevertheless, the potential enological value of these enzymes is limited by the strong negative effects of acidity and ethanol on the activities of Ram and Ram2. Therefore, a direct application in winemaking seems unlikely. Although both enzymes are members of the same glycosyl hydrolase family (GH 78), our results clearly suggest the distinct functionalities of Ram and Ram2, probably representing two subclasses within GH 78: Ram could efficiently hydrolyze only the synthetic substrate p-nitrophenyl-α-l-rhamnopyranoside (Vmax = 243 U mg−1). In contrast, Ram2 displayed considerable specificity toward hesperidin (Vmax = 34 U mg−1) and, especially, rutinose (Vmax = 1,200 U mg−1), a disaccharide composed of glucose and rhamnose. Both enzymes were unable to hydrolyze the flavanone glycoside naringin. Interestingly, both enzymes displayed indications of positive substrate cooperativity. This study presents detailed kinetic data on two novel rhamnosidases, which could be relevant for the further study of bacterial glycosidases.
INTRODUCTION
α-l-Rhamnosidases (EC 3.2.1.40) catalyze the hydrolysis of a wide spectrum of natural glycosides containing terminal l-rhamnose. The biotechnological interest in rhamnosidases includes the debittering of citrus fruit juices (naringinase and hesperidinase), the release of flavonoids from rhamnosylated precursors, and the production of l-rhamnose as starting material for chemical synthesis (52). Further, rhamnosidases may play a crucial role in wine making due to their contributions to releasing grape-derived aroma compounds (mainly terpenes and terpenic alcohols). Depending on the grape variety, rutinosides (α-l-rhamnopyranosyl-β-d-glucopyranoside) constitute 6 to 13% of the terpenic glycosides found in wine (29). The combination of β-d-glucosidase and α-l-rhamnosidase allows the sequential release of volatile compounds from these precursors (18). Several authors (9, 32, 46) have demonstrated the enological value of fungal (Aspergillus sp.) rhamnosidases. Based on their adaptation to the harsh wine milieu and their general hydrolytic abilities toward different glycosides (16, 17, 50), wine-related lactic acid bacteria (LAB) have attracted much interest as a potential source of novel glycosidases (34). Consequently, several (intracellular) glycosidases from different wine-related bacterial species have already been identified and characterized (11, 12, 36–38, 48). Another driving force for the identification and characterization of bacterial glycosidases, including rhamnosidases, is the observation that plant secondary metabolites (i.e., flavonoids) with potential health benefits have improved bioavailability in their aglycon forms (26). Accordingly, the potential of intestinal bacteria (enterococci, lactobacilli, clostridia, and bifidobacteria) to hydrolyze natural glycosides has been demonstrated (21, 42, 43).
For all these reasons, the isolation and characterization of glycosidases from food-related bacteria is a field of increasing research. However, only a few bacterial rhamnosidases have been characterized (15, 19, 20, 54) to date, and the properties of rhamnosidases derived from food LAB (Lactobacillus plantarum and Lactobacillus acidophilus) have only recently been reported (3, 7). In the present study, we characterized two α-l-rhamnosidases derived from the homofermentative lactic acid bacterium Pediococcus acidilactici. Pediococci can be found in a wide range of fermented products, including wine (41), and Pediococcus spp. have also been isolated from the human digestive tract and have been discussed as potential probiotics (5, 27). The potential of wine-related pediococci to hydrolyze synthetic glycosides has been demonstrated by Grimaldi et al. (16). Apart from a detailed biochemical characterization, the main objective of this study was to evaluate whether the two putative rhamnosidases from P. acidilactici DSM 20284 can release grape-derived terpenes from their natural precursors.
MATERIALS AND METHODS
DNA manipulation and sequence analysis.
PCR was performed with the Phusion Master Mix (New England BioLabs, Ipswich, MA). All restriction enzymes and the Quick Ligation kit were also purchased from New England BioLabs. PCR products were purified with the Wizard SV Gel and PCR Clean-UP Kit, and plasmid purification was done with the PureYield Plasmid Miniprep System (both from Promega, Madison, WI). The pET system (pET21 series; Novagen, Madison, WI) was used for gene overexpression in Escherichia coli. All procedures were performed according to the suppliers' recommendations.
Two putative rhamnosidase genes identified in the published genome of P. acidilactici DSM 20284 (GenBank accession no. NZ_AEEG00000000.1; locus tags HMPREF0623_0163 [ram] and HMPREF0623_0062 [ram2]) were amplified from the genomic DNA of P. acidilactici DSM 20248 (DSMZ GmbH, Braunschweig, Germany). Primers designed with specific restriction sites (Table 1) were used to introduce the genes into the expression vectors pET21a (ram) and pET21d (ram2) in frame with a C-terminal His6 tag. The restriction sites used (Table 1) were NdeI/XhoI (ram/pET21a) and NcoI/XhoI (ram2/pET21d). DNA sequencing (Agowa GmbH, Berlin, Germany) confirmed the correct formation of the constructs obtained, as well as the complete identity of the cloned genes with the published sequences of ram and ram2. Both constructs were used to transform E. coli T7 Express competent cells (New England BioLabs) according to the supplier's recommendations. Positive colonies were selected on Luria broth agar plates supplemented with ampicillin (100 μg ml−1).
Table 1.
Oligonucleotides used for amplification of rhamnosidase genes ram and ram2 from P. acidilactici
| Primer name | Sequence (5′-3′)a |
|---|---|
| ramF (NdeI) | GATATACATATGAATCAAACTTATTGGATC |
| ramR (XhoI) | GTGGTGCTCGAGCGCAATGGGAATTACTACG |
| ram2F (BsaI)b | GGTCTCACATGGCATTTACATTTCAAA |
| ram2R (XhoI) | GTGGTGCTCGAGAGATAAATACTTTCTCATTAAATA |
Restriction sites are underlined.
Digestion with BsaI creates an overhang for ligation with the NcoI site of pET21d.
The deduced protein sequences of ram and ram2 (GenBank accession no. ZP_07367044 and ZP_07366943, respectively) were analyzed with the following tools: Blastp (1) was used to search GenBank and to determine amino acid sequence similarities, and the subcellular locations of Ram (R) and Ram2 (R2) were predicted with SignalP (13), TMpred (25), and PSORTb (53).
Protein production and purification.
Protein production was carried out in Terrific broth (49) containing 100 μg ml−1 ampicillin at 25°C and 150 rpm. Expression was induced by addition of lactose (final concentration, 5 g liter−1). Following overnight cultivation, the cells were harvested by centrifugation at 6,000 × g for 20 min. After resuspension, the cells were disrupted in a French press at 8.3 MPa in 3 cycles. The homogenate was centrifuged at 100,000 × g for 30 min, and the supernatant was recovered as crude extract. The recombinant His6-tagged enzymes were purified by immobilized metal affinity chromatography (IMAC) on a column packed with Ni-charged chelating Sepharose (GE Healthcare, Uppsala, Sweden) according to the suppliers' recommendations.
Electrophoresis and molecular mass determination.
SDS-PAGE, including Coomassie blue staining, was performed using the Mini-Protean system with precast gels (4 to 20%) from Bio-Rad (Hercules, CA); the molecular mass marker used was High Precision Dual Color (10- to 250-kDa range).
Molecular masses were determined by size exclusion chromatography on a Sephacryl S-300 column (GE Healthcare) with 50 mM sodium phosphate buffer, pH 7.0, containing 150 mM NaCl at a flow rate of 0.15 cm min−1. The column was calibrated with protein standards from Sigma-Aldrich (kit for molecular weights 29,000 to 700,000).
Enzyme assays.
All substrates for enzyme assays were purchased from Sigma-Aldrich (St. Louis, MO). The substrates used were p-nitrophenol (pNP)-linked glycosides (pNP-α-l-rhamnopyranoside, pNP-β-d-glucopyranoside, pNP-β-d-xylopyranoside, pNP-β-d-mannopyranoside, and pNP-α-l-arabinofuranoside), naringin, hesperidin, rutin, and rutinose.
The enzyme solutions were stored in 0.02 M citrate phosphate buffer, pH 7.0, prepared according to the method of McIlvaine (35). Standard reaction conditions for all enzyme assays were 0.1 M McIlvaine buffer, pH 5.5, 37°C, and 10-min incubation time. All determinations were performed in triplicate.
Assays with p-nitrophenyl-glycosides were stopped with 0.5 M Na2CO3 (2-fold volumetric excess), and the absorbance of p-nitrophenol was measured at 400 nm (ε400 = 18.300 M−1 cm−1 at pH 10.2) in a Beckman DU 800 spectrometer (Paolo Alto, CA). One unit of rhamnosidase activity corresponds to 1 μmol of p-nitrophenol released per min at 37°C. The temperature dependence of enzyme activity was determined by variation of the temperature in the standard assay between 4 and 90°C. The pH dependence was determined using McIlvaine buffers ranging from pH 3.0 to 8.0 at 37°C.
Assays with nonchromogenic substrates (naringin, hesperidin, rutin, and rutinose) were conducted as described above but were stopped by heat inactivation at 80°C for 5 min. Rhamnose, glucose, and rutinose were quantified by HPLC analysis (Dionex [Sunnyvale, CA] DX500) with a CarboPac PA 1 column and pulsed amperometric gold electrode detection. The injection volume was 20 μl; 15 mM NaOH was used for elution, and the flow was 1 ml min−1 at 25°C and 115 × 105 Pa. One unit of rhamnosidase activity is expressed as released rhamnose (μmol) per min at 37°C and pH 5.5.
Kinetic constants for Ram and Ram2 (Table 2) were determined under the above-mentioned standard assay conditions (pH 5.5 and 37°C) by variation of the individual substrate (pNP-α-l-rhamnopyranoside, hesperidin, rutin, or rutinose) concentration. The resulting plots are shown in Fig. S1 and S2 in the supplemental material. All kinetic data were interpreted with the Enzyme Kinetics Module (version 1.3) of SigmaPlot 10.0. The regression models used were those of Michaelis-Menten and Hill (24, 44).
Table 2.
Kinetic constants of P. acidilactici rhamnosidases Ram and Ram2 determined at 37°C and pH 5.5
| Enzyme | Substrate | Kinetic constanta |
||||
|---|---|---|---|---|---|---|
| Km′ (mM)b | Vmax (U mg−1) | kcat (s−1) | kcat/Km (s−1 mM−1) | nhc | ||
| Ram | pNP-α-l-rhamnopyranoside | 16.2 ± 1.7 | 243 ± 14 | 311 | 19.2 | 1.6 ± 0.2 |
| Hesperidin | 3.88 ± 0.26 | 0.638 ± 0.022 | 0.817 | 0.211 | 1.5 ± 0.1 | |
| Rutin | 0.793 ± 0.050 | 1.45 ± 0.045 | 1.84 | 2.32 | 1.7 ± 0.2 | |
| Rutinose | 2.38 ± 0.20 | 0.0335 ± 0.0016 | 0.0423 | 0.0178 | 1.5 ± 0.2 | |
| Ram2 | pNP-α-l-rhamnopyranoside | 22.3 ± 0.69 | 3.62 ± 0.081 | 3.698 | 0.166 | 2.0 ± 0.1 |
| Hesperidin | 7.00 ± 0.40 | 33.0 ± 0.91 | 33.7 | 4.81 | 1.2 ± 0.04 | |
| Rutin | 0.332 ± 0.014 | 0.913 ± 0.023 | 0.930 | 2.82 | 2.0 ± 0.2 | |
| Rutinose | 2.54 ± 0.30 | 1200 ± 46 | 1224 | 482 | 1.0 ± 0.1 | |
All data represent the average of triplicate determinations ± SD. All results were interpreted with the Hill equation (24).
Equivalent to the Michaelis-Menten constant, Km.
Apparent Hill coefficient.
Release of terpenes from muscat wine.
White wine (Muskat Ottonel; Donauprinz, Hungary, 2009) was evaporated (Heidolph [Schwabach, Germany] rotovapor) at 35°C to remove ethanol and volatile compounds, and the pH was adjusted to 5.5 with KHCO3. The wine was sterilized by filtration (0.22 μm). The samples (50 ml) were treated with the rhamnosidases Ram and Ram2, a β-glucosidase (GL) from Lactobacillus brevis (36), and a commercial rhamnosidase preparation (naringinase [N] from Penicillium decumbens; Sigma-Aldrich) in the following combinations: GL, R, R2, GL plus R, GL plus R2, N, and GL plus N. All enzyme preparations were sterilized (0.22-μm filter) before application. The final enzyme concentrations were 2 U ml−1 as determined with p-nitrophenyl-glycosides. In the case of Ram2, the same protein concentration as for Ram was used. The samples were incubated for 7 days at 15°C, and all determinations were performed in triplicate.
Volatile compounds were analyzed by headspace gas chromatography-mass spectrometry (GC-MS). A 7890A GC system (Agilent Technologies, Palo Alto, CA) with a DB-5 capillary column (60 m by 0.25 mm; 0.25 μm), a CombiPal autosampler (CTC Analytics, Zwingen, Switzerland), and a 5975C MS detector (Agilent) were used. The samples were prepared by solid-phase microextraction (SPME). Five milliliters of sample and 50 μl of the internal standard (4-chlorobutyl acetate) were added to a vial containing 2 g NaCl. SPME fibers (100 μm polydimethylsiloxane) from Supelco (Bellefonte, PA) were used as an absorbant. Extraction was performed for 30 min at 50°C, followed by desorption for 5 min at 250°C. The samples were injected in splitless mode (3 min), and the carrier gas was helium (99.999%; Air Liquide, Vienna, Austria) with a flow rate of 1.2 ml min−1. The program for the oven temperature was as follows: initial temperature, 50°C for 3 min; temperature increase to 92°C (1°C min−1); holding time, 10 min; a further increase to 127°C (5°C min−1); then, an increase to 260°C (40°C min−1) with a holding time of 5 min. Ionization was performed at 70 eV. Ions were quantified by selected ion monitoring (SIM). The following standards were applied (the m/z ratios used for quantification are shown in parentheses): internal standard, 4-chlorobutyl acetate (54); cis/trans-linalool oxide (59); linalool (71); hotrienol (71); cis/trans-rose oxide (139); cis-limonene oxide (67); trans-limonene oxide (94); alpha-terpineol (93); beta-terpineol (71); gamma-terpineol (121); nerol (69); citronellol (69); geraniol (69); nerol oxide (68); and lavandulool (69).
The data were statistically analyzed with the software package SPSS 18. One-way analysis of variance was applied to test for significant differences between the individual treatments. The results were grouped into homogeneous groups by Student's t test (Student-Newman-Keuls) at a significance level of 95% (α = 0.05).
RESULTS
Heterologous expression and oligomeric structures.
Two putative rhamnosidase genes (ram and ram2) of P. acidilactici DSM 20284 were expressed in E. coli. The protein sequences deduced from ram and ram2 contain 653 and 525 amino acid residues, respectively. The highest sequence similarities were found in the already characterized glycosyl hydrolase family 78 rhamnosidases Ram1Lp and Ram2Lp of L. plantarum (7). Ram shares 56% identity with Ram1Lp, and Ram2 shares 61% with Ram2Lp. Ram and Ram2 share 29% sequence identity with each other. The topology prediction software TMpred (25) and PSORTb (53) indicated cytoplasmic localization of both enzymes; no signal peptides could be identified with SignalP (13).
As shown in Fig. 1, the His6-tagged enzymes were electrophoretically pure after immobilized metal affinity chromatography. Preliminary tests with pNP-α-l-rhamnopyranoside (1 mM) showed that both enzymes are functional α-l-rhamnosidases with specific activities of 8.7 U mg−1 (Ram) and 0.036 U mg−1 (Ram2). Molecular masses of 74 ± 1 kDa (Ram) and 241 ± 4 kDa (Ram2) were determined by size exclusion chromatography. Taking the calculated molecular masses of 76.8 kDa (Ram) and 61.3 kDa (Ram2) into account, these data suggest that Ram is a monomer and Ram2 is a tetramer.
Fig. 1.
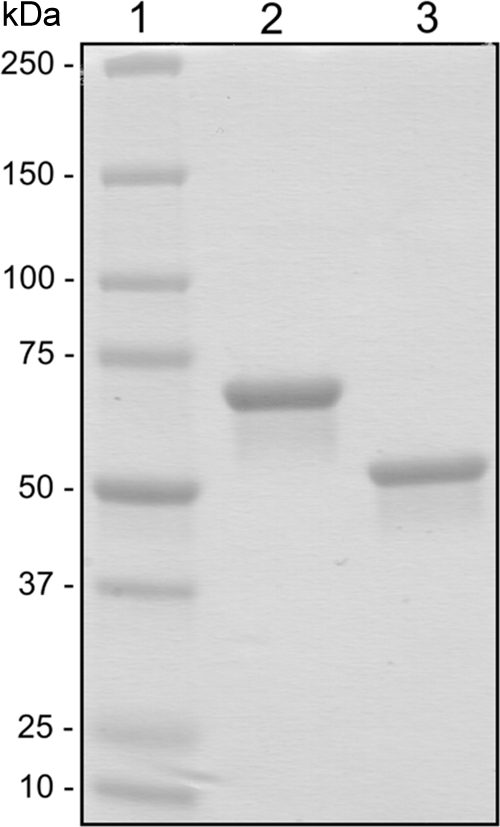
SDS-PAGE of the recombinant rhamnosidases from P. acidilactici after IMAC purification. Lane 1, Precision Plus protein standard (Bio-Rad); lane 2, Ram; lane 3, Ram2.
Substrate affinities of Ram and Ram2.
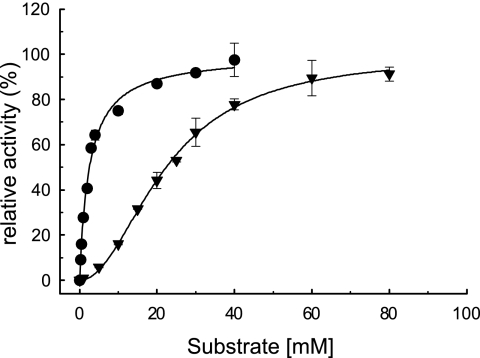
In assays with synthetic p-nitrophenyl-linked substrates, both enzymes could effectively hydrolyze only pNP-α-l-rhamnopyranoside (pNPR). Ram had low additional activity toward pNP-β-d-glucopyranoside (<0.01 U mg−1), and no other side activities (α-l-arabinosidase, β-d-xylosidase, and β-d-mannosidase) could be detected. Further, both enzymes were unable to liberate rhamnose (α-1,2 linked to glucose) from the flavanone rutinoside naringin. Table 2 displays the kinetic constants determined with the synthetic substrate pNPR and the natural substrates hesperidin, rutin, and rutinose (α-l-rhamnopyranosyl-1,6-β-d-glucopyranose). Hesperidin and rutin are the rutinosides of hesperetin and quercetin, respectively. These data clearly suggest distinct substrate specificities of Ram and Ram2. Ram showed its highest catalytic efficiency with pNPR but had low affinity for the other substrates. Ram2 was highly efficient with rutinose but not with pNPR. Unlike Ram, Ram2 also showed considerable activity toward hesperidin. Interestingly, both enzymes displayed sigmoidal responses to increasing substrate concentrations rather than following the hyperbolic Michaelis-Menten correlation (see Fig. S1 and S2 in the supplemental material). All results were interpreted with the Hill equation (24), where nh is the Hill coefficient reflecting the extent of substrate binding cooperativity (44, 51) and Km′ is equivalent to the Michaelis-Menten constant, Km. These results suggest that both enzymes are subject to allosteric regulation, exhibiting positive substrate binding cooperativity (nh > 1), a mechanism that implies an oligomeric quaternary structure (44). However, Ram showed rather weak positive cooperativity in all cases (nh = 1.5 to 1.7). Although using the Hill equation resulted in better estimates (i.e., curve fit) for Km and Vmax than the Michaelis-Menten equation (see Fig. S1 and S2 in the supplemental material), the fact that Ram is apparently a monomer makes the possibility of allosteric regulation unlikely. For Ram2, the results indicate clear positive cooperativity (nh = 2) with pNPR and rutin, the substrates for which the enzyme has the lowest affinity, and very low cooperativity (nh = 1.2) with hesperidin. Remarkably, no cooperativity was observed with rutinose; in this case (nh = 1), the Hill equation is reduced to the Michaelis-Menten equation. Consequently, interpretation of the results for rutinose yielded the same estimates for Km and Vmax with both models. Figure 2 shows the kinetic plots obtained with pNPR and rutinose, clearly demonstrating the different responses of Ram2 to these substrates.
Fig. 2.
Kinetic plots of the P. acidilactici rhamnosidase Ram2 with pNP-rhamnopyranoside (▾) and rutinose (•). The data were interpreted with the Hill equation (24); nh values were 2.0 ± 0.1 and 1.0 ± 0.1 for pNP-rhamnopyranoside and rutinose, respectively, and 100% relative activity refers to Vmax. All determinations were performed in triplicate; the error bars indicate standard deviations (SD).
Physicochemical properties.
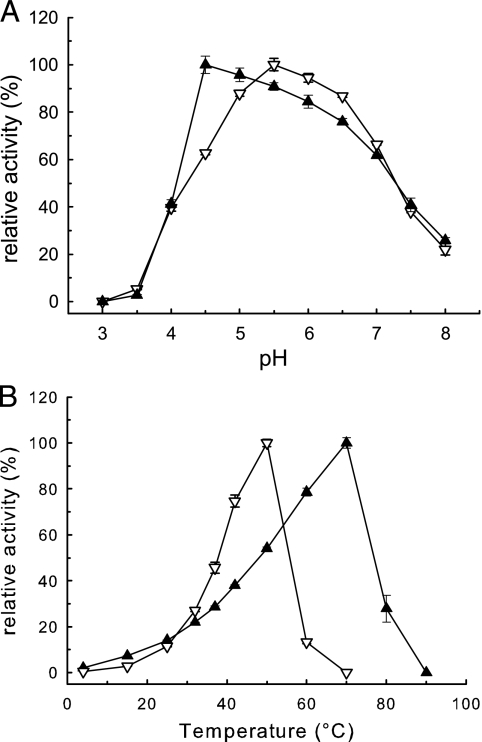
All assays were conducted with pNPR (1 mM). Figure 3 shows the influences of pH and temperature on the activities of Ram and Ram2. Ram displayed the highest activities at pH 5.5 and 50°C, Ram2 at pH 4.5 and 70°C. Both enzymes were rapidly inactivated below pH 4.
Fig. 3.
Influence of pH and temperature on the rhamnosidases from P. acidilactici determined with pNP-α-l-rhamnopyranoside (1 mM). ▿, Ram; ▴, Ram2. (A) Assays (37°C; 10-min reaction time) were performed with 0.1 M McIlvaine buffers, pH 3.0 to 8.0. (B) Standard assay (pH 5.5; 10 min) performed at 4 to 90°C. All data represent the averages of triplicate determinations. The error bars indicate SD; 100% relative activity refers to the maximal activity determined.
Table 3 shows the influences of solvents, sugars, and salts, showing that both ethanol and dimethyl sulfoxide (DMSO) reduced enzyme activities. The salts included (100 mM) had no detrimental effect; only the activity of Ram was reduced 16% by MgSO4. Both enzymes were significantly inhibited by glucose and rhamnose (250 mM each). While Ram was strongly inhibited by rhamnose and to a lesser degree by glucose, Ram2 was almost completely inhibited by glucose but retained 45% activity in the presence of rhamnose. In respect to the considerations of allosteric regulation presented above, further tests were conducted to determine whether the inhibition of Ram2 was subject to allosteric regulation (i.e., noncompetitive inhibition) as well. However, the results indicated that both rhamnose and glucose inhibited Ram2 competitively, as judged from increasing Km′ but no decrease in Vmax (data not shown).
Table 3.
Influences of solvents, sugars, and salts in the assay on activities of the P. acidilactici rhamnosidases Ram and Ram2 determined with pNP-α-l-rhamnopyranoside (1 mM)
| Additivea | Relative enzyme activity (%)b |
|
|---|---|---|
| Ram | Ram2 | |
| Solvents | ||
| EtOH (6% [vol/vol]) | 56 ± 0.5 | 78 ± 0.8 |
| EtOH (12% [vol/vol]) | 31 ± 1 | 74 ± 0.7 |
| DMSO (12.5% [vol/vol]) | 37 ± 0.7 | 11 ± 0.7 |
| DMSO (25% [vol/vol]) | 24 ± 1 | 4.7 ± 0.2 |
| Sugars | ||
| Rhamnose (250 mM) | 15 ± 1 | 45 ± 1 |
| Glucose (250 mM) | 76 ± 1 | 2.4 ± 0 |
| Salts | ||
| NaCl (100 mM) | 130 ± 2 | 121 ± 1 |
| KCl (100 mM) | 128 ± 6 | 142 ± 3 |
| K2SO4 (100 mM) | 115 ± 1 | 113 ± 6 |
| MgSO4 (100 mM) | 84 ± 2 | 111 ± 3 |
| MgCl2 (100 mM) | 127 ± 5 | 120 ± 2 |
| CaCl2 (100 mM) | 123 ± 1 | 102 ± 3 |
EtOH, ethanol.
Activity in the absence of additives (standard assay conditions) was set at 100%. All data represent the average of triplicate determination ± SD.
Release of terpenic compounds from muscat.
The abilities of Ram and Ram2 to release grape-derived terpenes were determined with a modified muscat wine (pH 5.5; ethanol removed) as a substrate. The samples were treated with enzymes in excess and incubated at 15°C for 7 days under sterile conditions. A β-glucosidase from L. brevis (GL) and a commercial fungal rhamnosidase preparation (N) were included as controls. As previously reported (36), GL is a bifunctional glucosidase/xylosidase with low arabinosidase side activity. In relation to its rhamnosidase activity (100%), naringinase had side activities of β-d-glucosidase (12.5%), β-d-xylosidase (6.4%), and α-l-arabinosidase (33%).
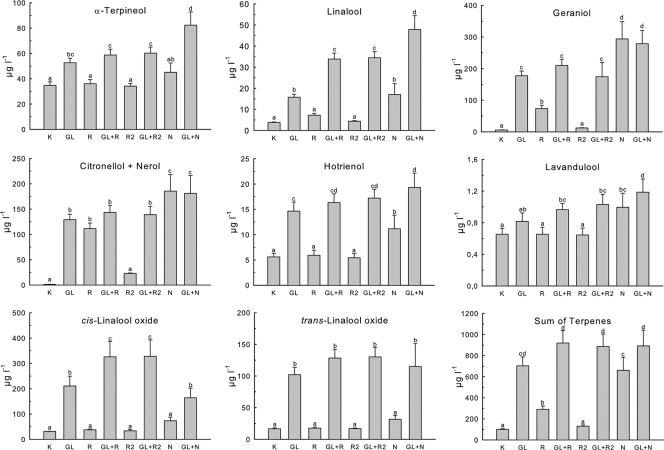
The results are shown in Fig. 4. As the peaks of citronellol and nerol were not sufficiently separable by the GC-MS method used (data not shown), the results are displayed as the sum of citronellol and nerol. Both GL and N demonstrated broad substrate specificities and were able to release large amounts of terpenes. On the other hand, it is obvious that both bacterial rhamnosidases had only a small impact on the overall release of terpenic compounds (Fig. 4, sum of terpenes). Although addition of Ram/Ram2 to GL could evidently increase the total free terpenes compared to treatment with GL alone, the differences are not statistically significant (Student-Newman-Keuls; α = 0.05). In detail, however, both Ram and Ram2 could significantly increase the concentrations of linalool and cis-linalool oxide in combination with GL in comparison to treatment with GL only, confirming the mode of sequential hydrolysis, as suggested by Gunata et al. (18). Further, Ram could also liberate statistically significant amounts of geraniol and citronellol/nerol without glucosidase, which is especially evident in the sum of citronellol/nerol. The same effect was not observed with Ram2. Although the β-glucosidase activity of Ram was low with pNPR, it is not inconceivable that Ram could display higher activity toward other glucosides (14). This seems plausible, since addition of GL to Ram did not result in the expected further increase of citronellol/nerol. An explanation for the differences between Ram and Ram2 could be different specificities toward aglycons with distinct chemical structures. Linalool and its oxides possess tertiary alcohol groups, while geraniol, citronellol, and nerol have primary OH groups (29, 50). A similar observation was made with GL/N: addition of GL to N could not further increase concentrations of geraniol and citronellol/nerol. On the other hand, the concentrations of α-terpineol, linalool, linalool oxides, and hotrienol could be significantly increased by the addition of GL to N. This suggests that the β-glucosidase of L. brevis has additional activity toward terpenols with tertiary hydroxyl groups, resulting in a synergistic effect in combination with the fungal enzyme preparation.
Fig. 4.
Release of monoterpenes from modified Muskat Ottonel (pH 5.5) by rhamnosidases of P. acidilactici. K, control without treatment. The samples were incubated with the enzymes at 15°C for 7 days. All experiments were performed in triplicate; the error bars represent SD. The results were grouped by t test (Student-Newman-Keuls; α = 0.05). Values labeled with the same letters are not significantly different.
DISCUSSION
At present, we have access to abundant genomic data on bacterial glycosidases in public databases (8, 10, 31). Unfortunately, only a small fraction of these putative enzymes have been characterized. Although the computational methods now available are invaluable in terms of identification, sequence analysis, and functional prediction of new enzymes, there is a strong scientific need to collect experimental data in order to confirm such in silico predictions. The aim of this study was to characterize two putative rhamnosidases derived from the same strain of P. acidilactici. While both enzymes are classified as members of glycosyl hydrolase family 78 (10, 22), our results indicate that Ram and Ram2 are quite distinct in functionality and therefore possibly in metabolic function, as well. In agreement with Gagné et al. (14), who argued that the mere use of synthetic substrates is insufficient to reflect the true potential of a given organism or enzyme, we sought to access the capabilities of Ram and Ram2 using both synthetic and natural substrates. In respect to the biotechnological interest in rhamnosidases summarized in the introduction, it is evident that the potential of the enzymes presented to hydrolyze flavonoid glycosides is limited. Though able to convert hesperidin and rutin, both catalysts demonstrated rather low affinity for these substrates. Further, no activity toward naringin could be detected. Ram2 was highly efficient with the disaccharide rutinose, indicating a possible importance in sugar metabolism, which might also be a reason for the strong inhibitory effect of glucose. However, since rutinose does not naturally occur in free form, the actual metabolic role of the enzyme is not obvious. A possible function of Ram2 might be the release of rhamnose from heteropolysaccharides containing rhamnose linked to glucose in α-1,6 configuration. Avila et al. (3) discussed the possible involvement of bacterial rhamnosidases in the breakdown of bacterial cell wall or extracellular polysaccharides.
Another interesting observation was that both enzymes displayed indications of positive substrate cooperativity to various degrees. This mechanism is usually related to oligomeric enzymes of metabolic importance, enabling the catalyst to exhibit a more sensitive response to physiological substrate concentrations (44). Although the effect has rarely been reported in association with glycosidases, Capaldo et al. (12) have recently described positive cooperativity in a bacterial phospho-β-glucosidase related to the phosphoenolpyruvate-dependent sugar-phosphotransferase system (PTS), which is probably involved in cellobiose degradation. However, in the present case, the kinetic data also suggest that the observed allosteric properties may not be a vital function in the regulation of Ram and Ram2 at all. Positive cooperativity seems to be related to poor substrate affinity, which is especially pronounced in the case of Ram2, where the hydrolysis of the obviously “true” substrate rutinose follows the typical Michaelis-Menten kinetics. This is in fact consistent with the sequential model of cooperative binding as postulated by Koshland et al., assuming that the catalyst's affinity to the ligand is initially poor but is enhanced by substrate binding (positive homotropic modulation) (28, 44).
Concerning the release of attractive wine aroma compounds, our results indicate that Ram is a true aryl-glycosidase that is able to hydrolyze grape-derived terpenyl-glycosides. Although Ram2 could liberate linalool and cis-linalool oxide, as well, it could not release terpenols with a primary OH group (geraniol, citronellol, and nerol), in contrast to Ram. In regard to these data, Ram might be an interesting candidate for the release of grape-derived aroma compounds. Nevertheless, in regard to a possible application of these rhamnosidases, it is important to state that the experiments conducted here were performed under optimal enzyme conditions (pH 5.5; no ethanol), which was mainly intended to determine both enzymes' biochemical characteristics. Due to the strong negative effect of acidity and ethanol on the activities of both rhamnosides, a direct application to wine or fruit juice seems unrealistic. Further, the strong inhibitory effect of glucose on Ram2 limits its possible application even in the early stages of wine making. Although immobilization has been reported to be a good option to enhance enzyme activity and stability under unfavorable conditions (4, 33, 47), an interesting alternative could be to overexpress such enzymes in lactic acid bacteria. Prominent expression systems developed for LAB are the NICE system for Lactococcus lactis (39) and pSIP for L. plantarum (45). Beekwilder et al. (7) overexpressed Ram1Lp in L. lactis (NICE) and used this strain to ferment tomato pulp. Although the strain showed increased activity toward pNPR, it failed to convert rutin, probably due to insufficient substrate internalization by L. lactis. Regarding the release of terpenes from wine, an interesting approach could be the overexpression of a glycosidase gene of interest in a LAB species suitable for malolactic fermentation (MLF). Although Oenococcus oeni, the preferred species for MLF, has often been reported to be nonsusceptible to DNA transformation (6), an electroporation protocol for O. oeni has been established recently (2). Further, recent developments showed promising results in the direction of true “food grade expression” by establishing selection markers that are not dependent on antibiotics (30, 40).
In conclusion, the observations made in this study should confirm that a detailed experimental characterization of putative glycosidases is indeed “more than adding to a stamp collection,” as stated by Henrissat et al. (23). Despite the possibly limited applications of the enzymes presented, our results give detailed insight into the biochemical properties of two bacterial rhamnosidases, which should be a step toward a better understanding of LAB glycosidases. Although an evolutionary trend toward metabolic simplification has been observed in lactic acid bacteria (31), there are indications that the glycosidase metabolism of LAB is of a rather complex nature. The research conducted in recent years has demonstrated that LAB are a versatile source of diverse glycosidases. Further, the current genomic databases (GenBank) indicate a wide occurrence of putative glycosidase genes in LAB, only a small fraction of which have been experimentally accessed so far. This includes a high redundancy of PTS-related phosphoglycosidases. The genome of P. acidilactici 20284 (GenBank accession no. NZ_AEEG00000000.1) alone contains at least 6 putative phospho-β-glucosidase genes. Unfortunately, we have only a little information about this class of enzymes. In summary, we are still far from understanding the complexity of bacterial glycoside metabolism. Therefore, it is clear that further meticulous work will be necessary to understand the functionality and metabolic relevance of LAB glycosidases. Apart from carefully conducted in vivo studies on gene regulation and expression, it is clear that the detailed elucidation of enzyme characteristics as attempted here is vital to lead to a better understanding of the principles of bacterial glycosidase metabolism.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant (FWF project 20246-B11) given to K.D.K. by the Austrian Science Fund.
We thank Viktoria Hell for analytical assistance. We also thank Konrad Domig and Paul Furtmüller for scientific advice.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Assad-García J. S., et al. 2008. An improved protocol for electroporation of Oenococcus oeni ATCC BAA-1163 using ethanol as immediate membrane fluidizing agent. Lett. Appl. Microbiol. 47: 333–338 [DOI] [PubMed] [Google Scholar]
- 3. Avila M., et al. 2009. Physiological and biochemical characterization of the two alpha-L-rhamnosidases of Lactobacillus plantarum NCC245. Microbiology 155: 2739–2749 [DOI] [PubMed] [Google Scholar]
- 4. Avnir D., Braun S., Lev O., Ottolenghi M. 1994. Enzymes and other proteins entrapped in sol-gel materials. Chem. Mater. 6: 1605–1614 [Google Scholar]
- 5. Barros R. R., Carvalho M. D. G. S., Peralta J. M., Facklam R. R., Teixeira L. M. 2001. Phenotypic and genotypic characterization of Pediococcus strains isolated from human clinical sources. J. Clin. Microbiol. 39: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartowsky E. J. 2005. Oenococcus oeni and malolactic fermentation: moving into the molecular arena. Aust. J. Grape Wine Res. 11: 174–187 [Google Scholar]
- 7. Beekwilder J., et al. 2009. Characterization of rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl. Environ. Microbiol. 75: 3447–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. 2011. GenBank. Nucleic Acids Res. 39: D32–D37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caldini C., Bonomi F., Pifferi P. G., Lanzarini G., Galante Y. M. 1994. Kinetic and immobilization studies on fungal glycosidases for aroma enhancement in wine. Enzyme Microbiol. Technol. 16: 286–291 [Google Scholar]
- 10. Cantarel B. L., et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Capaldo A., Walker M. E., Ford C. M., Jiranek V. 2011. β-Glucoside metabolism in Oenococcus oeni: cloning and characterisation of the phospho-β-glucosidase bglD. Food Chem. 125: 476–482 [Google Scholar]
- 12. Capaldo A., Walker M. E., Ford C. M., Jiranek V. 2011. β-Glucoside metabolism in Oenococcus oeni: cloning and characterization of the phospho-β-glucosidase CelD. J. Mol. Catal. B 69: 27–34 [Google Scholar]
- 13. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- 14. Gagné S., et al. 2011. Variety and variability of glycosidase activities in an Oenococcus oeni strain collection tested with synthetic and natural substrates. J. Appl. Microbiol. 110: 218–228 [DOI] [PubMed] [Google Scholar]
- 15. Gastón Orrillo A., Ledesma P., Delgado O. D., Spagna G., Breccia J. D. 2007. Cold-active α-L-rhamnosidase from psychrotolerant bacteria isolated from a sub-Antarctic ecosystem. Enzyme Microbiol. Technol. 40: 236–241 [Google Scholar]
- 16. Grimaldi A., Bartowsky E., Jiranek V. 2005. Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in oenology. J. Appl. Microbiol. 99: 1061–1069 [DOI] [PubMed] [Google Scholar]
- 17. Grimaldi A., Bartowsky E., Jiranek V. 2005. A survey of glycosidase activities of commercial wine strains of Oenococcus oeni. Int. J. Food Microbiol. 105: 233–244 [DOI] [PubMed] [Google Scholar]
- 18. Gunata Z., Bitteur S., Brillouet J.-M., Bayonove C., Cordonnier R. 1988. Sequential enzymic hydrolysis of potentially aromatic glycosides from grape. Carbohydr. Res. 184: 139–149 [Google Scholar]
- 19. Hashimoto W., Nankai H., Sato N., Kawai S., Murata K. 1999. Characterization of α-L-rhamnosidase of Bacillus sp. GL1 responsible for the complete depolymerization of gellan. Arch. Biochem. Biophys. 368: 56–60 [DOI] [PubMed] [Google Scholar]
- 20. Hashimoto W., Miyake O., Nankai H., Murata K. 2003. Molecular identification of an [alpha]-rhamnosidase from Bacillus sp. strain GL1 as an enzyme involved in complete metabolism of gellan. Arch. Biochem. Biophys. 415: 235–244 [DOI] [PubMed] [Google Scholar]
- 21. Hawksworth G., Drasar B. S., Hill M. J. 1971. Intestinal bacteria and the hydrolysis of glycosidic bonds. J. Med. Microbiol. 4: 451–459 [DOI] [PubMed] [Google Scholar]
- 22. Henrissat B., Davies G. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7: 637–644 [DOI] [PubMed] [Google Scholar]
- 23. Henrissat B., Sulzenbacher G., Bourne Y. 2008. Glycosyltransferases, glycoside hydrolases: surprise, surprise! Curr. Opin. Struct. Biol. 18: 527–533 [DOI] [PubMed] [Google Scholar]
- 24. Hill A. V. 1910. The combinations of haemoglobin with oxygen and with carbon monoxide. Int. J. Physiol. 40: iv–vii [Google Scholar]
- 25. Hofmann K., Stoffel W. 1993. TMBase: a database of membrane spanning proteins segments. Bio. Chem. Hoppe-Seyler 374: 166 [Google Scholar]
- 26. Hollman P. C. H. 2004. Absorption, bioavailability, and metabolism of flavonoids. Pharm. Biol. 42: 74–83 [Google Scholar]
- 27. Klaenhammer T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12: 39–85 [DOI] [PubMed] [Google Scholar]
- 28. Koshland D. E., Jr., Némethy G., Filmer D. 1966. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5: 365–385 [DOI] [PubMed] [Google Scholar]
- 29. Maicas S., Mateo J. J. 2005. Hydrolysis of terpenyl glycosides in grape juice and other fruit juices: a review. Appl. Microbiol. Biotechnol. 67: 322–335 [DOI] [PubMed] [Google Scholar]
- 30. Maischberger T., Mierau I., Peterbauer C. K., Hugenholtz J., Haltrich D. 2010. High-level expression of Lactobocillus β-galactosidases in Lactococcus lactis using the food-grade, nisin-controlled expression system NICE. J. Agric. Food Chem. 58: 2279–2287 [DOI] [PubMed] [Google Scholar]
- 31. Makarova K., et al. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103: 15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manzanares P., et al. 2003. Construction of a genetically modified wine yeast strain expressing the Aspergillus aculeatus rhaA gene, encoding an α-L-rhamnosidase of enological interest. Appl. Environ. Microbiol. 69: 7558–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martino A., Durante M., Pifferi P. G., Spagna G., Bianchi G. 1996. Immobilization of β-glucosidase from a commercial preparation. Part 1. A comparative study of natural supports. Process Biochem. 31: 281–285 [Google Scholar]
- 34. Matthews A., et al. 2004. Lactic acid bacteria as a potential source of enzymes for use in vinification. Appl. Environ. Microbiol. 70: 5715–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McIlvaine T. C. 1921. A buffer solution for colorimetric comparison. J. Biol. Chem. 49: 183–186 [Google Scholar]
- 36. Michlmayr H., Schümann C., Barreira Braz Da Silva N. M., Kulbe K. D., Del Hierro A. M. 2010. Isolation and basic characterization of a β-glucosidase from a strain of Lactobacillus brevis isolated from a malolactic starter culture. J. Appl. Microbiol. 108: 550–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Michlmayr H., Schümann C., Kulbe K. D., del Hierro A. M. 2011. Heterologously expressed family 51 alpha-L-arabinofuranosidases from Oenococcus oeni and Lactobacillus brevis. Appl. Environ. Microbiol. 77: 1528–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Michlmayr H., et al. 2010. A β-glucosidase from Oenococcus oeni ATCC BAA-1163 with potential for aroma release in wine: cloning and expression in E. coli. World J. Microbiol. Biotechnol. 26: 1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mierau I., Kleerebezem M. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68: 705–717 [DOI] [PubMed] [Google Scholar]
- 40. Nguyen T.-T., et al. 2011. A food-grade system for inducible gene expression in Lactobacillus plantarum using an alanine racemase-encoding selection marker. J. Agric. Food Chem. 59: 5617–5624 [DOI] [PubMed] [Google Scholar]
- 41. Pfannebecker J., Fröhlich J. 2008. Use of a species-specific multiplex PCR for the identification of pediococci. Int. J. Food Microbiol. 128: 288–296 [DOI] [PubMed] [Google Scholar]
- 42. Renwick A. G., Tarka S. M. 2008. Microbial hydrolysis of steviol glycosides. Food Chem. Toxicol. 46: S70–S74 [DOI] [PubMed] [Google Scholar]
- 43. Rodríguez H., Curiel J. A., Landete J. M., de las Rivas B., de Felipe F. L., Gómez-Cordovés C., Mancheño J. M., Muñoz R. 2009. Food phenolics and lactic acid bacteria. Int. J. Food Microbiol. 132: 79–90 [DOI] [PubMed] [Google Scholar]
- 44. Segel I. H. 1993. Enzyme kinetics. John Wiley & Sons, New York, NY [Google Scholar]
- 45. Sørvig E., Mathiesen G., Naterstad K., Eijsink V. G. H., Axelsson L. 2005. High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology 151: 2439–2449 [DOI] [PubMed] [Google Scholar]
- 46. Spagna G., Barbagallo R. N., Martino A., Pifferi P. G. 2000. A simple method for purifying glycosidases: α-L-rhamnopyranosidase from Aspergillus niger to increase the aroma of Moscato wine. Enzyme Microb. Technol. 27: 522–530 [DOI] [PubMed] [Google Scholar]
- 47. Spagna G., Barbagallo R. N., Greco E., Manenti I., Pifferi P. G. 2002. A mixture of purified glycosidases from Aspergillus niger for oenological application immobilised by inclusion in chitosan gels. Enzyme Microbiol. Technol. 30: 80–89 [Google Scholar]
- 48. Spano G., et al. 2005. A beta-glucosidase gene isolated from wine Lactobacillus plantarum is regulated by abiotic stresses. J. Appl. Microbiol. 98: 855–861 [DOI] [PubMed] [Google Scholar]
- 49. Tartoff K. D., Hobbs C. A. 1987. Improved media for growing plasmid and cosmid clones. Bethesda Res. Lab. Focus 9: 12–14 [Google Scholar]
- 50. Ugliano M., Moio L. 2006. The influence of malolactic fermentation and Oenococcus oeni strain on glycosidic aroma precursors and related volatile compounds of red wine. J. Sci. Food Agric. 86: 2468–2476 [Google Scholar]
- 51. Weiss J. N. 1997. The Hill equation revisited: uses and misuses. FASEB J. 11: 835–841 [PubMed] [Google Scholar]
- 52. Yadav V., Yadav P. K., Yadav S., Yadav K. D. S. 2010. α-L-Rhamnosidase: a review. Process Biochem. 45: 1226–1235 [Google Scholar]
- 53. Yu N. Y., et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zverlov V. V., et al. 2000. The thermostable α-L-rhamnosidase RamA of Clostridium stercorarium: biochemical characterization and primary structure of a bacterial α-L-rhamnoside hydrolase, a new type of inverting glycoside hydrolase. Mol. Microbiol. 35: 173–179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.