Abstract
This study presents the suitability of D1/D2 domain of large-subunit (LSU) ribosomal DNA (rDNA) for differentiation of Orpinomyces joyonii and Orpinomyces intercalaris based on PCR-restriction fragment length polymorphism (RFLP). A variation of G/T in O. intercalaris created an additional restriction site for AluI, which was used as an RFLP marker. The results demonstrate adequate heterogeneity in the LSU rDNA for species-level differentiation.
TEXT
Anaerobic fungi constitute nearly 20% of the total microbial biomass in the rumen (17). These are regarded as the primary colonizers (1) and most active lignocellulose degraders in the biological world (23, 25). Hence, their role is very critical in the digestion, especially in tropical regions, where forage is generally fibrous and of low quality (10). Taxonomically, these fungi belong to the phylum Neocallimastigomycota, class Neocallimastigomycetes, and order Neocallimastigales (9). Based on morphological features like growth pattern (monocentric or polycentric), thallus morphology (filamentous or bulbous), and number of flagella per zoospore, 6 genera divided into 20 species have been described (7). Within the genus Orpinomyces, two species, O. joyonii and O. intercalaris, have been identified, based on the position of sporangia, but the paucity of morphological features presents a big problem in resolving their taxonomy (2). Often, the polycentric taxa (including Orpinomyces) fail to produce sporangia, making their identification and differentiation very difficult. Therefore, various molecular approaches employing small subunit (SSU; 18S) ribosomal DNA (rDNA) or internal transcribed spacer (ITS) regions have been tried (2, 5, 24), but the highly conserved nature of the SSU rDNA and abundant intraindividual variations of ITS makes these regions inappropriate for diversity studies (3). Hence, the present study was conducted to explore the D1/D2 domain of large subunit (LSU; 28S) rDNA region for its potential application in species-level differentiation in Orpinomyces spp.
For this purpose, rumen liquor was collected from fistulated Murrah buffaloes maintained at the cattle yard of the National Dairy Research Institute (NDRI), Karnal, India, into O2-free, CO2-flushed, and preautoclaved bottles after 30 to 40 min of feeding (12). The liquor was immediately brought to the laboratory, flushed with O2-free CO2 (1 min), and diluted (10−1 dilution) in anaerobic diluent (13) containing 2% (vol/vol) antibiotic solution from a stock of benzylpenicillin, streptomycin sulfate, and chloramphenicol (each at 5 mg/ml). Roll tubes were prepared by injecting 0.5 ml of a 10−1 dilution into 50-ml serum bottles (14) containing 5 ml cellobiose agar medium (pH 6.7 ± 1) with antibiotics (1, 15). The medium composition was slightly modified (i.e., tryptone and cellobiose at 5 g/liter, cysteine-HCl at 1 g/liter, and hemin at 2 ml from a stock of 0.05% hemin mixed in 1:1 ethanol and 0.05 M NaOH). Serum bottles were incubated at 39 ± 1°C for 2 to 3 days for the development of colonies. Morphologically distinct colonies were transferred to a fresh medium using a bent needle and purified through repeated roll tube culturing. Cultures were maintained by routine subculture in 50-ml serum bottles containing 10 ml medium with wheat straw (5 g/liter) as the sole carbon source every fifth day. Features such as thallus morphology, growth patterns, and positions of sporangia were examined by phase-contrast and fluorescence microscopy using bisbenzimide (4). Genomic DNA was isolated from the 3-day-old culture (grown in cellobiose broth) by the cetyltrimethylammonium bromide (CTAB) method (7). PCR amplification of the D1/D2 domains of the LSU was performed with NL1 (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and NL4 (5′-GGT CCG TGT TTC AAG ACG G-3′) according to reference 5. The amplified PCR products were visualized on 1.5% agarose gel and purified using the MinElute PCR purification kit (Qiagen, India), before sequencing (Xcelris Genomic Centre, Ahmadabad, Gujarat). The sequences obtained were edited, compiled, and aligned using BioEdit software (8). Sequence similarity searches were performed using GenBank Blastn. In silico digestion was done using Cleaver (11). A phylogenetic tree was generated using the neighbor-joining algorithm in MEGA5 (22). For restriction fragment length polymorphism (RFLP) analysis, 12 μl (≈1 μg) of amplified PCR product was digested overnight at 37°C using 2.5 U of AluI (recognition sequence, AG′CT) and 1× buffer in a 20-μl reaction volume. The digested products were separated by electrophoresis (65 V for 4 h; Biometra Power Pack P25) on 3% agarose gel containing ethidium bromide (2.0 μg/ml) in 1× Tris-borate-EDTA (TBE) buffer.
The Orpinomyces spp. were identified by polycentric growth pattern (many sporangia on a single rhizoid) and nucleated rhizoids (Fig. 1A and B). Sporangia were mostly spherical, developing on the terminal end of a simple or branched sporangiophore in O. joyonii (Fig. 2A), while the sporangial position was intercalary in the case of O. intercalaris (Fig. 2B). The zoospores of both species were polyflagellated, and no distinct morphological differences were observed. Five isolates each of O. joyonii and O. intercalaris were chosen for further study. The amplified region showed a product of ≈780 bp (Fig. 3) in both species. The sequence similarity searches showed various similarities of 98 to 99% with the Orpinomyces sp. strain OUS1 partial 18S rRNA gene, ITS1, 5.8S rRNA gene, ITS2, and partial 28S rRNA, clone V5-1 (AJ864475.1), the only sequence available with related matching sequence and which, therefore, was taken as a reference. The DNA sequence alignment showed a total of 10 nucleotide variations between both species (Fig. 4). Four variations each were observed for both A/G and T/C, while two variations were found for G/T. In silico digestion showed restriction sites for AfaI, BseRI, AluI, DdeI, HpyF3I, MaeII, MnlI+, RsaI, and TscI. Based on the availability, AluI was selected. The AluI digestion showed common bands of approximately 379, 79, 57, 40, and 13 bp, while discriminatory bands of 212 bp and 138 bp were found in O. joyonii and O. intercalaris, respectively. It was observed that the variation of G/T created an extra restriction site for AluI in O. intercalaris, which resulted in cleavage of 212-bp regions in O. joyonii into two regions of 138 bp and 74 bp in O. intercalaris. A phylogenetic tree of all of the isolates grouped these into two major clusters (Fig. 5), thereby confirming there was sufficient heterogeneity of this region for their differentiation. The reference sequence was found to be clustered with O. joyonii, suggesting their similar evolutionary relationship. Restriction patterns (Fig. 6) generated after digestion with AluI clearly differentiated both species. Two types of banding patterns were observed among the isolates examined. The bands were identical for the isolates pertaining to the same species. In both species, two bands of 379 bp and ≈79 bp were common, while the discriminatory bands were 212 bp and 138 bp in O. joyonii and O. intercalaris, respectively. The bands below the size of ≈79 bp were not visible in the gel. The highly pleomorphic morphological features (10) and unsuitability of SSU and ITS regions (3, 16, 24) for the genus- and species-level differentiations have necessitated the need for better alternative genetic markers like the LSU, RNA polymerase II subunits, actin-coding genes, translation elongation factor 1-α, β-tubulin, or mitochondrial cytochrome c oxidase. Since the anaerobic fungi lack mitochondria (26), design of mitochondrially based genetic markers is not feasible. In addition, many of the above markers are single copy; therefore, their amplification from low-quantity samples is problematic, while for others, universal primer designing is very difficult (16). On the other hand, the multicopy LSU gives a possibility to design universal primers that can be easily amplified and has been shown to possess less intraindividual polymorphism (20). Moreover, this region has also been used for genus-level differentiation in anaerobic fungi (5) and species-level differentiation in yeasts (6, 18). In this study, the sequenced products of morphologically distinct Orpinomyces spp. showed proper alignment, rare intraindividual polymorphism, and a high degree of variation, sufficient enough to differentiate at the species level.
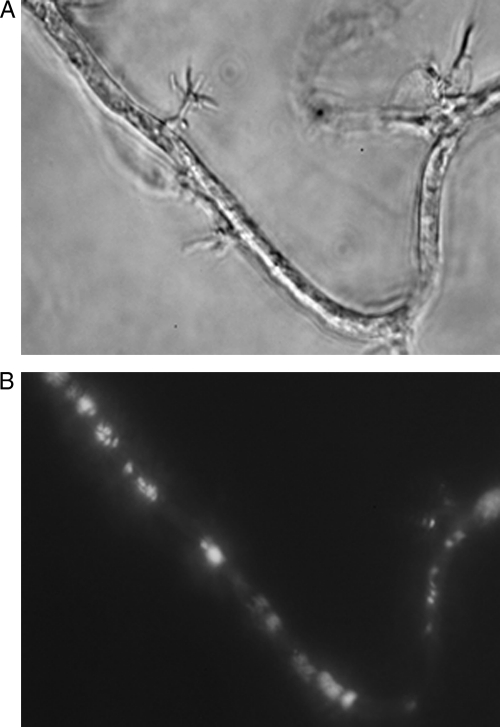
Fig. 1.
Nucleated rhizoids of Orpinomyces intercalaris by phase-contrast microscopy (A) and fluorescence microscopy (B). Magnification, ×400.
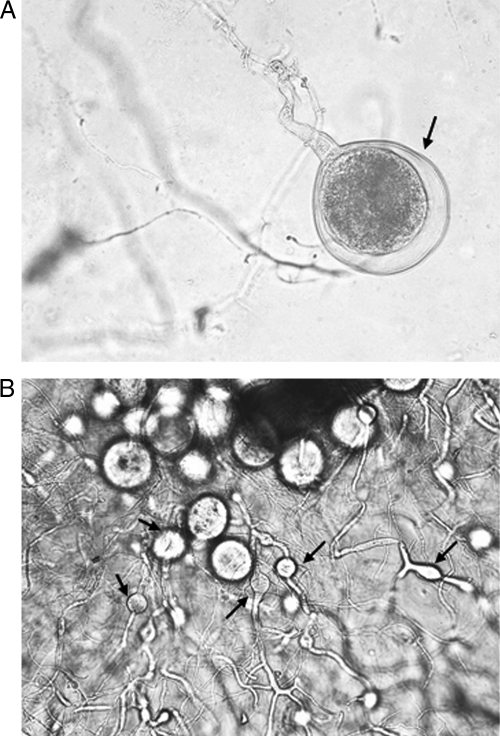
Fig. 2.
Different patterns (as indicated by arrows) of sporangial development. (A) Terminal end in O. joyonii. Magnification, ×400. (B) Intercalary position in O. intercalaris. Magnification, ×200.
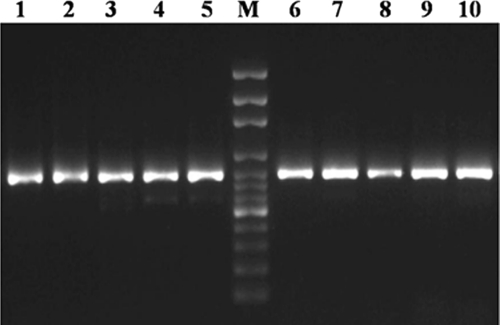
Fig. 3.
PCR-amplified product (≈780 bp) of the LSU region of O. joyonii (lanes 1 to 5) and O. intercalaris (lanes 6 to 10). M, 100-bp DNA marker.
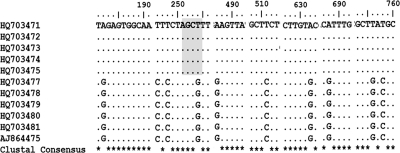
Fig. 4.
Patterns of variation obtained after multiple sequence alignment of isolates with an extra restriction site (highlighted area) for AluI in O. intercalaris.
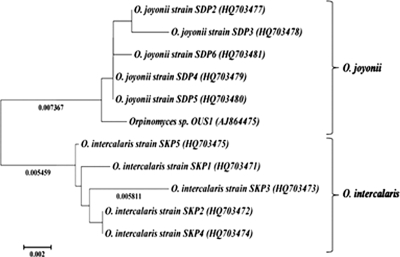
Fig. 5.
Neighbor-joining phylogenetic tree of Orpinomyces spp. obtained from the sequences of the LSU. The evolutionary history was inferred using the neighbor-joining method (19). The optimal tree with the sum of branch length of 0.03107997 is shown. The tree is drawn to scale, with branch lengths (below the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (21) and are in the units of the number of base substitutions per site. The analysis involved 11 nucleotide sequences. The codon positions included were the 1st, 2nd, 3rd, and noncoding positions. All positions containing gaps and missing data were eliminated. There were a total of 743 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (22).
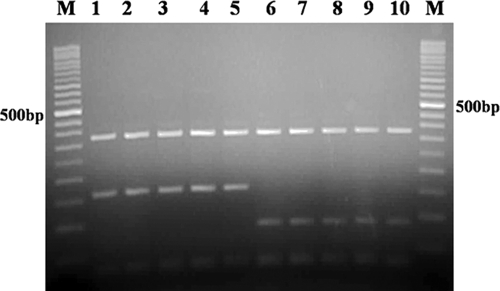
Fig. 6.
AluI-digested PCR product of the LSU regions of O. joyonii (lanes 1 to 5) and O. intercalaris (lanes 6 to 10). M, 50-bp DNA marker.
It can be concluded that the LSU has the potential to be used as a taxonomic marker for species-level differentiation in anaerobic fungi. We also report that AluI can be used for differentiating species of Orpinomyces, suggesting that PCR-RFLP can be used for carrying out genetic heterogeneity studies without the need for sequencing.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under accession no. HQ703471, HQ703472, HQ703473, HQ703474, and HQ703475 for O. intercalaris and HQ703477, HQ703478, HQ703479, HQ703480, and HQ703481 for O. joyonii.
Acknowledgments
The work was conducted as a part of the Ph.D. project of S. S. Dagar, supported by the NDRI (ICAR) fellowship and project “Veterinary Type Culture—Rumen Microbes.”
We are grateful to Ugur Comlekcioglu, Gareth Wyn Griffith, Katerina Fliegerova, and Tony Callaghan for valuable suggestions during the work and B. R. Yadav and S. K. Sirohi for providing some of the facilities.
Footnotes
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Bauchop T. 1979. Rumen anaerobic fungi of cattle and sheep. Appl. Environ. Microbiol. 38: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brookman J. L., Mennim G., Trinci A. P., Theodorou M. K., Tuckwell D. S. 2000. Identification and characterization of anaerobic gut fungi using molecular methodologies based on ribosomal ITS1 and 185 rRNA. Microbiology 146: 393–403 [DOI] [PubMed] [Google Scholar]
- 3. Eckart M., Fliegerová K., Hoffmann K., Voigt K. 2010. Molecular identification of anaerobic rumen fungi, p. 297–313 In Gherbawy Y., Voigt K. (ed.), Molecular identification of fungi. Springer, Berlin, Germany [Google Scholar]
- 4. Fliegerova K., Hodrova B., Voigt K. 2004. Classical and molecular approaches as a powerful tool for the characterization of rumen polycentric fungi. Folia Microbiol. (Praha) 49: 157–164 [DOI] [PubMed] [Google Scholar]
- 5. Fliegerova K., Mrazek J., Voigt K. 2006. Differentiation of anaerobic polycentric fungi by rDNA PCR-RFLP. Folia Microbiol. (Praha) 51: 273–277 [DOI] [PubMed] [Google Scholar]
- 6. Garner C. D., Starr J. K., McDonough P. L., Altier C. 2010. Molecular identification of veterinary yeast isolates by use of sequence-based analysis of the D1/D2 region of the large ribosomal subunit. J. Clin. Microbiol. 48: 2140–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Griffith G. W., Ozkose E., Theodorou M. K., Davies D. R. 2009. Diversity of anaerobic fungal populations in cattle revealed by selective enrichment culture using different carbon sources. Fungal Ecol. 2: 87–97 [Google Scholar]
- 8. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98 [Google Scholar]
- 9. Hibbett D. S., et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111: 509–547 [DOI] [PubMed] [Google Scholar]
- 10. Ho Y. W., Barr D. J. S. 1995. Classification of anaerobic gut fungi from herbivores with emphasis on rumen fungi from Malaysia. Mycologia 87: 655–677 [Google Scholar]
- 11. Jarman S. N. 2006. Cleaver: software for identifying taxon specific restriction endonuclease recognition sites. Bioinformatics 22: 2160–2161 [DOI] [PubMed] [Google Scholar]
- 12. Kostyukovsky V. A., Okunev O. N., Tarakanov B. V. 1991. Description of two anaerobic fungal strains from the bovine rumen and influence of diet on the fungal population in vivo. J. Gen. Microbiol. 137: 1759–1764 [DOI] [PubMed] [Google Scholar]
- 13. McSweeney C., Denman S., Mackie R. 2005. Rumen bacteria, p. 23–37 In Makkar H., McSweeney C. (ed.), Methods in gut microbial ecology for ruminants. Springer, Dordrecht, Netherlands [Google Scholar]
- 14. Miller T. L., Wolin M. J. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Environ. Microbiol. 27: 985–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nam I. S., Garnsworthy P. C. 2007. Biohydrogenation of linoleic acid by rumen fungi compared with rumen bacteria. J. Appl. Microbiol. 103: 551–556 [DOI] [PubMed] [Google Scholar]
- 16. Nilsson R. H., Kristiansson E., Ryberg M., Hallenberg N., Larsson K.-H. 2008. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. Online 4: 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rezaeian M., Beakes G. W., Parker D. S. 2004. Distribution and estimation of anaerobic zoosporic fungi along the digestive tracts of sheep. Mycol. Res. 108: 1227–1233 [DOI] [PubMed] [Google Scholar]
- 18. Romanelli A. M., Sutton D. A., Thompson E. H., Rinaldi M. G., Wickes B. L. 2010. Sequence-based identification of filamentous basidiomycetous fungi from clinical specimens: a cautionary note. J. Clin. Microbiol. 48: 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- 20. Sonnenberg R., Nolte A., Tautz D. 2007. An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Front. Zool. 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamura K., Nei M., Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101: 11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura K., et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods Mol. Biol. Evol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thareja A., et al. 2006. In vitro degradation of wheat straw by anaerobic fungi from small ruminants. Arch. Anim. Nutr. 60: 412–417 [DOI] [PubMed] [Google Scholar]
- 24. Tuckwell D. S., Nicholson M. J., McSweeney C. S., Theodorou M. K., Brookman J. L. 2005. The rapid assignment of ruminal fungi to presumptive genera using ITS1 and ITS2 RNA secondary structures to produce group-specific fingerprints. Microbiology 151: 1557–1567 [DOI] [PubMed] [Google Scholar]
- 25. Wood T. M., Wilson C. A. 1995. Studies on the capacity of the cellulase of the anaerobic fungus Piromonas communis P to degrade hydrogen bond ordered cellulose. Appl. Microbiol. Biotechnol. 43: 572–578 [DOI] [PubMed] [Google Scholar]
- 26. Yarlett N., Orpin C. G., Munn E. A., Yarlett N. C., Greenwood C. A. 1986. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem. J. 236: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]