Abstract
Infections caused by members of the Chlamydiaceae family have long been underestimated due to the requirement of special laboratory facilities for the detection of this group of intracellular pathogens. Furthermore, new studies of this group of intracellular pathogens have revealed that host specificity of different species is not as clear as recently believed. As most members of the genus Chlamydophila have shown to be transmissible from animals to humans, sensitive and fast detection methods are required. In this study, SYBR green-based real-time assays were developed that detect all members of Chlamydiaceae and differentiate the most prevalent veterinary Chlamydophila species: Cp. psittaci, Cp. abortus, Cp. felis, and Cp. caviae. By adding bovine serum albumin to the master mixes, target DNA could be detected directly in crude lysates of enzymatically digested conjunctival or pharyngeal swabs or tissue specimens from heart, liver, and spleen without further purification. The assays were evaluated on veterinary specimens where all samples were screened using a family-specific PCR, and positive samples were further tested using species-specific PCRs. Cp. psittaci was detected in 47 birds, Cp. felis was found in 10 cats, Cp. caviae was found in one guinea pig, and Cp. abortus was detected in one sheep. The screening assay appeared more sensitive than traditional microscopical examination of stained tissue smears. By combining a fast, robust, and cost-effective method for sample preparation with a highly sensitive family-specific PCR, we were able to screen for Chlamydiaceae in veterinary specimens and confirm the species in positive samples with additional PCR assays.
INTRODUCTION
The family Chlamydiaceae comprises two genera, Chlamydia and Chlamydophila, with nine different species, all of which cause disease in humans and animals (8). Although the different species show a high degree of host specificity, other hosts may also be susceptible to infection as these organisms have been isolated from a variety of mammals (4, 6, 17), birds (10, 11), and reptiles (3, 6). Chlamydia trachomatis (C. trachomatis) and Chlamydophila pneumoniae (Cp. pneumoniae) mainly cause disease in humans, while the other species have nonhuman hosts. However, it is within the genus Chlamydophila that we find species with serious zoonotic potential, where Cp. psittaci may cause severe pneumonia and systemic disease in humans, and Cp. abortus may cause human abortion (20).
Members of the family Chlamydiaceae mainly cause infection of the mucosal lining in the respiratory system, the gastrointestinal tract, or in the reproductive system. The location and the severity of infection are highly dependent on the species and the host. Frequently, the infection is subclinical and in most animals causes mild conjunctivitis and rhinitis, or it may infect the reproductive system leading to abortion or infertility (20). Birds, especially psittacine, may be subclinically infected with Cp. psittaci, but the infection may be activated upon stress and lead to clinical disease with severe pneumonia and organ failure. Humans are susceptible to Cp. psittaci from shedding birds and may develop a disease with influenza-like symptoms, which can be misinterpreted with fatal consequences (18).
The diagnosis of infection with Chlamydiaceae has long been hampered by the obligate intracellular nature of this pathogen, requiring growth in embryonated eggs or tissue culture for detection. Furthermore, there are no reliable biochemical tests to differentiate species, which has complicated research in this field. However, the introduction of molecular methods in the detection of Chlamydiaceae has significantly improved the possibilities for detection and our understanding of the pathogenesis and transmission of this group of pathogens. Several PCR assays have been developed based on the ompA gene, the 16S-23S rRNA operon, and the pmp gene, using traditional and real-time assays (21). However, as the host range for Chlamydiaceae is widening, new species variants are identified which may not be detected by previously published primers due to sequence polymorphism in the target gene (13). The majority of assays have been made as species-specific assays, but for a routine diagnostic veterinary laboratory with a broad range of specimens from various animals, this is not appropriate or economically feasible as fast and reliable diagnosis is needed in order to select a suitable treatment (6). However, as many infections are subclinical or without characteristic symptoms, it is important also to know the species as it may give additional information for tracing the source of infection. The aim of this study was to establish a simple, fast, and reliable diagnostic method for Chlamydiaceae in various clinical specimens that enables detection of all members of Chlamydiaceae and identification of the most relevant veterinary species in a routine laboratory.
MATERIALS AND METHODS
Bacterial reference strains.
Reference DNA from the following strains was used for testing the PCR assays: Chlamydophila felis (FP), Chlamydophila caviae (GPIC), Chlamydophila abortus (B577), Chlamydophila pecorum (IPA), Chlamydia suis (R19), and Chlamydia muridarum (EP12) were kindly provided by Arthur A. Anderson, USDA, and Chlamydophila psittaci (Cal10), Chlamydia trachomatis (L2), and Chlamydophila pneumoniae (VR1310) were from Gunna Christiansen, University of Aarhus. The reference strains were received as allantoic fluid harvested from embryonated egg cultures. A panel of veterinary bacterial pathogens and viruses which may constitute differential diagnosis to chlamydiosis in birds is listed in Table 1. These were either reference strains or cultures that have been characterized in our laboratory using the API identification system (bioMérieux, Herlev, Denmark).
Table 1.
Listing of nontarget microorganisms which were used for testing of cross-reactivity toward the primers
| Bacterium or virus | Strain identification |
|---|---|
| Bacteria | |
| Escherichia coli | CCUG 17620 |
| Citrobacter freundii | SVS9740947 |
| Salmonellaenterica serovar Typhimurium | SVS9762491 |
| Ornithobacterium rhinotracheale | CCUG 23171 |
| Yersinia pseudotuberculosis | SVS9906990 |
| Yersinia enterocolitica | SVS9988486 |
| Mannheimia haemolytica | SVS7406196 |
| Pasteurella multocida | SVS7406206 |
| Pseudomonas aeruginosa | CCUG 17619 |
| Aeromonas hydrophila | SVS7406192 |
| Bordetella bronchiseptica | SVS9641022 |
| Klebsiella pneumoniae | SVS9882353 |
| Riemerella anatipestifer | NCTC11014T |
| Erysipelothrix rhusiopathiae | SVS9964278 |
| Staphylococcus intermedius | SVS9906870 |
| Streptococcus bovis | CCUG 17828 |
| Clostridium perfringens | ATCC 3624 |
| Enterococcus faecalis | CCUG 19916 |
| Hafnia alvei | SVS9908255 |
| Alcaligenes faecalis | SVS9907473 |
| Viruses | |
| Infectious laryngotracheitis | SVS7272860 |
| Chicken anemia virus | 975-982 |
| Marek's disease virus | CE-2249-51 |
| Avian adenovirus | SVS9676386 |
DNA preparation from cultures.
DNA from bacterial strains or egg cultures was prepared by enzymatic and detergent lysis of cells. Five colonies from reference cultures grown on blood agar (CM 271; Oxoid, Greve, Denmark) supplemented with 5% calf blood or 200 μl of allantoic fluid were harvested by centrifugation (13,000 × g for 5 min). DNA was released by adding 200 μl of a lysis buffer (0.5% SDS [L4390; Sigma-Aldrich, Bøndby, Denmark], 0.1 M Tris-HCl, pH 8.0) and 10 μl of proteinase K (20 mg/ml; Roche, Hvidovre, Denmark). Lysates were incubated at 56°C for 30 min, followed by 5 min at 97°C. Instead of purifying DNA, all samples were serially diluted (1:10 and 1:100) by transferring 10-μl lysates to 90 μl of 10 mM Tris-EDTA (10:1; 10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0 [Sigma-Aldrich]) before PCR. Diluted or nondiluted samples were stored at −20°C until analysis.
Primer design.
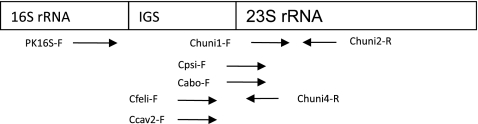
DNA sequences from the ribosomal genes of Chlamydiaceae and closely related families deposited at the GenBank database (www.ncbi.nlm.nih.gov/GenBank) were retrieved and aligned using the DNA Workbench from CLC (CLC bio, Aarhus N, Denmark). In the rRNA operon, the intergenic spacer (IGS) region and the initial part of the 23S ribosomal gene were selected for designing family- and species-specific primers. A family-specific primer set targeting all Chlamydiaceae was selected (Chuni1-F and Chuni2-R), and another four species-specific forward primers were selected targeting Cp. psittaci (Cpsi-F), Cp. felis (Cfeli-F), Cp. abortus (Cabo-F), and Cp. caviae (Ccav-F). These species-specific primers were used in combination with another family-specific reverse primer, Chuni4-R (Fig. 1 and Table 2). The theoretical performance of each primer pair was evaluated using the Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm). The specificity of the primers was tested toward all sequences in the GenBank using the BLAST function at NCBI (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi).
Fig. 1.
Schematic presentation of the primers used in this study.
Table 2.
Primer sequences used in this study
| Primer | Sequence 5′ → 3′ | Fragment length (bp) | Target gene | Specificity |
|---|---|---|---|---|
| Chuni1-F | GGG CTA GAC ACG TGA AAC CTA | 356 | 23S rRNA | Chlamydiaceae |
| Chuni2-R | CCA TGC TTC AAC CTG GTC ATA A | 23S rRNA | Chlamydiaceae | |
| Cpsi-F | GAT TAA TCA TCT ACC ATT ATA CGT | 301 | 23S rRNA | Cp. psittaci |
| Cfeli-F | CGT TGT TAA GCG TGG GTT | 583 | IGSa | Cp. felis |
| Ccav2-F | TGC ATT GTT AAG CGC TCG T | 585 | IGS | Cp. caviae |
| Cabo-F | AAT CAT CTA TCA TTG TAC GC | 299 | 23S rRNA | Cp. abortus |
| Chuni4-R | AGA CTA GGT TTC ACG TGT CTA G | 23S rRNA | Chlamydiaceae | |
| PK16S-F | GGG CTG GAT CAC CTC CTT | 16S rRNA | Bacteria |
IGS, 16S-23S ribosomal intergenic spacer.
SYBR green-based real-time PCR assay.
Primers were designed to have comparable melting temperature (Tm) values around 60°C, thereby allowing all primer sets to be used with the same master mix and analyzed using the same PCR protocol. In this way, all five reactions could be done in parallel on the same machine. The master mix was prepared in large batches, and volumes of 23 μl were prealiquoted in PCR tubes to be stored at −20°C until use. In each tube, the master mix consisted of 12.5 μl of Promega PCR Mastermix (catalog number M7505; Promega, Madison, WI), 2 mM MgCl2, 25 μg of bovine serum albumin (BSA) (catalog number A9647; Sigma-Aldrich), 400 nM each forward and reverse primer, 1 μl of a 1:10,000 dilution of SYBR green I (catalog number S7563; Invitrogen, Taastrup, Denmark), and nuclease-free water to a total volume of 23 μl. Before PCR was performed, 2 μl of diluted sample DNA was added, and to samples that served as inhibition controls, 1 μl of positive-control DNA was also added. PCR was carried out on an MX3005P real-time PCR machine (Stratagene, CA) with the following cycling conditions: 95°C for 5 min and 45 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30s. A melting curve analysis of generated amplicons was performed immediately after completion of the PCR cycling, using 0.3°C s−1 as a ramping temperature in the interval of 75 to 90°C. Calculation of threshold cycle (CT) values was based on automatic adaptive baseline settings, and the melting curve analyses were performed using the MxPro software, version 4.10 (Stratagene). To verify the sizes of generated amplicons, these were visualized in a 1.5% agarose gel containing ethidium bromide (Roche), and photos were captured using a GelDoc-IT Imaging Systems (UVP, CA).
Validation of the specificity of primers on reference DNA and spiked samples.
Using the optimized PCR assay, the sensitivities of all primer sets were evaluated on reference DNA from Chlamydiaceae strains. To test for cross-reactivity/specificity, all primer pairs were tested against a reference panel of veterinary bacterial pathogens and viruses, selected as agents that may cause syndromes similar to chlamydiosis in birds. To verify the specificity of all five of the different primer pairs, amplicons from each PCR were purified using a commercial kit (Promega) and submitted for sequencing at LGCgenomics, Berlin, Germany. Generated sequence data were aligned against reference sequences using the CLC DNA Workbench. The performance of each assay to detect the target genes directly in clinical samples was evaluated by PCR analysis of spiked samples that were constructed by adding 2 μl of a 1:10 diluted Chlamydiaceae-negative crude spleen lysate from a parrot with 1 μl of diluted reference DNA in each PCR.
Selection of clinical samples.
From birds, clinical samples (heart, liver, and spleen) were obtained from carcasses submitted to the laboratory for autopsy. Carcasses were typically sent by ordinary mail to the laboratory, and due to storage at ambient temperature during transportation, the sample quality showed large variations, with some carcasses being in less favorable conditions. The majority of the specimens were from parrots (order Psittaciformes) or land fowl (order Galliformes), but passerines (order Passeriformes), doves (order Columbiformes), and waterfowl (order Anseriformes) were also sampled (Table 3). Samples were selected on the basis of a clinical history comparable to chlamydiosis (sudden death, respiratory disease, or emaciation), but samples from subclinically infected birds from aviaries where there had been pervious outbreaks of ornithosis or where the owner had been diagnosed with ornithosis were also included. The majority of submissions comprised 1 to 3 birds from minor breeders, but samples were also included from a larger outbreak of psittacosis in a farm raising game birds (grey partridge, Perdix perdix). From cases of abortion in sheep, placenta tissue was received for analysis. From cats and guinea pigs with conjunctivitis, conjunctival swab samples were collected in veterinary clinics. The cotton swab samples were submitted for laboratory analysis in screw-cap tubes containing 1 ml of 10 mM Tris (pH 8.0) using ordinary mail. Samples were processed immediately after sampling or stored at −20°C.
Table 3.
Clinical samples from birds and animals with suspected chlamydiosis screened with a Chlamydiaceae-specific PCR assay and positive samples verified with species-specific assays
| Sample source | Total no. of samples | No. of PCR-positive cases by primer pair |
||||
|---|---|---|---|---|---|---|
| Chuni1-F/Chuni2-R | Cpsi-F/Chuni4-R | Cfeli-F/Chuni4-R | Ccav2-F/Chuni4-R | Cabo-F/Chuni4-R | ||
| Birds (description) | ||||||
| Anseriformes (waterfowl) | 15 | 4 | 4 | |||
| Galliformes (land fowl) | 109 | 17 | 17 | |||
| Columbiformes (doves) | 6 | 0 | ||||
| Psittaciformes (parrots) | 167 | 26 | 26 | |||
| Passeriformes (passerines) | 15 | 0 | ||||
| Not specified | 15 | 0 | ||||
| Cats | 73 | 10 | 10 | |||
| Guinea pigs | 2 | 1 | 1 | |||
| Sheep | 3 | 1 | 1 | |||
| Total | 405 | 59 | 47 | 10 | 1 | 1 |
Validation of the PCR assay on clinical samples.
Total DNA from the clinical specimens was extracted using the same lysis method as described for pure cultures, with minor modifications. Approximately 100 mg of tissue was transferred to a 1.5-ml Eppendorf tube containing 190 μl of lysis buffer (0.5% SDS, 0.1 M Tris-HCl, pH 8.0) and 10 μl of proteinase K (20 mg/ml). Cells from the swab specimens were collected by washing them in 1 ml of 10 mM Tris-EDTA (10:1; pH 8.0), followed by centrifugation at 10,000 × g for 5 min. The pellet was resuspended in 95 μl of lysis buffer and 5 μl of proteinase K (20 mg/ml). After incubation at 56°C for 30 min and at 97°C for 5 min, the DNA lysate was diluted 10-fold by transferring 10-μl lysates to 90 μl of 10 mM Tris-EDTA (10:1; pH 8.0). Complete lysis was achieved only for cells from swab samples while the digest of the tissue sample was only partial. Following sample preparation and dilution, DNA was used directly in the PCR or stored at −20°C. PCR analysis was conducted using the same master mix as previously described, but all samples were tested in triplicate; two PCR tubes each received 2 μl of the 1:10 DNA solution, and one tube received 2 μl of a 1:100 solution. To test for inhibitors, 1 μl of positive-control DNA was further added to one of the two reaction mixtures with the 1:10 dilution. Samples were first screened using the family-specific primer pair, and positive samples were further tested using the species-specific primers. Only cases positive by both the family-specific assays and one of the species-specific assays were counted as positive samples. Cases from birds were regarded as positive if at least one of the three organ samples was positive.
Nucleotide sequencing of the IGS.
A sample from a guinea pig which was positive with the family-specific primers but not with any of the species-specific primers was further investigated by amplifying the IGS and the initial part of the 23S rRNA using the clinical sample as a template and the primer combination PK16S-F/Chuni4-R. The generated amplicon was purified using a commercial kit for purifying PCR amplicons (Promega) and sent for sequencing at LGCgenomics (Berlin, Germany). Generated data files were assembled and aligned against different reference DNA sequences using the CLC DNA Workbench. On the basis of this alignment, a new Cp. caviae-specific primer (Ccav2-F) was selected and validated.
Comparison of microscopical methods and PCR for detecting Cp. psittaci.
To compare our PCR assay with classical methods for detecting Cp. psittaci, a part of the collected bird samples were also tested using two different microscopic examination techniques. For a period of time, cell smears of all collected samples were also examined using a modified Ziehl-Neelsen staining technique (MZN), or immunofluorescence (IF). The samples comprised all ornithosis-suspected cases from birds in a period of time, thus representing different bird species and degrees of clinical signs. The MZN method was previously described by Stamp et al. (23); however, we used a slightly modified method. Briefly, tissue cell smears were heat fixed on glass slides and stained with a 1% solution of carbol fuchsin (VWR, Herlev, Denmark) for 15 min, followed by flushing with deionized water for 15 s; samples were counterstained with 1% malachite green solution (Merck, Darmstadt, Germany) for 3 min and finally washed briefly with deionized water. Slides were examined by light microscopy at a ×1,000 magnification. Samples with red intracellular inclusion bodies were regarded as positive. For immunofluorescence (IF) detection, a commercial kit (Chlamydia Direct IF, bioMérieux, Herlev, Denmark) was used according to the manufacturer's directions. The detection is based on a fluorescence-labeled Chlamydiaceae monoclonal antibody which is specific for all developmental stages of Chlamydiaceae. Samples were examined by fluorescence microscopy at a ×630 magnification, and those with fluorescently labeled intracellular inclusion bodies were considered positive.
Nucleotide sequence accession number.
The sequence of the IGS and the initial part of the 23S rRNA from the guinea pig isolate has been deposited in GenBank under accession number JF313242.
RESULTS
Selection of primer sequences.
More than 80 published sequences of the rRNA operon from all members of Chlamydiaceae and closely related families were aligned, and family- or species-specific signature sequences were identified. The IGS between the 16S and 23S rRNA genes and the first segment part of the 23S gene were of interest as they contained highly conserved parts interrupted by minor sections with species-specific sequence polymorphisms or deletions (7). This combination allowed selection of five primer pairs: four species-specific primers targeting Cp. psittaci, Cp. felis, Cp. abortus, and Cp. caviae, all working together with a family-specific reverse primer (Chuni4-R), and a family-specific primer pair (Chuni1-R/Chuni2-R) that targets all species in Chlamydiaceae (Fig. 1). Primers were designed to have similar Tm values around 60°C, allowing all primer sets to be used with the same master mix and PCR protocol.
Validation of primer specificity.
The primer pairs were evaluated using the BLAST function in GenBank (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). The family-specific primers had exact matches with all published ribosomal gene sequences from Cp. psittaci, Cp. abortus, Cp. caviae, and C. muridarum (Table 4). However, interspecies polymorphism was observed in sequences from Cp. felis, Cp. pneumoniae, C. trachomatis, and C. suis, where a single nucleotide polymorphism (SNP) in the 5′ end of the forward primer was observed. In sequences from Chlamydophila pecorum, an SNP in the 5′ end was observed in both primers. For the species-specific primers, exact matches were observed for the reference strains of Cp. felis, Cp. caviae, Cp. psittaci, and Cp. abortus. However, identical SNPs were observed in 2 out of 16 published sequences of Cp. psittaci and in 2 out of 10 sequences of Cp. abortus, where sequences contained a single mismatch in the center of the primer. The observed differences were unique for these strains and were not found in any of the other published sequences.
Table 4.
Evaluation of the theoretical specificity of the primer pairs using the BLAST search engine at GenBank
| Organism | No. of sequences with primer match/total no. of sequencesa |
||||
|---|---|---|---|---|---|
| Chuni1-F/Chuni2-R | Cpsi-F/Chuni4-R | Cfeli-F/Chuni4-R | Ccav2-F/Chuni4-R | Cabo-F/Chuni4-R | |
| Cp. psittaci | 16/16 | 14/16d | − | − | − |
| Cp. felis | 4/4b | − | 4/4 | − | − |
| Cp. caviae | 2/2 | − | − | 2/2 | − |
| Cp. abortus | 5/5 | − | − | − | 8/10e |
| Cp. pneumoniae | 11/11b | − | − | − | − |
| Cp. pecorum | 6/6b,c | − | − | − | − |
| C. trachomatis | 20/20c | − | − | − | − |
| C. muridarum | 3/3 | − | − | − | − |
| C. suis | 7/7c | − | − | − | − |
A match is defined as no mismatches in the 3′ end or more than two in the 5′ end of the primer. −, no primer match (any mismatches in the 3′ end or more than 2 in the rest of the primer).
All sequenced strains had one SNP in the 5′ end of the forward primer.
All sequenced strains had one SNP in the 5′ end of the reverse primer.
The primers were evaluated by PCR on reference DNA representing the different members of the Chlamydiaceae. With the family-specific primer pair, amplicons of the expected sizes were generated from all nine Chlamydiaceae species. No difference in amplification efficiencies was observed between strains with a perfect match and strains with the SNP in the 5′ end. In the species-specific assays, amplification was observed in samples with homologous DNA, and no cross-reaction was found with nonhomologous DNA. Generated amplicons from each primer pair were further verified by sequencing, confirming the specificity of the primers. The assays were further tested for cross-reactivity against a panel of DNA samples from veterinary pathogens that may cause diseases resembling those caused by Chlamydophila spp., but there was no match (Table 1).
Development of real-time assays for clinical specimens.
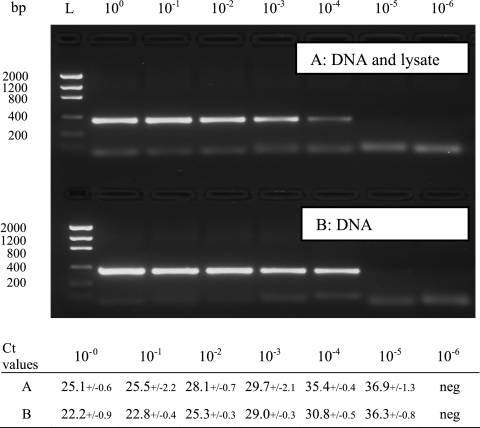
A real-time PCR method based on SYBR green detection of Chlamydiaceae in crude tissue lysates was established. By selecting appropriate concentrations of SDS and proteinase K in the lysis buffer, diluting the DNA lysate, and including BSA in the master mix, specific amplification of the target could be achieved without the need for further purification steps. The performance of the master mix was evaluated by real-time PCR testing of a dilution series of target DNAs with or without the addition of 2 μl of a negative tissue lysate, and the products were visualized by gel analysis (Fig. 2). With the addition of tissue lysate to the PCR, the PCR efficiency was lowered, as visualized by higher CT values and weaker bands in gel analysis than obtained with pure DNA. However, it was possible to detect target in the same dilutions. By comparing the results from the melting curves with gel analysis, we observed that the melting curve analysis allowed up to 10-fold higher sensitivity than gel electrophoresis (Fig. 3).
Fig. 2.
A 10-fold dilution series of Cp. psittaci DNA was tested with Chuni1-F/Chuni2-R primers either together with 2 μl of a negative heart sample lysate diluted 1:10 (A) or alone (B). Mean CT values of three repeated runs are presented in the table below. The highest PCR yield was observed in samples with pure DNA. Although no gel bands were visualized in the 10−5 dilution, melt curve analysis revealed specific peaks (3/3) in both dilution series. Lane L, DNA ladder.
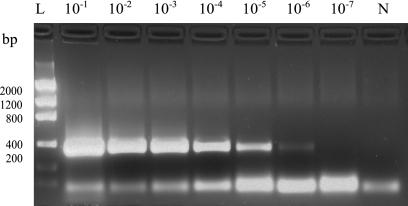
Fig. 3.
A 10-fold dilution series of a positive liver sample was tested with the Chuni1-F/Chuni2-R primers. The positive reaction at the 10−6 dilution corresponded to a CT value of 36. A small weak band (<100 bp) of primer dimer is observed below the specific band. Lane L, DNA ladder; lane N, no-template control.
In negative tissue lysates spiked with reference DNA, homologue DNA was detected with all primer pair combinations in tissue lysates of heart, liver, or spleen. No cross-reaction was observed between the species-specific primers. Although a general increase in fluorescence signal was observed in all samples during a PCR run, true-positive samples were distinguished by melt curve analysis of the PCR amplicons afterwards. Apart from the Cp. felis PCR assay where two melting peaks were generated, single peaks were generated in the other assays. The melting point of amplicons obtained with the family-specific primers on pure DNA showed only minor variations between species: Cp. psittaci, 85.2°C; Cp. felis, 85.2°C; Cp. caviae, 86.5°C; and Cp. abortus, 85.8°C. However, intraspecies variations in temperatures in the range of ±0.25°C were observed when crude DNA lysates from birds and cats were tested.
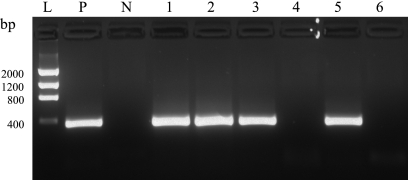
As an example of the dynamic range of the PCR assay, a dilution series of a positive liver sample is shown in Fig. 3. Samples were generally tested in the 10−1 and 10−2 dilutions, but some clinical samples could even be diluted to 10−6 and still be positive. However, we observed large variations in the target amount between each case, with CT values ranging from 18 to 35. The setup for testing clinical samples is shown in Fig. 4. The use of a control tube for each sample, containing both positive DNA and sample DNA, verified that inhibition had not prevented amplification in any sample. As an additional inhibition reduction step, each sample was tested at two dilutions.
Fig. 4.
Agarose gel setup showing positive (sample A) and negative (sample B) clinical samples. Lane L, DNA ladder; lane P, positive PCR control; lane N, no-template control; lane 1 sample A at a dilution of 10−1; lane 2, sample A at 10−1 plus Cp. psittaci DNA; lane 3, sample A at 10−2; lane 4, sample B at 10−1; lane 5, sample B at 10−1 plus Cp. psittaci DNA; lane 6, sample B at 10−2.
Validation of the PCR assay on clinical samples.
The PCR assays were applied on selected clinical samples from birds, cats, guinea pigs, and sheep. All samples were first screened for Chlamydiaceae using the family-specific primers, and positive samples were further tested with the species-specific primers. Results are listed in Table 3. With only one exception, all positive samples were also identifiable to species level using the species-specific assays. All bird samples were positive for Cp. psittaci, samples from cats were positive for Cp. felis, and Cp. abortus was detected in the positive ovine sample. However, the positive sample from a guinea pig reacted only with the family-specific primer and not with the first selected specific primer (Ccav-F) in the Cp. caviae assay or any of the others.
In positive bird cases, Cp. psittaci was detected in all three organs but in different concentrations. As indicated by the CT values, spleen and liver samples contained the largest amount of target, while heart samples usually showed CT values 2 to 5 cycles higher. The quality of the postmortem samples showed considerable variation; however, samples with a high degree of autolysis were also among the PCR-positive samples.
Sequencing of the IGS in a clinical sample.
The first selected Cp. caviae-specific primer (Ccav-F) did not generate any amplicons when applied on a Chlamydiaceae-positive guinea pig sample. An amplicon spanning the IGS and the first part of the 23S rRNA was generated directly from the diluted sample and sequenced. The obtained sequence showed highest similarity (99%) to the sequence of the published Cp. caviae GPIC reference strain (U68451); however, an SNP in the 3′ end of the Ccav-F primer was discovered. A new primer, Ccav2-F, was selected and validated against reference DNA and the clinical sample with satisfactory results.
Comparative testing of three different methods for detecting Chlamydiaceae in tissue specimens.
The sensitivity of the PCR assay was compared with microscopical examination of smears stained with either fluorescence-labeled monoclonal antibodies (IF) or the MZN staining technique. In Table 5, the results from the comparison are listed. The PCR-positive samples were detected with both the family- and species-specific assays. All samples that were positive with IF or MZN were also positive by PCR. The interpretations of the microscopical methods were quite easy in samples containing large amounts of elementary bodies (EB) packed in macrophages. However, in low-quality samples (due to autolysis) or in samples with a low number of EB, it was difficult to discriminate specific signal from background staining. The PCR method performed significantly better in all three sample types, with the MZN method being the least sensitive method, detecting only between 26 and 56% of PCR-positive samples. Though no difference in detection was observed between sample types by PCR, more positive samples were detected by the microscopy of samples from spleen and liver than from heart.
Table 5.
Comparison of three methods for detecting Cp. psittaci in samples from birds: Chlamydiaceae-specific PCR, direct IF detection, and modified Ziehl-Neelsen staining
| Tissue | Total no. of samples | No. of positive samples by: |
||
|---|---|---|---|---|
| PCR | IF assay | MZN staininga | ||
| Heart | 272 | 35 | 14 | 9 |
| Liver | 283 | 36 | 19 | 13 |
| Spleen | 278 | 32 | 21 | 18 |
MZN, modified Ziehl-Neelsen.
DISCUSSION
The family Chlamydiaceae comprises nine different species, all of which, with the exception of C. trachomatis, may infect and cause disease in animals. Due to the large variety in specimens that a veterinary diagnostic laboratory may receive, it is essential to use sensitive and general methods for screening samples and to be able to identify the species in positive samples for epidemical purposes to clarify transmission routes (6). The aim of this study was to establish an easy and reliable PCR protocol for identification of Chlamydiaceae in swab and tissue specimens. By using real-time detection and SYBR green as the intercalating dye, a method was developed for fast preparation, detection, and identification of a number of Chlamydophila species of veterinary interest. Furthermore, the simplicity of this approach makes it applicable for many veterinary laboratories. With the family-specific primer pair, it was possible to detect all nine species, and with species-specific primer pairs, Cp. psittaci, Cp. abortus, Cp. felis, and Cp. caviae could be confirmed. No purification of DNA was performed, but a combination of a robust master mix containing BSA and dilution of the enzymatically digested lysate allowed generation of specific amplicons. The real-time PCR approach gave several benefits: first, we were able to increase the sensitivity up to 10-fold compared to traditional gel analysis; second, the CT value gave a relative quantitative measure of the target concentration; and finally specific amplicons could be identified as distinct peaks in the melt curve analysis, thus eliminating the need for gel analysis.
Diagnostic PCR methods for Chlamydiaceae have generally been developed on the basis of 16S-23S rRNA genes or the ompA gene, which codes for a major surface protein. These genes are believed to be highly conserved among Chlamydiaceae species, but with molecular methods, new variants of well-known species have been described which might not be detected by previously published species-specific primers. In one study, Kutlin et al. (13) found that the ompA and 16S rRNA genes in Cp. pneumonia isolated from bandicoots and koalas show sequence variations compared to human and horse strains and that one of the published primer/probe sets based on ompA had several mismatches toward these strains. However, intraspecies variation was less in the ribosomal genes.
The primers selected in this study are located in the IGS region and domain I of 23S rRNA gene, which previously has been shown to be highly conserved and with well-defined segments of species-specific sequences appropriate for classification of Chlamydiaceae (7). For this reason, we selected primers to detect members of Chlamydiaceae, and in spite of an SNP in the 5′ end of the forward primer in Cp. felis, Cp. pneumoniae, C. trachomatis, C. suis, and Cp. pecorum, efficient amplification was observed for all nine species on reference DNA. For the species-specific assays, we did find some intraspecies variations in the primer sites among published sequences as two Cp. psittaci (24) and two Cp. abortus (22) strains shared the same two SNPs. The similarity of these four strains has led other investigators to suggest that these strains may represent an intermediate group between the two species, linking these two together with a common ancestor (24). It is likely that the location of these SNPs may prevent amplification from the two species-specific assays, but they should be detected by the family-specific primers during screening. In our case, all positive specimens from birds were positive with both the family and the species assay primers, thus indicating that the intermediate group of psittaci strains represents a rare group. However, the prevalence of the intermediate Cp. abortus group needs to be established as we tested only a very limited number of strains.
The value of screening with a family-specific primer was evident in the positive guinea pig sample that did not react with any of the species assays. Due to the evolution of species from a common ancestor, a higher sequence polymorphism is expected in species-specific signature sequences than in family-specific signature sequences. The selection of species primers should preferentially be based on sequences from several different strains. In our case, the low number of published sequences from Cp. caviae strains led us initially to select a primer that turned out not to be species specific; however, after sequencing of the isolate, a new specific primer was selected and applied.
Two real-time PCR detection systems for identification of Chlamydiaceae have been described (17, 19). One, applying a family-specific PCR targeting the ribosomal genes, was able to distinguish six species by subjecting generated amplicons to high-resolution melt curve analysis (19). However, as this is a highly sensitive method allowing discrimination on the basis of a single base pair polymorphism, it also requires highly standardized conditions for preparing the master mix and clean DNA to be reproducible. When trying to differentiate Chlamydiaceae amplicons based on melting temperature, we found in our assay that the intraspecies variation was too large to ensure sufficient identification of species. The crude lysate and variations in master mixes may explain the observed differences. The second method was based on a panel of species-specific primers and probes targeting the ompA gene which were used to differentiate Chlamydiaceae species of veterinary interest in samples that previously tested positive with family-specific PCR methods (17). However, this assay may not be economically feasible in a routine laboratory, due to the high costs for labeled probes, and a higher level of strain variation in sequence of the ompA gene may affect specificity of the assay.
Based on the CT values, we found a correlation between the target DNA concentration in the sample and clinical observations. A large amount of target was found in postmortem samples of birds with a clinical history comparable to ornithosis, where samples could be diluted more than 10−5, in comparison to pharyngeal swabs from subclinically infected live birds, which allowed only a 10−1 dilution.
The gold standard for Chlamydophila detection methods is still cell culture; however, as the method is laborious, many other methods are used in routine laboratories. In this study, we have compared the sensitivity of using classical microscopical detection of stained cell smears with our PCR assays. To ensure the specificity of the PCR assay, all screened positive samples were also tested using the species-specific PCR. The PCR method showed superior sensitivity to microscopy using either IF or MZN. Although these methods are straightforward and rapid to perform, they require skilled staff, and evaluation is subjective (21). In samples from clinical cases, elementary bodies of Chlamydiaceae can easily be found in intracellular vacuoles in macrophages; however, interpretation is difficult when the sample is partly degraded by autolysis or in subclinical cases when the amount of target is low (21). We observed that samples that were not positive by microscopy generally had high CT values in the real-time PCR, corresponding to a small amount of target DNA.
Inhibition is one of the major problems in PCR analysis of clinical samples, giving rise to weak or false-negative results. Especially blood components, such as hemoglobin, have been shown to be highly inhibitory (2). It is widely believed that inhibition is mediated by organic compounds that may either inactivate the DNA polymerase (12) or compete with the template (2). To overcome these problems a polymerase that is less sensitive to inhibition (1) or compounds that bind inhibitors such as BSA (12) or resins can be added. In the bloodstream, BSA serves as a transport molecule by binding molecules with low solubility, and this action may be exploited in the PCR. Enzymatically digested tissue samples, as used in this study, may contain a lot of different inhibitory substances. However, we were able to reduce the inhibitory effect of these to such a magnitude that spiked samples treated for inhibitory substances showed comparable sensitivity in samples with pure DNA, but the CT values were higher and the gel band intensity was lower (Fig. 2). The binding properties of BSA seem to be diverse and do not apply only to tissue substances as BSA may also be beneficial in PCR testing of poultry fecal samples (14).
The majority of the veterinary submissions used for testing of the primer assays were from psittacine birds and cats as we had access only to a limited number of samples from sheep and guinea pigs. Although the primers may be specific for Chlamydophila species in general when tested on pure DNA, we have tested tissue lysates only from a limited number of animal species. It remains to be established if other animals may contain inhibitors which are not inactivated by this protocol.
In the setup for testing clinical specimens, each lysate was added to three PCR tubes, where one worked as an inhibition control by also including Chlamydophila DNA. A positive signal from this indicated that conditions were appropriate for PCR (Fig. 4). To ensure that the inhibition control was a sensitive indicator, it was adjusted to a concentration which in negative samples resulted in a CT value of approximately 30. The addition of two different dilutions of lysate to the test tubes served two purposes: each sample was tested in duplicate, and if the lowest dilution was inhibited (no signal from the internal control), the next dilution had a fair chance of revealing positivity without the need for retesting.
With PCR methods, detection of pathogens can be fast and accurate; however, it is cumbersome and expensive if the master mix has to be prepared regularly for small numbers of diagnostic samples. To make a more convenient assay, a master mix containing all reagents can be prepared in large volumes and stored as PCR tubes at −20 Co until use (14, 15). Furthermore, this also allows performance testing of each batch before use, thus providing a measure of quality assurance. When a sample is received at our routine diagnostic laboratory, a lysate is quickly prepared from the sample, added to thawed, prealiquoted PCR tubes, and tested by real-time PCR.
The most recent taxonomic reclassification based on sequencing of the rRNA operon (5, 8) and the improvement in detecting Chlamydiaceae have significantly increased our knowledge of this obligate intracellular pathogen. The Chlamydiaceae species are known to be very host specific, and in our study we identified Chlamydophila species only in their expected hosts. However, other studies have shown that one species may be isolated from various hosts (4, 6, 9, 13, 16, 17). Host specificity may be viewed in terms of primary or principal hosts and occasional hosts (20), where the infection usually spreads from the primary host to occasional hosts living in close proximity. This is illustrated in Cp. caviae isolated from the primary host (a guinea pig) but also detected in the animal's owner, as well as in a cat and a rabbit in the household (16). Usually, primary hosts are sampled because they show typical clinical signs, but the larger host diversity applicable for Chlamydophila sp. infections may call for broadening the range of relevant animals and samples to be tested. Although other species-specific PCR assays have been developed, these may be of limited value if the infecting species is difficult to predict. In accordance with Condon and Oakey (6), we applied a Chlamydiaceae-specific primer pair to screen for positive samples that can be subjected to species identification. Although the immediate treatment of infections by all Chlamydophila species is based on administration of the same group of antibiotics (20), the zoonotic potential of the different species is not equal, and a species determination is therefore required for adequate disease control.
In conclusion, we have developed and applied a fast, robust, and cost-effective method that detects Chlamydiaceae species and differentiates the most important Chlamydophila species causing disease problems in animals. To reduce processing time, the assay has been developed as a real-time PCR method, but primers may also be used for conventional gel-based PCR detection. However, in this study the real-time analysis provided higher sensitivity, the possibility for quantitative measurement of the target DNA, and in-tube detection of positive samples by melt curve analysis. It is also adaptable to the detection of new variants or species as it detects Chlamydiaceae based on the IGS and domain I in the 23S rRNA gene, which shows conserved interspecies sequences; the method also has scope for variations that may be applicable for generating new species-specific primers. Furthermore, the principle of enzymatic treatment coupled to dilution of prepared template DNA may be a platform from which protocols for other pathogens in various specimens may be developed.
ACKNOWLEDGMENTS
We sincerely thank Arthur A. Anderson, National Animal Disease Center, USDA Agriculture Research Service, and Gunna Christiansen, University of Aarhus, Denmark, for providing DNA from Chlamydiaceae reference strains for this study.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Abu Al-Soud W., Radstrom P. 1998. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 64:3748–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abu Al-Soud W., Radstrom P. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bodetti T. J., et al. 2002. Molecular evidence to support the expansion of the host range of Chlamydophila pneumoniae to include reptiles as well as humans, horses, koalas and amphibians. Syst. Appl. Microbiol. 25:146–152 [DOI] [PubMed] [Google Scholar]
- 4. Bodetti T. J., et al. 2003. Wide range of Chlamydiales types detected in native Australian mammals. Vet. Microbiol. 96:177–187 [DOI] [PubMed] [Google Scholar]
- 5. Bush R. M., Everett K. D. 2001. Molecular evolution of the Chlamydiaceae. Int. J. Syst. Evol. Microbiol. 51:203–220 [DOI] [PubMed] [Google Scholar]
- 6. Condon K., Oakey J. 2007. Detection of Chlamydiaceae DNA in veterinary specimens using a family-specific PCR. Lett. Appl. Microbiol. 45:121–127 [DOI] [PubMed] [Google Scholar]
- 7. Everett K. D., Andersen A. A. 1997. The ribosomal intergenic spacer and domain I of the 23S rRNA gene are phylogenetic markers for Chlamydia spp. Int. J. Syst. Bacteriol. 47:461–473 [DOI] [PubMed] [Google Scholar]
- 8. Everett K. D., Bush R. M., Andersen A. A. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415–440 [DOI] [PubMed] [Google Scholar]
- 9. Gaede W., et al. 2010. Detection of Chlamydophila caviae and Streptococcus equi subsp. zooepidemicus in horses with signs of rhinitis and conjunctivitis. Vet. Microbiol. 142:440–444 [DOI] [PubMed] [Google Scholar]
- 10. Herrmann B., et al. 2006. Chlamydophila psittaci in Fulmars, the Faroe Islands. Emerg. Infect. Dis. 12:330–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaleta E. F., Taday E. M. 2003. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian. Pathol. 32:435–461 [DOI] [PubMed] [Google Scholar]
- 12. Kreader C. A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kutlin A., et al. 2007. Molecular characterization of Chlamydophila pneumoniae isolates from Western barred bandicoots. J. Med. Microbiol. 56:407–417 [DOI] [PubMed] [Google Scholar]
- 14. Lund M., et al. 2003. Evaluation of PCR for detection of Campylobacter in a national broiler surveillance programme in Denmark. J. Appl. Microbiol. 94:929–935 [DOI] [PubMed] [Google Scholar]
- 15. Lund M., Nordentoft S., Pedersen K., Madsen M. 2004. Detection of Campylobacter spp. in Chicken Fecal Samples by Real-Time PCR. J. Clin. Microbiol. 42:5125–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lutz-Wohlgroth L., et al. 2006. Chlamydiales in guinea-pigs and their zoonotic potential. J. Vet. Med. A Physiol. Pathol. Clin. Med. 53:185–193 [DOI] [PubMed] [Google Scholar]
- 17. Pantchev A., Sting R., Bauerfeind R., Tyczka J., Sachse K. 2010. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp. Immunol. Microbiol. Infect. Dis. 33:473–484 [DOI] [PubMed] [Google Scholar]
- 18. Petrovay F., Balla E. 2008. Two fatal cases of psittacosis caused by Chlamydophila psittaci. J. Med. Microbiol. 57:1296–1298 [DOI] [PubMed] [Google Scholar]
- 19. Robertson T., et al. 2009. Characterization of Chlamydiaceae species using PCR and high resolution melt curve analysis of the 16S rRNA gene. J. Appl. Microbiol. 107:2017–2028 [DOI] [PubMed] [Google Scholar]
- 20. Rodolakis A., Yousef Mohamad K. 2010. Zoonotic potential of Chlamydophila. Vet. Microbiol. 140:382–391 [DOI] [PubMed] [Google Scholar]
- 21. Sachse K., et al. 2009. Recent developments in the laboratory diagnosis of chlamydial infections. Vet. Microbiol. 135:2–21 [DOI] [PubMed] [Google Scholar]
- 22. Siarkou V., Lambropoulos A. F., Chrisafi S., Kotsis A., Papadopoulos O. 2002. Subspecies variation in Greek strains of Chlamydophila abortus. Vet. Microbiol. 85:145–157 [DOI] [PubMed] [Google Scholar]
- 23. Stamp J. T., McEwen A. D., Watt J. A., Nisbet D. I. 1950. Enzootic abortion in ewes; transmission of the disease. Vet. Rec. 62:251–254 [DOI] [PubMed] [Google Scholar]
- 24. Van Loock M., et al. 2003. Missing links in the divergence of Chlamydophila abortus from Chlamydophila psittaci. Int. J. Syst. Evol. Microbiol. 53:761–770 [DOI] [PubMed] [Google Scholar]