Abstract
Lactococcus lactis is a prokaryotic microorganism with great importance as a culture starter and has become the model species among the lactic acid bacteria. The long and safe history of use of L. lactis in dairy fermentations has resulted in the classification of this species as GRAS (General Regarded As Safe) or QPS (Qualified Presumption of Safety). However, our group has identified several strains of L. lactis subsp. lactis and L. lactis subsp. cremoris that are able to produce putrescine from agmatine via the agmatine deiminase (AGDI) pathway. Putrescine is a biogenic amine that confers undesirable flavor characteristics and may even have toxic effects. The AGDI cluster of L. lactis is composed of a putative regulatory gene, aguR, followed by the genes (aguB, aguD, aguA, and aguC) encoding the catabolic enzymes. These genes are transcribed as an operon that is induced in the presence of agmatine. In some strains, an insertion (IS) element interrupts the transcription of the cluster, which results in a non-putrescine-producing phenotype. Based on this knowledge, a PCR-based test was developed in order to differentiate nonproducing L. lactis strains from those with a functional AGDI cluster. The analysis of the AGDI cluster and their flanking regions revealed that the capacity to produce putrescine via the AGDI pathway could be a specific characteristic that was lost during the adaptation to the milk environment by a process of reductive genome evolution.
INTRODUCTION
Lactic acid bacteria (LAB) play an essential role in the production of fermented dairy products, with Lactococcus lactis, Streptococcus thermophilus, and Lactobacillus helveticus being the species most frequently used in starter cultures. LAB are mainly used as primary fermentation starters for the production of a large diversity of dairy products all over the world. Their principal role in dairy fermentations is the rapid production of lactic acid from lactose, resulting in a lowering of the pH, thus inhibiting the growth of spoilage and pathogenic microorganisms. In addition, they possess other metabolic activities that confer the flavor and texture characteristics of the final product.
The long and safe history of use of LAB in dairy fermentations has resulted in the majority of species being classified as GRAS (Generally Regarded as Safe) in the United States. A similar concept is used in the EU by the European Food Safety Authority (EFSA), whereby the majority of LAB species are assigned a Qualified Presumption of Safety (QPS) status. Nevertheless, some properties or enzymatic activities can generate undesirable flavors (32) or even toxic compounds as a result of, e.g., the production of biogenic amines (BA) (22).
BA are nitrogenous compounds that are formed mainly in foodstuffs through the decarboxylation of certain amino acids by particular bacteria. The ingestion of BA-rich foods may lead to several toxicological problems such as tachycardia, hypotension, or respiratory disorders (for a review, see reference 16). In nonfermented foods, such as fish, BA are formed by the action of contaminating Gram-negative bacteria, while in fermented products (wine, meat, cheese, and cider), the production is principally due to LAB (2, 14, 17, 19, 20). The BA-producing LAB can be present in the raw material or introduced during the production process (4, 18, 28). The BA most frequently found in fermented dairy products are histamine and tyramine, but putrescine is also commonly detected (8). A great variability in the levels of putrescine in cheeses was previously reported, with concentrations of up to 0.9 g per kg (8, 16) noted in some cases. Although nondirect toxic effects have been described for putrescine itself, it is also able to increase the toxic effect of other BA. In addition, putrescine can participate directly in the promotion of malignancy, due to its role in the regulation of cell growth and the transformation of cells, or indirectly, since it can give rise to secondary amines that can combine with nitrites to generate nitrosamines (16). Putrescine is produced from arginine through two successive catabolic reactions: decarboxylation and deamination. Depending on the order of these reactions, two different production routes have been described: the agmatine deiminase (AGDI) pathway, in which arginine is first decarboxylated to agmatine and then deiminated to putrescine, and the ornithine decarboxylase (ODC) pathway, in which arginine is first deiminated to ornithine and then decarboxylated to putrescine. In wine and cider studies, putrescine-producing LAB that use the ODC pathway have been previously isolated (17, 25) and the ODC enzyme has been extensively characterized (25, 27). However, no putrescine producers using the ODC pathway have been isolated from dairy products. In contrast, there are a large number of dairy LAB and enterococci belonging to different species such as Lactobacillus brevis, Lactobacillus curvatus, and Enterococcus faecalis (15, 17, 22) that have been previously identified as putrescine producers operating via the AGDI pathway. Therefore, the main pathway for putrescine biosynthesis in dairy products would appear to be the AGDI pathway (Fig. 1A). To date, no putrescine-producing L. lactis strain has been identified. L. lactis is the LAB fermentation starter with the greatest importance, as it is used as a primary starter, in either pure or mixed cultures, for the production of a large diversity of cheeses and fermented milks. Moreover, L. lactis has become a model organism among the LAB and it is the second-best-studied Gram-positive bacterial species (12). L. lactis is used for cheese and fermented milk production due to its high growth rate and rapid production of lactic acid in the dairy environment. In nature, this species is found on plant surfaces, from which it can reach the milk after being swallowed by ruminants. It is believed that it is through this route that L. lactis has adapted to the dairy environment. L. lactis is divided in two subspecies, L. lactis subsp. lactis and L. lactis subsp. cremoris, with the former subspecies distinguished from the latter by its ability to grow at 40°C and in the presence of 4% NaCl and by the production of ammonia from arginine (7). Nevertheless, several strains are atypical and cannot be distinguished from each other based on these simple phenotypic assays. However, they can be classified at the genetic level by 16S rRNA gene sequence comparisons, since they present 0.7% dissimilarity (30).
Fig. 1.
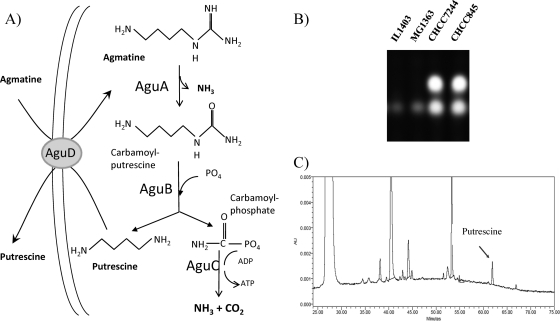
(A) Scheme for the conversion of agmatine to putrescine via the AGDI metabolic pathway; the reactions catalyzing each enzyme of the route are indicated. (B) TLC analysis of the production of putrescine from agmatine in the IL1403, MG1363, CHCC7244, and CHCC845 strains. (C) HPLC analysis of the production of putrescine from agmatine overnight supernatants of CHCC7244 in M17+A medium. AU, absorbance units.
In this work, L. lactis strains from artisanal cheeses and industrial collections were screened for putrescine production. The identification and characterization of several L. lactis subsp. lactis and L. lactis subsp. cremoris strains able to produce putrescine from agmatine are described. The genes responsible for putrescine biosynthesis were sequenced and their transcription analyzed. A PCR-based test was developed for the identification of strains with a functional AGDI cluster that could be used for the selection of non-putrescine-producing L. lactis strains.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of 72 strains of L. lactis (45 classified as L. lactis subsp. lactis and 27 as L. lactis subsp. cremoris), 30 obtained from the Christian Hansen Culture Collection (CHCC, Hørsholm, Denmark) and 42 from the collection of the Instituto de Productos Lácteos de Asturias (IPLA, Asturias, Spain), were included in this work (see Table S1 in the supplemental material). The strains were identified and genotypically classified to the subspecies level (subsp. lactis or subsp. cremoris) by comparing the 16S rRNA gene sequences with those in the public databases by the use of the universal primers 27F and 1492R (21). In addition, the four L. lactis genomes available in databases were also included in this study.
All L. lactis strains were grown at 30°C without aeration in M17 medium (Oxoid, UK) supplemented with 5 g liter−1 lactose and glucose. Where indicated, 20 mM agmatine (Sigma-Aldrich, St. Louis, MO) was added to the medium (M17+A). In both cases, the initial pH of the medium was 6.8 ± 0.2. E. faecalis BA62 (a putrescine producer) was grown at 30°C in M17 (Oxoid, UK) supplemented with 5 g liter−1 lactose and glucose, while L. brevis CECT 3810, L. curvatus VI14, and L. collinoides IPLA11000 (putrescine producers) were grown at 37°C in MRS broth (Oxoid, UK) (pH 6.2 ± 0.2).
Screening for putrescine-producing L. lactis strains.
Inocula for subsequent experiments were prepared from stock cultures of the above-mentioned collections as follows. Vials were thawed, 20 μl of the contents was used to inoculate 2 ml of M17+A, and cultures were incubated at 30°C for 24 h. Cultures were then centrifuged at 2,250 × g for 10 min, and 1 ml of the resulting supernatants was filtered through a 0.2-μm-pore-size Supor membrane (Pall, UK) and analyzed in an initial screening for the presence of putrescine by thin-layer chromatography (TLC) according to the method of García-Moruno et al. (9). Subsequently, the presence of putrescine in the TLC-positive samples was confirmed by reversed-phase high-performance liquid chromatography (RP-HPLC) as previously described (18). Briefly, supernatants were derivatized using dabsyl chloride and injected onto a Waters Nova-pack C18 column in a Waters liquid chromatograph system controlled by Millenium 32 software (Waters). The gradient and detection conditions were as described previously (13).
DNA manipulation and PCR amplification.
Total DNA from all the strains used in this work was isolated using a genomic DNA purification kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions.
One microliter of the DNA was added to the PCR mix containing 5 μl of supplied PCR buffer (10× Dreamtaq buffer; Fermentas, Lithuania), 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.2 mM each oligonucleotide primer, 2 U of DreamTaq DNA polymerase (Fermentas, Lithuania), and water in a final volume of 50 μl. PCR amplification was performed using a DNA thermal cycler (iCycler; Bio-Rad, Hercules, CA) for 35 cycles with the general program (94°C for 30 s, 25 s at the suitable annealing temperature of the primers used, and 72°C for 1 min per kb of DNA to be amplified). DNA was separated on a 0.8% agarose gel in TAE buffer (40 mM Tris–acetate–1 mM EDTA; pH 8.0) and visualized after ethidium bromide staining under UV light conditions. PCR products were purified from an agarose gel by the use of a QIAquick gel extraction kit (Qiagen, Germany).
The primer pair PTC2 and AgDdr (unpublished results) (see Table S2 in the supplemental material) was used to amplify by PCR (35 cycles of 94°C for 30 s, 50°C for 25 s, and 72°C for 3 min) a 2.9-kb fragment within the AGDI cluster of the L. lactis strains studied in this work (see Table S1 in the supplemental material). Subsequently, the amplified fragments were sequenced using the PCR primers. The obtained sequences were used together with the sequences of AGDI clusters available in the public genome databases as a basis for further primer design for a progressive “genome walking” sequencing approach. The various PCR fragments generated were sequenced by Macrogen Inc. (Korea) and assembled using Vector NTI Advance software, version 9.1 (Invitrogen, CA). Sequence analysis was performed using the University of Wisconsin Genetics Computer Group software package. BLAST and BLASTP programs were used to determine the similarities of the deduced amino acid sequences to those present in the databases. Analysis of transmembrane elements was performed with the TMHMM tool (http://www.cbs.dtu.dk/services/TMHMM/; Danish Technicalt University, Denmark).
The AgmSq1 and AgmSq2 primer pair designed in the present work was used to screen for the presence of the AGDI cluster in a battery of LAB strains (Table 1) by the use of the PCR mix composition mentioned above and the following PCR program: 35 cycles of 94°C for 30 s, 52°C for 25 s, and 72°C for 2 min.
Table 1.
Strains used for validation of the AGDI-PCR testa
| Species | Strain(s) | Origin | Putrescine production (TLC/HPLC) | Detection of AGDI cluster by primer pair AgmSq1 and AgmSq2 | Source(s) |
|---|---|---|---|---|---|
| Lactococcus lactis subsp. lactis | IL1403, CHCC1915, T2-26 | Dairy | All − | All +/IS | 3a, CHCC, IPLA |
| CHCC845, CHCC7244, T1-48 | Dairy | All + | All + | CHCC, IPLA | |
| Lactococcus lactis subsp. cremoris | MG1363, CHCC3370, LC144 | Dairy | All − | All − | 9q, CHCC, IPLA |
| 2A22, GE214, 3AA9 | Dairy | All + | All + | IPLA | |
| Enterococcus faecalis | CNRZ1535, IPLA148T, BA62 | Dairy | All + | All + | CNRZ, IPLA |
| V583, IPLA67, IPLA55 | Human | All + | All + | 29a, IPLA | |
| Enterococcus hirae | 268 | Dairy | All + | All + | IPLA |
| Lactobacillus brevis | CECT 3810, CECT 3811 | Dairy | All + | All + | CECT |
| CLC23 | Human | All + | All + | J. M. Rodríguez | |
| Lactobacillus curvatus | IPLAVI14, IPLAVI16 | Dairy | All + | All + | IPLA |
| Lactobacillus collinoides | IPLA11000 | Cider | + | + | IPLA |
| Enterococcus faecium | IPLA29, IPLA56 | Human | All − | All − | IPLA |
| Enterococcus durans | IPLA655, IPLA833, IPLA984 | Dairy | All − | All − | IPLA |
| Streptococcus thermophilus | CHCC1524, LMG18311 | Dairy | All − | All − | CHCC, LMG |
| Lactobacillus ruminis | IPLA44 | Human | − | − | IPLA |
| Lactobacillus buchneri | B301 | Dairy | − | − | NIZO |
| Lactobacillus plantarum | LL441 | Dairy | − | − | IPLA |
| Lactobacillus casei | ATCC393 | Human | − | − | ATCC |
Origin, origin of the sequences and strains used for the design and testing of the AGDI-PCR assay. The presence of the AGDI cluster, the capacity of the strains to produce putrescine in broth, and the results of the specific AGDI-PCR assays are indicated. +/IS indicates a positive PCR result but a nonfunctional AGDI cluster due to the presence of the IS element (see text for details).
Northern blot analysis.
L. lactis subsp. lactis T1-48 and CHCC1915 cells were grown in M17 or M17+A to the stationary phase and harvested by centrifugation in a refrigerated benchtop microcentrifuge (Eppendorf, Germany) at maximum speed. Total RNA was purified using an RNA kit (Qiagen, Germany). Cells were mechanically disrupted using glass beads (Sigma, St. Louis, MO) (≤106 μm) and the recommended Qiagen buffer (Qiagen, Germany). The tubes were then vigorously shaken three times for 1 min at high speed in a bead beater (FastPrep-24 system; MP Biomedicals, Illkirch, France). RNA was subjected to electrophoresis as previously described (3). Transfers and hybridizations were performed using standard protocols (31). DNA probes, obtained by PCR using the primers described in Table S2 in the supplemental material, were radiolabeled by nick translation, incorporating an [α-32P]dATP nucleotide (Perkin-Elmer, Covina, CA), and the signal was obtained from a Typhoon Trio system (GE Healthcare, Piscataway, NJ) after exposure to a storage phosphor screen.
RT-PCR.
L. lactis cells were grown in M17 and M17+A and harvested under the conditions described previously. Total RNA was extracted using TRI reagent (Sigma-Aldrich, St. Louis, MO) as previously described (23). RNA samples (2 μg of total RNA) were treated with 2 U of DNase I (Fermentas, Lithuania) to eliminate any DNA contamination. Then, cDNA was synthesized from total RNA by the use of a high-capacity cDNA reverse transcription (RT) kit (Applied Biosystems, Foster City, CA) with specific primers for each of the analyzed genes (see Table S2 in the supplemental material). PCRs were performed using 2 μl of the cDNA preparation and a 0.4 μM concentration of each gene-specific primer (see Table S2 in the supplemental material). Amplifications were performed for 35 cycles (94°C for 30 s, 50°C for 25 s, and 72°C for 1 min), and samples were analyzed on a 0.8% agarose gel in TAE buffer. The absence of contaminating DNA was checked by non-reverse-transcribed PCR, which was performed under the conditions described above, using the corresponding RNA as the template.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper are available at GenBank (http://www.ncbi.nlm.nih.gov) under accession numbers FR8586582 to FR8586602 (these are detailed in Table S1 in the supplemental material).
RESULTS
Screening for putrescine-producing L. lactis strains.
From a total of 20 L. lactis strains (10 L. lactis subsp. lactis and 10 L. lactis subsp. cremoris) from the CHCC collection that were subjected to screening, two L. lactis subsp. lactis strains, CHCC7244 and CHCC845, gave a positive spot in the TLC analysis of the overnight M17+A culture supernatants (Fig. 1B), indicating that they were able to produce putrescine from agmatine through deamination. The capability of these two strains to produce putrescine was confirmed and quantified by HPLC analysis (Fig. 1C); the CHCC845 and CHCC7244 strains produced 0.351 mM and 2.07 mM putrescine, respectively. This is the first report of strains of L. lactis being able to produce the biogenic amine putrescine.
In view of this surprising result, we decided to check for the presence of the agmatine deiminase gene (aguA) in the genomes of the sequenced strains of L. lactis of dairy origin: L. lactis subsp. lactis IL1403 (GenBank accession no. AE005176.1), L. lactis subsp. cremoris MG1363 (GenBank accession no. AM406671.1), and L. lactis subsp. cremoris SK11 (GenBank accession no. CP000425.1). None of these genomes had annotations of the agmatine deiminase genes, although its presence in L. lactis subsp. lactis IL1403 had been previously suggested (12, 24). A BLAST search using known aguA sequences revealed the presence of the gene in the L. lactis subsp. lactis IL1403 strain but not in the two L. lactis subsp. cremoris strains, MG1363 and SK11. In addition, the aguA gene is also present in the genome of L. lactis subsp. lactis KF147 (GenBank accession no. CP001834.1), a strain of vegetable origin. A sequence analysis of that genome region in L. lactis subsp. lactis IL1403 showed the presence of all the genes necessary for the production of putrescine via AGDI pathway, although the results also showed an insertion (IS) element in the cluster in the opposite orientation (Fig. 2A).
Fig. 2.
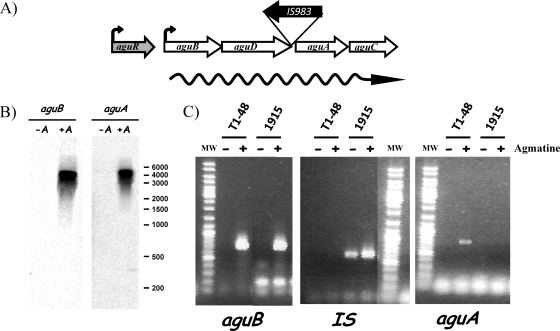
Genetic organization and transcriptional analysis of the AGDI cluster in L. lactis. (A) Scheme for the genetic organization of the AGDI cluster in L. lactis. The regulatory gene (aguR), the catabolic genes (aguB, aguD, aguA, and aguC), the insertion element (IS) found in some strains, and the putative promoters (wavy arrow) are indicated. (B) Northern blot analysis of the catabolic genes in the AGDI cluster in the absence (−A) and presence (+A) of agmatine. Internal fragments of the aguB and aguA genes were used as probes. The positions of RiboRuler high-range RNA ladder (Fermentas, Germany) fragments are indicated. (C) RT-PCR amplification with primers designed to amplify the internal regions of the aguB, aguA, and IS element genes in the absence (−) and presence (+) of agmatine in the T1-48 and CHCC1915 L. lactis subsp. lactis strains. Lanes MW, molecular weight markers (GeneRuler DNA ladder mix; Fermentas, Lithuania).
To confirm the presence of the cluster in the putrescine-positive strains, PCR amplifications using PTC2 and agDdr primers (see Table S2 in the supplemental material) previously designed in our laboratory based on the E. faecalis AGDI cluster genes (unpublished) were performed. The two producer strains (L. lactis subsp. lactis CHCC7244 and CHCC845) yielded a PCR product of the expected size (2.9 kb). L. lactis subsp. lactis IL1403 yielded a 1-kb-larger band (3.9 kb), which is consistent with the presence of the IS element. In the eight nonproducing L. lactis subsp. lactis CHCC strains, the same 3.9-kb band was obtained. The nonproducing L. lactis subsp. cremoris CHCC strains and L. lactis subsp. cremoris MG1363 gave no amplification. As expected, the IL1403 and MG1363 strains did not produce putrescine from agmatine (data not shown).
In view of these results, we decided to increase the number of lactococcus strains analyzed by combining phenotypical (TLC) and molecular (PCR with primer pair PTC2 and agDdr) screening strategies. In this new survey, we added 10 strains from the CHCC collection (for a total of 30 strains) and included 42 strains from the IPLA collection (isolated from different artisanal cheeses) (see Table S1 in the supplemental material). Nineteen strains, all identified as L. lactis subsp. cremoris, did not produce any PCR product, and 24 strains, all identified as L. lactis subsp. lactis, produced a 3.9-kb PCR band similar to that of the IL1403 strain; none of the 43 strains produced putrescine. However, 29 out of the 72 analyzed strains produced the 2.9-kb PCR band corresponding to the AGDI cluster. Putrescine production from agmatine was confirmed by TLC and HPLC for all but one of these PCR-positive strains (the exception was strain 3AA11; see Table S1 in the supplemental material). Among these AGDI-positive strains, 21 (3 from the CHCC collection and 18 from the IPLA collection) were classified as L. lactis subsp. lactis and 7 (all from the IPLA collection) as L. lactis subsp. cremoris. Interestingly, the agmatine deiminase genes were not present in the previously sequenced genomes of L. lactis subsp. cremoris MG1363 and SK11.
Sequence analysis of the AGDI cluster of L. lactis.
To obtain the sequence of the AGDI cluster, some primers were designed based on the sequence of L. lactis subsp. lactis IL1403 to render several overlapping PCR products that were sequenced and assembled. The whole AGDI clusters of 6 of the putrescine-producing isolates were sequenced (see Table S1 in the supplemental material). All the clusters had the same genetic organization, composed of five genes in the same orientation (Fig. 2A).
aguR is the first gene in the cluster.
The translated protein showed similarities to the transcriptional regulators of the LuxR family and had the highest amino acid identity (41%) with the product of the orthologous gene present in the AGDI clusters of several E. faecalis strains. The aguR gene is preceded by a putative promoter and shows a putative transcriptional terminator.
aguB.
This gene encodes a protein that showed similarity to putrescine carbamoyl transferases present in several bacterial species such as E. faecalis and Lb. brevis. The closest sequence was that of E. faecalis V583 (NP_814483), which showed 80% identity. It is preceded by a putative promoter composed of very similar consensus sequences.
aguD.
This gene showed similarity to several genes encoding amino acid/biogenic amine antiporters. Analyses of the deduced amino acid sequence performed using the TMHMM tool predicted the presence of 12 transmembrane elements (data not shown).
aguA.
This gene encodes the putative agmatine deiminase, and its amino acid sequence presented the highest identity (83%) to that of E. faecalis V583 (NP_814483).
aguC.
The deduced protein of the last gene of the cluster has similarities to carbamate kinases belonging to AGDI clusters (56% amino acid identity with that of Lb. brevis ATCC27305) but also to those associated with the arginine deamination pathway (ADI). In fact, the highest amino acid identity (71%) was found with the carbamate kinase associated with the ADI cluster that is present in each of the available L. lactis subsp. lactis genomes.
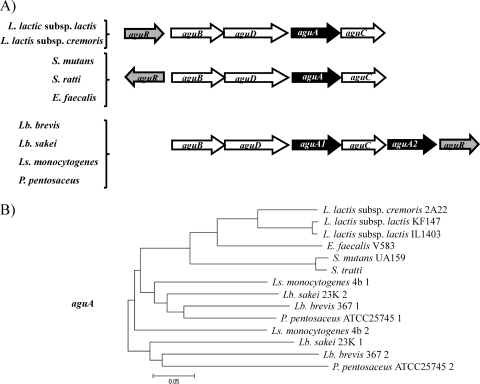
The organization of the L. lactis AGDI cluster is unique (Fig. 3 A), although it did show similarities to those of other Gram-positive cocci such as E. faecalis, Enterococcus gallinarum, and Streptococcus mutans. These species have the same number and order of the genes, but aguR is in the opposite orientation (Fig. 3A). In the case of Lactobacillus, Pediococcus, and Listeria, the order is different, with the putative regulator located at the end of the cluster in the same orientation. In addition, this group has a second putative agmatine deiminase gene (aguA2) (Fig. 3A).
Fig. 3.
(A) Comparison of the genetic organizations of the AGDI clusters from different bacteria. Grey arrow, regulator gene; white arrows, structural genes; black arrows, structural genes duplicated in the same genomes. Lb., Lactobacillus; Ls., Listeria. (B) Phylogenetic tree of the aguA genes from different species. Nucleotide sequences were aligned using ClustalW, and the phylogenetic tree was constructed with MEGA version 4 software, using the p-distance as the nucleotide substitution model. Those species with duplicated aguA genes are identified with a number (1 or 2) at the end of the strain name.
These observed differences in the cluster organization are a reflection of the genetic groups; the differences are also evident in comparisons of the nucleotide sequences of the aguA genes of different species (aguA1 and aguA2) (Fig. 3B). We can observe that L. lactis forms a separate branch, and although they are very closely related, there are differences between the aguA sequences from L. lactis subsp. lactis and those from L. lactis subsp. cremoris (Fig. 3B).
IS983 interrupts the transcription of the AGDI cluster.
The presence of the IS element in the 24 L. lactis subsp. lactis strains that produced a 3.9-kb PCR band seems to be the explanation for the inability of these strains to produce putrescine from agmatine. As has been mentioned before, IS983 is located in the middle of the AGDI catabolic genes of L. lactis subsp. lactis IL1403. In addition, there are 14 other copies of IS983 spread in different regions of the chromosome. A BLAST database search revealed that IS983 is present only in L. lactis subsp. lactis sequences of dairy origin. We have sequenced the complete AGDI clusters of three of these non-putrescine-producing L. lactis subsp. lactis strains (see Table S1 in the supplemental material). The analysis of the obtained sequences revealed that, in all cases, the IS983 gene is located between the aguD and aguA genes and is in the opposite orientation.
The fact that the IS element is located between the aguD and aguA genes but in the opposite orientation suggests that transcription of the downstream genes may be interrupted if the catabolic cluster is transcribed as an operon (an mRNA covering aguB, aguD, aguA, and aguC; see Fig. 1A and 2A), as seems to be the situation based on the sequence analysis. In order to verify whether the catabolic genes are expressed in the same mRNA, Northern blot analysis was performed. After hybridization of the total RNA from CHCC7244 (growth in the presence of agmatine) with two probes matching the first catabolic gene (aguB) and the first gene after the IS (aguA), a unique band of a size corresponding to the four genes was obtained (Fig. 2B), confirming that they constitute a single operon. In contrast, when the total RNA was obtained from the same strain grown in the absence of agmatine, no hybridization signal was obtained, indicating that transcription of the polycistronic mRNA is induced by the presence of the substrate of the AGDI pathway.
Subsequently, to test that whether the presence of the IS element is responsible for the inactivation of the AGDI cluster, we used RT-PCR to analyze transcription of two genes upstream (aguB) and downstream (aguA) of the IS element in two strains: T1-48, a putrescine-producing strain, and CHCC1915, a nonproducing strain harboring the IS element. To avoid synthesis of cDNA from other copies of the IS element from different regions of the genome or interference from transcription from the IS promoter, the cDNA was synthesized with specific primers located in front of each gene (see Table S2 in the supplemental material).
Figure 2C clearly shows that transcription of aguB is induced in the presence of agmatine in both strains and that transcription of aguA was detected in the presence of agmatine in the producer strain, whereas no transcription was detected in the IS-harboring strain. aguA expression is interrupted by the presence of the IS element, which is in fact transcribed in the CHCC1915 strain in both the presence and the absence of agmatine (Fig. 2C).
Chromosomal surrounding regions of the AGDI cluster.
The presence of BA-producing genes in LAB has been postulated to be a strain-specific trait acquired through horizontal transfer (5, 26). However, in Enterococcus species, the capacity to produce tyramine and putrescine appears to be species dependent (unpublished). In order to establish whether this is the case in L. lactis, a deeper look into the genomic information and the surrounding sequences was undertaken for the genomes of the IL1403, KF147, MG1363, and SK11 strains.
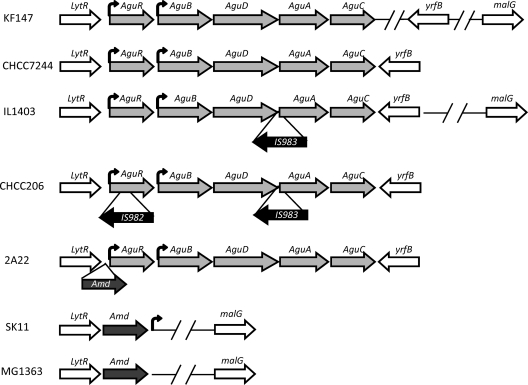
In L. lactis subsp. lactis IL1403 (dairy strain) and KF147 (vegetable origin), an open reading frame (ORF) that presents similarities to the transcriptional regulators of the LytR family was located upstream of the aguR gene (Fig. 4). In the case of L. lactis subsp. cremoris MG1363 and SK11 (both dairy strains) without the AGDI cluster, we also found the lytR gene. A search in the four genomes showed that the first common gene downstream of lytR is malG, which encodes an ABC maltose transporter (Fig. 4). The two genes are separated by two and three orf genes in L. lactis subsp. cremoris strains SK11 and MG1363, respectively, and by 17 and 18 orf genes—including the AGDI cluster—in L. lactis subsp. lactis strains KF147 and IL1403, respectively. In the two latter strains, an NADH-dependent oxidoreductase gene (yrfB) was found to be located downstream of the AGDI cluster. In the KF147 strain, however, there are 6 additional small ORFs between this gene and aguC (Fig. 4).
Fig. 4.
Scheme for the different genetic organizations of the AGDI clusters found in L. lactis. The insertion elements (IS) in the L. lactis subsp. lactis strains that possess them and the amidase gene in the L. lactis subsp. cremoris strain that possesses the AGDI cluster are indicated as insertions to facilitate comparisons between clusters. Common and flanking genes within all strains are shown. Truncated horizontal lines indicate that other ORFs have been located in that region. Putative promoters are indicated as bent arrows.
PCR amplification of the adjacent regions was performed with the 53 strains analyzed in this study that have the AGDI cluster (with or without IS983). Two primers were designed to investigate the presence of the lytR gene upstream of the AGDI cluster (see Table S2 in the supplemental material). The first primer was designed based on the internal sequence of the lytR gene (orf20), and a second primer (agur1c) was designed within the aguR gene. All the strains gave lytR-positive amplification upstream of the AGDI cluster. However, all the L. lactis subsp. cremoris strains showing productive results yielded a band larger than expected. The sequence of this band revealed the presence of an amidase gene between lytR and aguR. This gene is present in the genomes of SK11 and MG1363, just downstream of the lytR gene, indicating that, in L. lactis subsp. cremoris putrescine-producing strains, the AGDI cluster is located after the amidase gene (Fig. 4). Two L. lactis subsp. lactis strains (CHCC206 and CHCC3052), which have the AGDI cluster interrupted by IS983, also produced a band larger than expected. The sequence of this band revealed the presence of a gene encoding a putative transposase (IS982) that can also be found in other locations in the L. lactis subsp. lactis and subsp. cremoris sequenced genomes.
A second pair of primers (see Table S2 in the supplemental material) was designed to test for the presence of yrfB downstream of the AGDI cluster; one was designed within aguC (ck3), and the second (orf3c) was designed from an internal sequence of yrfB from the L. lactis subsp. lactis IL1403 genome. When PCR with these primers was performed using the 53 AGDI-positive strains, 42 strains—including L. lactis subsp. lactis and L. lactis subsp. cremoris strains from both the CHCC and the IPLA collections—yielded a band of the expected size (see Table S1 in the supplemental material). However, 11 strains (9 L. lactis subsp. lactis and 2 L. lactis subsp. cremoris, all from the IPLA collection) did not produce a PCR band. In order to analyze these strains, a new primer (Lcorf4C) was designed based on the sequence of the downstream gene (locus tag LLKF_1858) of L. lactis subsp. lactis KF147 and used in combination with the ck3 primer (see Table S2 in the supplemental material). All but 2 of these strains yielded a band (see Table S1 in the supplemental material), indicating that these strains isolated from artisanal cheeses showed higher similarity to the strains of vegetable origin. All these results together indicate that the genomic regions surrounding the AGDI cluster are highly conserved.
One of the characteristics of heterologous acquired sequences is that they have G+C content different from that of the surrounding regions of the host species. The G+C content of the AGDI cluster and that of its surrounding regions ranged from 35 to 38%, a percentage range that is similar to that of the L. lactis species (around 35%, as calculated from the available sequenced genomes). There were only two exceptions: (i) aguR, which showed lower G+C content (28% in L. lactis subsp. lactis and 30% in L. lactis subsp. cremoris) and (ii) IS983, which showed higher content (44%), suggesting a possible different origin.
PCR assay for detection of putrescine producers in dairy products.
Different PCR methods have been described and optimized for detection of putrescine-producing LAB via the AGDI pathway (6, 24). All of them use the aguA gene as the target for detection. However, in dairy products this target would lead to the results representing false positives due to those L. lactis subsp. lactis strains that have the AGDI cluster interrupted by IS983. To avoid this problem, we designed a new set of primers located in aguD and aguA, the genes surrounding the IS element, in order to distinguish the strains carrying the functional AGDI cluster from those with the nonfunctional AGDI cluster. In order to extend the method to other strains described as putrescine producers and belonging to different species present in fermented foods, sequences from aguD and aguA genes present in databases and those obtained in this work were used (Table 1). The alignment of the corresponding regions of the AGDI clusters from Lb. brevis IOEB 9809 (GenBank accession no. AF446085) and ATCC 367 (NCBI accession no. NC_008497), L. lactis subsp. lactis IL1403 (NCBI accession no. NC_002662), KF147 (GenBank accession no. NC_013656), and T3-17 (GenBank accession no. FR856588), L. lactis subsp. cremoris IPLA2A22 (GenBank accession no. FR856590) and GE2-14 (GenBank accession no. FR856601), and Enterococcus faecalis V583 (GenBank accession no. AE016830), JH2-2 (GenBank accession no. AF354231), and CNRZ1535 (GenBank accession no. FN392111) allowed the design of degenerate primers (AgmSq1 [5′-CAAGATTTDTTCTGGGCHTTYTTCTC-3′] and AgmSq2 [5′-TTGGHCCACARTCACGAACCCT-3′]). These primers were tested against a battery of LAB belonging to different genera with or without the ability to produce putrescine from agmatine (Table 1). A PCR fragment of around 700 bp was obtained with those strains known to produce putrescine. As expected, those L. lactis subsp. lactis strains classified as non-putrescine producers but presenting a nonfunctional AGDI cluster rendered a larger PCR band (1,700 bp) corresponding to the addition of the IS element (see Table S1 in the supplemental material). No amplification was obtained from the nonproducers without the AGDI cluster (Table 1). Since this method could be used to screen large culture collections, the assay was also performed using overnight liquid cultures and isolated colonies picked from plates without the need for a DNA purification step. In these cases, 1 μl from an overnight culture, or from a colony previously resuspended in 50 μl of sterile water, was directly used in the PCR assay. The same amplification profile was obtained as when purified DNA was used as the template (data not shown). The results demonstrate that this test could be used for the screening of microbial collections and other isolates for putrescine producers.
DISCUSSION
Society is increasingly concerned with the issue of food safety, with ever-increasing demands from the consumer for higher quality and safer foods. Thus, a great deal of effort has been made in the development of techniques to detect contaminants such as biogenic amines in foodstuffs. The synthesis and accumulation of BA in food requires the presence of microorganisms with the capacity to produce these toxic compounds. In this work, we report the identification of several putrescine-producing L. lactis strains belonging to L. lactis subsp. lactis and subsp. cremoris. We describe the genetic determinants and their organization and surrounding regions. Based on this knowledge, a PCR test for the detection of LAB producing putrescine via the AGDI pathway has been developed.
Putrescine is one of the BA most frequently detected in dairy products (8) and can be formed by the decarboxylation of arginine to produce agmatine, which in turn yields putrescine by deamination. Several species of LAB have this pathway and thus are able to produce putrescine from agmatine. Two different genetic organizations of the AGDI clusters in those species have been described; it is noteworthy that the organization found in L. lactis strains, although resembling that of other cocci such as Streptococcus mutans, Streptococcus ratti, or E. faecalis, is unique (Fig. 3A). Several authors have suggested that the capacity to produce BA by LAB has been acquired recently through horizontal gene transfer (5, 26). However, in the case of Lactococcus species, most of the data obtained from the analysis of AGDI cluster organization and sequences indicate that the capacity to produce putrescine from agmatine is not a recent trait acquired from horizontal gene transfer, as it would have been present in the genomes even before the differentiation between L. lactis subsp. lactis and subsp. cremoris occurred. The data from the AGDI cluster organization in L. lactis reveal that it is unique among LAB, with the regulatory gene located upstream of the cluster and orientated in the same direction as the catabolic genes (Fig. 3A). A phylogenetic tree, based on aguA sequences, clustered the genes from L. lactis subsp. lactis and L. lactis subsp. cremoris together and next to those of other cocci (Fig. 3B). Sequence comparisons showed 85% identity between the AGDI clusters from L. lactis subsp. lactis and those from L. lactis subsp. cremoris. This value is the same as that found in comparisons of the two subspecies at the genomic level (33), and it is in accordance with the hypothesis that L. lactis subsp. lactis and subsp. cremoris constitute two different genetic lineages that diverged a long time ago (11, 29). In addition, the overall G+C content of the cluster is similar to that of the genome (around 35%). All these data support our hypothesis that the AGDI cluster has been present in the genome of L. lactis for a long time rather than having been recently acquired. In some strains, it would have been lost during the adaptation to the milk environment or even due to an empirical selection of fermentation starters unable to produce putrescine, a compound that confers undesirable flavor. Moreover, the cluster is located in a conserved region present in all L. lactis strains (of the available genomes analyzed), including dairy and nondairy strains, although in different positions of the genomic map, mainly due to the chromosome inversion observed in the MG1363 strain and the different genome sizes (11). The surrounding regions of the AGDI are very well conserved in all the analyzed strains. Two common genes (lytR and malG) were located in all the available L. lactis genomes that comprise the AGDI cluster. The lytR gene was located upstream in all the analyzed strains, including the L. lactis subsp. lactis and L. lactis subsp. cremoris strains, although, in the latter case, an amidase gene was found between lytR and aguR. It is remarkable that, in L. lactis subsp. cremoris SK11, a remnant of the putative promoter of the aguR gene is still present in the chromosome after the amidase gene (Fig. 4), thus indicating the loss of the AGDI cluster. The downstream region of the AGDI cluster is also well conserved, although greater variability of that region has been observed. In fact, the distance between the cluster and the malG gene differs between the genomes and is lower in the case of the L. lactis subsp. cremoris strains. Interestingly, there was a group of strains isolated from artisanal cheeses in which the downstream sequence was more similar to that of L. lactis subsp. lactis KF147, a strain of vegetable origin, than to other strains of dairy origin. This sequence was not detected in any of the strains of industrial origin, again indicating that these strains are better adapted to the milk environment. This adaptation has been related to the acquisition of enzymatic properties, usually through the acquisition of plasmids, and, more importantly in the present case, by a process of reductive genome evolution, as indicated by the smaller chromosome size of the dairy strains (11).
The analysis of the sequence revealed (i) a small distance (16 nt on average) between the adjacent catabolic genes of the cluster (aguB, aguD, aguA, and aguC), (ii) the absence of clear promoters, and (iii) the presence of consensus ribosome binding sites (AGGAGG) in the intergenic regions. This suggested that all these genes are cotranscribed in the form of a single mRNA molecule, as was shown by the results of Northern blot and RT-PCR analysis in this work (Fig. 2B). The transcription of the catabolic genes of the AGDI cluster as a single mRNA molecule in a study of S. mutans UA159 has also been previously reported (10).
The genetic difference found between the putrescine-producing and nonproducing L. lactis subsp. lactis strains was due to the presence of an insertion element between aguD and aguA genes. In addition, this IS element was orientated in the opposite direction, suggesting the existence of transcription interference. RT-PCR experiments showed that the presence of the IS element impeded the transcription of the aguA gene (Fig. 2C) and resulted in an inability to produce putrescine. The fact that, in some L. lactis subsp. lactis strains, the AGDI cluster is silenced by the occurrence of one or two IS elements and that some L. lactis subsp. cremoris strains have even lost the cluster completely indicates that this cluster is not necessary in dairy environments. The differences between strains such as the presence or absence of a functional AGDI cluster, of an inactivated IS element, or of two inactivated IS elements, evidence of a deleted AGDI cluster together with the presence of a residual promoter, and evidence of a completely deleted AGDI cluster reflect different steps in the evolution of L. lactis dairy strains.
The presence of BA-producing microorganisms is a negative trait that can affect the safety and quality of dairy products; therefore, methods that can rapidly detect BA producers in raw material or foodstuffs are required. Microbiological methods have traditionally been used for detection of this capability (1). However, in the case of the detection of putrescine producers from agmatine (deamination reaction), no microbiological methods have been described so far, since all of them are based on detection of the decarboxylation reactions (1). Culture-independent methods based on microbial DNA detection for identification of AGDI-positive strains have been previously described (6, 24). However, these methods are not appropriate for dairy products, since they target the aguA gene and therefore give positive results for the L. lactis subsp. lactis strains with the AGDI cluster inactivated by IS983. The PCR method described in this work targets the intergenic region between the aguD and aguA genes, allowing discrimination of those strains that possess the AGDI cluster but do not produce putrescine.
In summary, this work describes the existence of L. lactis subsp. lactis and L. lactis subsp. cremoris strains with the capability to produce putrescine from agmatine. The gene cluster of this pathway has been characterized and was shown to possess a unique organization, suggesting that, together with other characteristics of the sequence, it has not been recently acquired by L. lactis. It seems that the adaptation to the milk environment and/or the selective pressure exerted because of the use of the strains as fermentation starters resulted in IS inactivation or even the loss of the AGDI cluster in dairy strains. The AGDI-PCR test described here would help to discriminate putrescine-producing strains, not only for dairy products but also for any LAB species, independently of the presence of the fermented food.
Supplementary Material
ACKNOWLEDGMENTS
This research was performed with financial support from the Ministry of Science and Innovation, Spain (AGL2010-01024), and the European Community's Seventh Framework Programme (KBBE-CT-2007-211441).
The skillful technical assistance of Sussi Hansen, Chr. Hansen A/S, and Elena Cañedo, IPLA-CSIC, is gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Bover-Cid S., Holzapfel W. H. 1999. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 53:33–41 [DOI] [PubMed] [Google Scholar]
- 2. Burdychova R., Komprda T. 2007. Biogenic amine-forming microbial communities in cheese. FEMS Microbiol. Lett. 276:149–155 [DOI] [PubMed] [Google Scholar]
- 3. Calles-Enríquez M., et al. 2010. Sequencing and trancriptional analysis of Streptococcus thermophilus histamine biosynthesis gene cluster: factors that affect differential hdcA expression. Appl. Environ. Microbiol. 76:6231–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a. Chopin A., Chopin M.-C., Moillot-Batt A., Langella P. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260–263 [DOI] [PubMed] [Google Scholar]
- 4. Costantini A., Vaudano E., Prete V. D., Danei M., García-Moruno E. 2009. Biogenic amine production by contaminating bacteria found in starter preparations used in winemaking. J. Agric. Food Chem. 57:10664–10669 [DOI] [PubMed] [Google Scholar]
- 5. Coton E., Coton M. 2009. Evidence of horizontal transfer as origin of strain to strain variation of the tyramine production trait in Lactobacillus brevis. Food Microbiol. 26:52–57 [DOI] [PubMed] [Google Scholar]
- 6. Coton M., et al. 2010. Occurrence of biogenic amine-forming lactic acid bacteria in wine and cider. Food Microbiol. 27:1078–1085 [DOI] [PubMed] [Google Scholar]
- 7. Courtney P. D. 2000. Lactococcus lactis subspecies lactis and cremoris, p. 1166–1171In Robinson R. K. (ed.), Encyclopedia of food microbiology, 1st ed. Elsevier, Amsterdam, the Netherlands [Google Scholar]
- 8. Fernández M., Linares D. M., del Río B., Ladero V., Alvarez M. A. 2007. HPLC quantification of biogenic amines in cheeses: correlation with PCR-detection of tyramine-producing microorganisms. J. Dairy Res. 74:276–282 [DOI] [PubMed] [Google Scholar]
- 9. García-Moruno E., Carrascosa A. V., Muñoz R. 2005. A rapid and inexpensive method for the determination of biogenic amines from bacterial cultures by thin-layer chromatography. J. Food Prot. 68:625–629 [DOI] [PubMed] [Google Scholar]
- 9a. Gasson M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Griswold A. R., Chen Y. Y., Burne R. A. 2004. Analysis of an agmatine deiminase gene cluster in Streptococcus mutans UA159. J. Bacteriol. 186:1902–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kelly W. J., Ward L. J., Leahy S. C. 2010. Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. Evol. 2:729–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kok J., Buist G., Zomer A. L., van Hijum S. A. F. T., Kuipers O. P. 2005. Comparative and functional genomics of lactococci. FEMS Microbiol. Rev. 29:411–433 [DOI] [PubMed] [Google Scholar]
- 13. Krause I., Bockhardt A., Neckermann H., Henleand T., Klostermeyer H. 1995. Simultaneous determination of amino acids and biogenic amines by reversed phase high-performance liquid chromatography of the dabsyl derivatives. J. Chromatogr. A 715:67–79 [Google Scholar]
- 14. Ladero V., Linares D. M., Fernández M., Alvarez M. A. 2008. Real time quantitative PCR detection of histamine-producing lactic acid bacteria in cheese: relation with histamine content. Food Res. Int. 41:1015–1019 [Google Scholar]
- 15. Ladero V., Sánchez-Llana E., Fernández M., Alvarez M. A. 2011. Survival of biogenic amine-producing dairy LAB strains at pasteurisation conditions. Int. J. Food Sci. Technol. 46:516–521 [Google Scholar]
- 16. Ladero V., Calles-Enríquez M., Fernández M., Alvarez M. A. 2010. Toxicological effects of dietary biogenic amines. Curr. Nutr. Food Sci. 6:145–156 [Google Scholar]
- 17. Ladero V., et al. 2011. Biogenic amines content in Spanish and French natural ciders: application of qPCR for quantitative detection of biogenic amine-producers. Food Microbiol. 28:554–561 [DOI] [PubMed] [Google Scholar]
- 18. Ladero V., Fernández M., Alvarez M. A. 2009. Effect of post-ripening processing on the histamine and histamine-producing bacteria contents of different cheeses. Int. Dairy J. 19:759–762 [Google Scholar]
- 19. Ladero V., Fernández M., Cuesta I., Alvarez M. A. 2010. Quantitative detection and identification of tyramine-producing enterococci and lactobacilli in cheese by multiplex qPCR. Food Microbiol. 27:933–939 [DOI] [PubMed] [Google Scholar]
- 20. Landete J. M., Ferrer S., Pardo I. 2005. Which lactic acid bacteria are responsible for histamine production in wine? J. Appl. Microbiol. 99:580–586 [DOI] [PubMed] [Google Scholar]
- 21. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–147In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY [Google Scholar]
- 22. Linares D. M., Martín M., Ladero V., Alvarez M. A., Fernández M. 2011. Biogenic amines in dairy products. Crit. Rev. Food. Sci. Nutr. 51:691–703 [DOI] [PubMed] [Google Scholar]
- 23. Linares D. M., Fernández M., Martín M. C., Alvarez M. A. 2009. Tyramine biosynthesis in Enterococcus durans is transcriptionally regulated by the extracellular pH and tyrosine concentration. Microb. Biotechnol. 2:625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lucas P. M., et al. 2007. Agmatine deiminase pathway genes in Lactobacillus brevis are linked to the tyrosine decarboxylation operon in a putative acid resistance locus. Microbiology 153:2221–2230 [DOI] [PubMed] [Google Scholar]
- 25. Marcobal A., de las Rivas B., Moreno-Arribas M. V., Muñoz R. 2004. Identification of the ornithine decarboxylase gene in the putrescine-producer Oenococcus oeni BIFI-83. FEMS Microbiol. Lett. 239:213–220 [DOI] [PubMed] [Google Scholar]
- 26. Marcobal A., de las Rivas B., Moreno-Arribas M. V., Muñoz R. 2006. Evidence for horizontal gene transfer as origin of putrescine production in Oenococcus oeni BIFI-83. Appl. Environ. Microbiol. 72:7954–7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Momany C., Ernst S., Ghosh R., Chang N. L., Hackert M. L. 1995. Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 Å resolution. J. Mol. Biol. 252:643–655 [DOI] [PubMed] [Google Scholar]
- 28. Novella-Rodríguez S., Veciana-Nogués M. T., Roig-Sagués A., Trujillo-Mesa A., Vidal-Carou M. C. 2002. Influence of starter and non starter bacteria on the formation of biogenic amines in goat cheese during ripening. J. Dairy Sci. 85:2471–2478 [DOI] [PubMed] [Google Scholar]
- 29. Passerini D., et al. 2010. Genes but not genomes reveal bacterial domestication of Lactococcus lactis. PLoS One 5:e15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a. Sahm D. F., et al. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salama M. S., Sandine W. E., Giovannoni S. J. 1991. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl. Environ. Microbiol. 57:1313–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Smit G., Smit B. A., Engels W. J. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591–610 [DOI] [PubMed] [Google Scholar]
- 33. Wegmann U., et al. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.