Abstract
Plasmid pSEUDO and derivatives were used to show that llmg_pseudo_10 in Lactococcus lactis MG1363 and its homologous locus in L. lactis IL1403 are suitable for chromosomal integrations. L. lactis MG1363 and IL1403 nisin-induced controlled expression (NICE) system derivatives (JP9000 and IL9000) and two general stress reporter strains (NZ9000::PhrcA-GFP and NZ9000::PgroES-GFP) enabling in vivo noninvasive monitoring of cellular fitness were constructed.
TEXT
Lactococcus lactis is a widely used Gram-positive model organism that belongs to the clade of lactic acid bacteria (LAB). Advances in the genomics research of L. lactis, such as the sequencing of the genomes of the most commonly studied subspecies (1, 14, 23) and the various genome-wide transcriptome studies (see, e.g., reference 4), provide a broad view of the mechanisms of genetic regulation in this economically important organism. This knowledge is invaluable for strain selection and for the improvement of lactococcal strains by rational design. However, a standard locus in the chromosome of L. lactis for integration of DNA fragments, whether for genetic complementation (single-copy or merodiploid-like situations) or cloning of a reporter gene or promoter, is lacking. Whereas, for example, in Bacillus subtilis the amyE locus is often used for these purposes (19), in L. lactis various loci in the chromosome have been chosen as targets for integration. Unfortunately, to the best of our knowledge none of the proposed strategies exclude the possibility of phenotypic consequences. The leuA locus was shown to suffer from active read-through from the native leuA promoter (10), whereas the choice of the sex factor locus might interfere with the biology of L. lactis and, in addition, may have consequences with respect to possible conjugational transfer of the inserted DNA. Also, bacteriophage sequences have been used to drive site-specific integration of plasmids in the chromosome of L. lactis (2, 22). However, these methods do not allow making strains without resistance markers, while some require the use of a second plasmid to provide the bacteriophage integrase in trans. Furthermore, the localization in the chromosome of L. lactis MG1363 of sequences with high similarity to that at a given attB, e.g., in the comGC gene for the TP901-1 attB and in rex for the TUC2009 attB (data not shown), implies that the integration process might lead to the simultaneous disruption of potentially relevant processes.
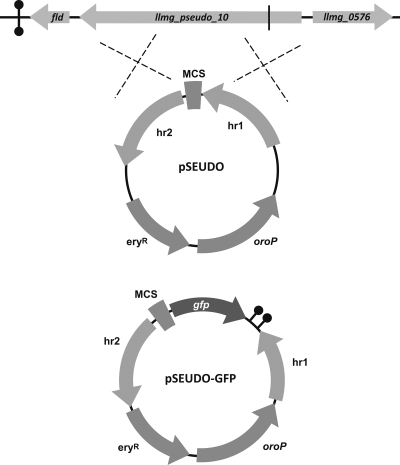
Here it was examined whether the llmg_pseudo_10 locus of L. lactis MG1363 or its corresponding region in L. lactis IL1403 is a suitable neutral region for chromosomal integrations (Fig. 1). In L. lactis IL1403, this locus contains yfjF, a gene of 1,506 bp whose product exhibits homology to transport proteins from the major facilitator superfamily. In L. lactis MG1363, translation is halted prematurely due to the presence of a stop codon at position 303, hence its annotation as a pseudogene (23). By cloning and resequencing, the nucleotide sequence of this region in L. lactis MG1363, originally described by Wegmann et al. (23), was confirmed. The loss of function of the locus in L. lactis MG1363 suggests that yfjF is nonessential. The llmg_pseudo_10 locus has been shown to be silent throughout the growth of L. lactis MG1363 in batch cultures in M17 medium (R. W. W. Brouwer, J. P. C. Pinto, A. Zeyniyev, J. Kok, and O. P. Kuipers, submitted for publication) and milk (A. de Jong, personal communication). In addition, llmg_pseudo_10 and yfjF display low nucleotide sequence similarity with other regions in the L. lactis genome, minimizing the possibility of illegitimate recombination.
Fig. 1.
Genomic context of llmg_pseudo_10 of L. lactis MG1363 and its relation to the homologous regions (hr's) present in pSEUDO and pSEUDO-GFP. The multiple cloning site (MCS) contains, in clockwise order, EcoRI, XmaI/SmaI, SphI, ScaII, SalI, HindIII, BglII, XhoI, and BamHI restriction enzyme sites. The gfp gene for superfolder GFP (17) was cloned in pSEUDO using the XhoI and BamHI sites, yielding pSEUDO-GFP. Terminators are indicated by “lollipop” structures. The vertical line on llmg_pseudo_10 depicts the stop codon that prematurely halts translation of the gene in L. lactis MG1363. eryR, erythromycin resistance gene. oroP encodes the L. lactis orotate transporter (3).
Construction and applicability of pSEUDO and pSEUDO-GFP.
To be able to perform unmarked integrations in the chromosome of L. lactis, the chromosomal integration vector pCS1966 was employed, allowing positive selection of cells in which the plasmid had been excised from the genome (20). Two DNA fragments were amplified from L. lactis MG1363 chromosomal DNA by PCR, one of 529 bp, obtained with the primer pair P1_pseudo10/P2_pseudo10, and another of 804 bp, obtained using P3_pseudo10/P4_pseudo10, and sequentially inserted into pCS1966 using the restriction enzymes indicated in Table 1 and Escherichia coli DH5α as the cloning host. Selection was performed on tryptone-yeast extract (TY)–agar plates with 150 μg/ml erythromycin. The custom-made multiple cloning site GAATTCCCCGGGCATGCCGCGGTCGACAAGCTTAGATCTCGAGGATCC was introduced between the BamHI and EcoRI sites, thus producing the plasmid pSEUDO (Fig. 1). This vector can be used to insert DNA fragments into the llmg_pseudo_10 locus of L. lactis MG1363 using positive selection for resistance to the toxic pyrimidine analog 5-fluoroorotate by methods described before (20), with minor modifications (18). Although it is not possible to screen for integrants via loss of function through gene disruption, pSEUDO allows for a quick and efficient positive survival strategy to monitor both integration (erythromycin resistance) and excision of the vector backbone from the chromosome (5-fluoroorotate resistance), thus enabling the production of unmarked strains in an easy and fast manner.
Table 1.
Oligonucleotides used in this study
| Name | Nucleotide sequence (5′–3′)a | Restriction enzyme |
|---|---|---|
| P1_pseudo10 | GCTCTAGACAATTGCTCCCATGCTTGATTCC | BglII |
| P2_pseudo10 | CGCGGATCCTGCTTTTGGATTAAAAGGTTTGAAAG | BamHI |
| P3_pseudo10 | CGGAATTCGGCGGCTCTGTTGGATTAATATATGG | EcoRI |
| P4_pseudo10 | GCGGTACCCAATTGAAGAGACAAGAAAACC | KpnI |
| P1_yfjF | GCGAAATCTAGACTTCAAACATAAGAGACCTCG | XbaI |
| P2_yfjF | GCGAAAGGATCCTTTAGCTTTAGGGTTGAAAGG | BamHI |
| P3_yfjF | GCGAAAGGATCCTTTGGCTGGCGGTTCTGTGGG | BamHI |
| P4_yfjF | GCGAAAGGTACCAATCGAAGAGACAAGAAAGCC | KpnI |
| nisRK_Forw | TAAAGGGATCCGCTTAGATACAGATAAAGGTCAGG | BamHI |
| nisRK_Rev | AGATTGGATCCCAAAACTGATATCTTGTAGCACCTGC | BamHI |
| GFP-SF_Forw | ATAGTCTCGAGTAAGGAGGCAAATATGAAACATCTTCGTAAAGGCGAAGAGCTGTTCACTGG | XhoI |
| GFP-SF_Rev | ACTATGGATCCTATAAACGCAGAAAGGCCCACC | BamHI |
| PhrcA_forw | ATCTGGAATTCATCCAAAGATTCTAATCTTTTATAACAG | EcoRI |
| PhrcA_rev | GCTCCCTCGAGTATCTCTAAGTTTTTTCTTTTAGCACTC | XhoI |
| PgroES_forw | GGAACGAATTCTTGAAGCTGATGAGCTCCCTTTCTG | EcoRI |
| PgroES_rev | ATACTCTCGAGCATTTTTTATTTTTAGCACTCTTAATAG | XhoI |
Restriction enzyme sites are underlined.
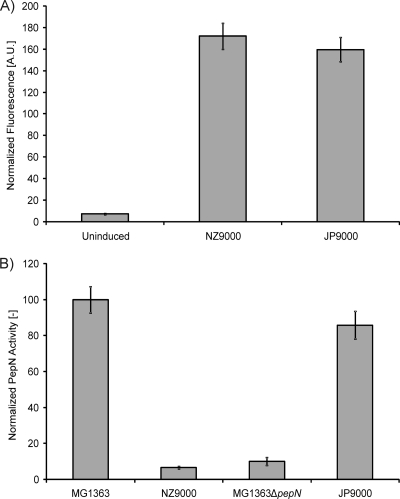
As a proof of principle, the applicability of pSEUDO is illustrated through the integration, internal and in opposite orientation to llmg_pseudo_10, of the genes of the two-component nisin sensor, NisRK (11), in the chromosome of L. lactis MG1363. A fragment containing the nisRK genes with their own promoter was amplified by PCR from chromosomal DNA of L. lactis NZ9000 (19) using the primers nisRK_Forw and nisRK_Rev. The PCR product and pSEUDO were digested with BamHI and ligated after dephosphorylation of the digested vector, producing pSEUDO::nisRK. The presence and orientation of the insert were checked with PCR and restriction endonuclease digestions. The integration of nisRK into L. lactis MG1363 using pSEUDO::nisRK generated the L. lactis strain JP9000. Expression of nisRK and the applicability of the nisin-induced controlled expression (NICE) system (6, 11) using L. lactis JP9000 are the same as for L. lactis NZ9000. As an example, the NICE system was used for the overproduction of a green fluorescent protein (GFP)- and hexahistidine-tagged membrane protein of L. lactis, BcaP-GFP-H6, using pNZbcaP-GFP-H6, a pNZ8048 derivative in which the bcaP-gfp-H6 gene is driven by the nisin-inducible PnisA promoter (12). L. lactis strains NZ9000 and JP9000 carrying this plasmid produced equivalent levels of the tagged protein since similar fluorescent signals from the overproduced BcaP-GFP-H6 were obtained using either strain (Fig. 2). Contrary to previous observations (10), the native nisRK promoter was sufficient to yield significant amounts of NisRK and read-through from the neighboring genes was not required for a functional NICE system. In addition to pSEUDO::nisRK, a similar vector was constructed for nisRK integration in the yfjF locus of L. lactis IL1403. The llmg_pseudo_10 homologous regions of pSEUDO were replaced by homologous regions of the yfjF locus, amplified from chromosomal DNA of IL1403 using primers P1_yfjF, P2_yfjF, P3_yfjF, and P4_yfjF (Table 1). The nisRK genes were inserted in the BamHI site in opposite direction to the yfjF gene, resulting in plasmid pSEUDO-IL::nisRK. The integration of nisRK into L. lactis IL1403 resulted in strain IL9000. To test functionality, IL9000 was transformed with a pNZ8048-derived plasmid in which gfp was inserted downstream the nisin-inducible PnisA promoter (H. Trip, unpublished results). GFP fluorescence levels upon induction with nisin were similar to those obtained with L. lactis NZ9000 harboring the same plasmid (data not shown), demonstrating that the NICE system is identically functional in L. lactis IL9000. The plasmids pSEUDO::nisRK and pSEUDO-IL::nisRK can be used to introduce the NICE system into any L. lactis MG1363- or L. lactis IL1403-derived strain, respectively. This is thought to be very useful for the LAB research community since L. lactis MG1363 and L. lactis IL1403 are by far the most commonly used strains in the L. lactis applied and fundamental research fields. The use of pSEUDO::nisRK or pSEUDO-IL::nisRK circumvents the use of combinations of vectors in strains that do not carry nisRK on the chromosome. In those cases, pNZ9530 (11) is usually required to provide nisRK in trans for complementation analysis. L. lactis JP9000 is preferable to standard strain NZ9000, since pepN is intact in the former whereas it is disrupted in NZ9000 (pepN::nisRK) (11) (Fig. 2). L. lactis IL9000 is the first IL1403 derivative in which the NICE system for nisin-inducible gene expression can be employed. The general aminopeptidase N (PepN) (21), while generally assumed not to be relevant under most conditions, is likely to have an effect on nitrogen metabolism through the influence of the products of peptide hydrolysis on, e.g., the major pleiotropic regulator CodY (5). Also, the availability of specific peptides in the medium has been correlated with the ability of L. lactis to overproduce membrane proteins (15).
Fig. 2.
Heterologous protein production and activity of aminopeptidase PepN in L. lactis. (A) Expression of the membrane protein BcaP-GFP-H6 was induced in L. lactis strains NZ9000 and JP9000, both carrying plasmid pNZbcaP-GFP-H6 (12). The strains were grown in GM17 until an optical density (OD) at 600 nm of 0.5 was reached, after which they were induced for 1 h with 5 ng/ml of nisin. Mean fluorescence, as measured by flow cytometry, of the plasmid-carrying strains normalized to that of plasmid-free L. lactis NZ9000 is plotted. The uninduced bar depicts the fluorescence of the noninduced JP9000 (pNZbcaP-GFP-H6) culture. (B) PepN activity in L. lactis strains MG1363 (9), NZ9000 (11), MG1363ΔpepN (16), and JP9000 (this work) was determined in cell extracts of cultures grown in GM17 until an OD at 600 nm of 0.5, by monitoring the hydrolysis of the PepN substrate lysyl-p-nitroanilide, as described previously (8). Data (A and B) are the averages of 4 biological replicates, and the error bars are the associated standard deviations.
To facilitate the integration of promoter-gfp reporters in the chromosome of L. lactis, pSEUDO-GFP was also constructed (Fig. 1). This vector was made by cloning the gene of the superfolder variant of GFP (17) together with the two terminator sequences from the iGEM Biobrick I746909 (http://partsregistry.org/Part:BBa_I746909) (13) to block read-through into llmg_0576, a putative transcriptional regulator of the TetR family. The PCR fragment obtained with primers GFP-SF_Forw and GFP-SF_Rev was inserted in pSEUDO using BamHI and XhoI. The first primer was used to add 4 well-translated codons and a strong ribosome binding site (RBS) sequence to the 5′ end of gfp (7).
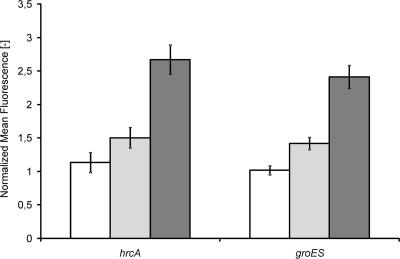
As an example of the applicability of pSEUDO-GFP, promoter-gfp fusions were constructed using the upstream regions of two general stress response genes, hrcA and groES. The promoter region of hrcA was amplified using the primer pair PhrcA_forw/PhrcA_rev, and that of groES was amplified with the primers PgroES_forw/PgroES_rev. EcoRI and XhoI were used to digest these DNA fragments and pSEUDO-GFP, in which the two promoter fragments were separately inserted. Insertion of the promoter-gfp fusions in the llmg_pseudo_10 locus of the chromosome of L. lactis NZ9000 yielded the strains NZ9000::PhrcA-gfp and NZ9000::PgroES-gfp. By exposing both strains to high temperatures and making use of the stress-induced activity of PhrcA and PgroES, it was demonstrated that they are able to reliably monitor, in a noninvasive manner (unlike, e.g., promoter-lacZ fusions) and in real time, the effect of growth and environmental conditions on the general fitness of cells (Fig. 3).
Fig. 3.
Induction of GFP production in L. lactis NZ9000::PgroES- GFP and in L. lactis NZ9000::PhrcA-GFP grown in GM17 medium at 30°C. Both cultures were shifted to 60°C when they had reached an optical density at 600 nm of 0.5. White, 0 min after the temperature shift; light gray, 15 min after the temperature shift; dark gray, 60 min after the temperature shift. The fluorescence was measured over time using an Epics XL-MCL flow cytometer (Coulter, Fullerton, CA). Values were normalized to the fluorescence of L. lactis NZ9000 undergoing the same heat treatment. Twenty thousand cells were measured per experiment, and 4 biological replicates were obtained per strain and per time point. The error bars are the associated standard deviations.
The use of pSEUDO-GFP enables application of high-throughput screening strategies (e.g., using microtiter plates) for conditions in which putative promoters are expected to be active. Furthermore, the use of flow cytometry or other single-cell analysis techniques allows characterization of phenomena such as gene expression heterogeneity.
Altogether, pSEUDO, pSEUDO-GFP, and the derived plasmids and strains add value to the lactococcal research community in that they establish an improved working standard for the effective and efficient integration of DNA fragments into the chromosome of L. lactis.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Bolotin A., et al. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brøndsted L., Hammer K. 1999. Use of the integration elements encoded by the temperate lactococcal bacteriophage TP901-1 to obtain chromosomal single-copy transcriptional fusions in Lactococcus lactis. Appl. Environ. Microbiol. 65:752–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Defoor E., Kryger M.-B., Martinussen J. 2007. The orotate transporter encoded by oroP from Lactococcus lactis is required for orotate utilization and has utility as a food-grade selectable marker. Microbiology 153:3645–3659 [DOI] [PubMed] [Google Scholar]
- 4. de Jong A., Kok J., Kuipers O. P. 2011. Data resources and mining tools for reconstructing gene regulatory networks in Lactococcus lactis. Jpn. J. Lactic Acid Bacteria 22:1–14 [Google Scholar]
- 5. den Hengst C. D., et al. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Ruyter P. G., Kuipers O. P., de Vos W. M. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eberhardt A., Wu L. J., Errington J., Vollmer W., Veening J.-W. 2009. Cellular localization of choline-utilization proteins in Streptococcus pneumoniae using novel fluorescent reporter systems. Mol. Microbiol. 74:395–408 [DOI] [PubMed] [Google Scholar]
- 8. Exterkate F. A. 1984. Location of peptidases outside and inside the membrane of Streptococcus cremoris. Appl. Environ. Microbiol. 47:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gasson M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Henrich B., et al. 2002. Food-grade delivery system for controlled gene expression in Lactococcus lactis. Appl. Environ. Microbiol. 68:5429–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuipers O. P., de Ruyter P. G. G. A., Kleerebezem M., de Vos W. M. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15–21 [Google Scholar]
- 12. Linares D. M., Geertsma E. R., Poolman B. 2010. Evolved Lactococcus lactis strains for enhanced expression of recombinant membrane proteins. J. Mol. Biol. 401:45–55 [DOI] [PubMed] [Google Scholar]
- 13. Lou C., et al. 2010. Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Mol. Syst. Biol. 6:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Makarova K., et al. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marreddy R. K. R., et al. 2010. Amino acid accumulation limits the overexpression of proteins in Lactococcus lactis. PLoS One 5:e10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mierau I., et al. 1996. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J. Bacteriol. 178:2794–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88 [DOI] [PubMed] [Google Scholar]
- 18. Pinto J. P. C., Kuipers O. P., Marreddy R. K. R., Poolman B., Kok J. 2011. Efficient overproduction of membrane proteins in Lactococcus lactis requires the cell envelope stress sensor/regulator couple CesSR. PLoS One 6:e21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shimotsu H., Henner D. J. 1986. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene 43:85–94 [DOI] [PubMed] [Google Scholar]
- 20. Solem C., Defoor E., Jensen P. R., Martinussen J. 2008. Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis. Appl. Environ. Microbiol. 74:4772–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Alen-Boerrigter I. J., Baankreis R., de Vos W. M. 1991. Characterization and overexpression of the Lactococcus lactis pepN gene and localization of its product, aminopeptidase N. Appl. Environ. Microbiol. 57:2555–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van de Guchte M., Daly C., Fitzgerald G. F., Arendt E. K. 1994. Identification of int and attP on the genome of lactococcal bacteriophage Tuc2009 and their use for site-specific plasmid integration in the chromosome of Tuc2009-resistant Lactococcus lactis MG1363. Appl. Environ. Microbiol. 60:2324–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wegmann U., et al. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]