Abstract
The organism Acinetobacter sp. RKJ12 is capable of utilizing 2-chloro-4-nitrobenzoic acid (2C4NBA) as a sole source of carbon, nitrogen, and energy. In the degradation of 2C4NBA by strain RKJ12, various metabolites were isolated and identified by a combination of chromatographic, spectroscopic, and enzymatic activities, revealing a novel assimilation pathway involving both oxidative and reductive catabolic mechanisms. The metabolism of 2C4NBA was initiated by oxidative ortho dehalogenation, leading to the formation of 2-hydroxy-4-nitrobenzoic acid (2H4NBA), which subsequently was metabolized into 2,4-dihydroxybenzoic acid (2,4-DHBA) by a mono-oxygenase with the concomitant release of chloride and nitrite ions. Stoichiometric analysis indicated the consumption of 1 mol O2 per conversion of 2C4NBA to 2,4-DHBA, ruling out the possibility of two oxidative reactions. Experiments with labeled H218O and 18O2 indicated the involvement of mono-oxygenase-catalyzed initial hydrolytic dechlorination and oxidative denitration mechanisms. The further degradation of 2,4-DHBA then proceeds via reductive dehydroxylation involving the formation of salicylic acid. In the lower pathway, the organism transformed salicylic acid into catechol, which was mineralized by the ortho ring cleavage catechol-1,2-dioxygenase to cis, cis-muconic acid, ultimately forming tricarboxylic acid cycle intermediates. Furthermore, the studies carried out on a 2C4NBA− derivative and a 2C4NBA+ transconjugant demonstrated that the catabolic genes for the 2C4NBA degradation pathway possibly reside on the ∼55-kb transmissible plasmid present in RKJ12.
INTRODUCTION
Chloronitroaromatic compounds (CNACs) have been distributed in the environment largely due to their application in agriculture as broad-specificity pesticides, fungicides, and seed-dressing chemicals. Chlorinated nitroaromatic compounds also have been used as precursors for the chemical synthesis of industrial chemicals, dyes, pigments, pharmaceuticals, rubber chemicals, etc. (45, 50, 51, 54). The worldwide production of 2-chloro-4-nitrobenzoic acid (2C4NBA) for use as an intermediate in the industrial production of dyes and as a raw material in the production of drugs and pesticides is ∼175,000 tons (7).
The International Agency Research on Cancer (IARC) reported persistent toxic concentrations of 2C4NBA along with other CNACs worldwide, including in the United States, China, Germany, Italy, the Netherlands, and Belgium, at a level of 0.5 ng/liter to 37 μg/liter (21a). High concentrations of this compound were detected in the groundwater samples collected from Degrémont, France (44), treated effluents from the Advanced Wastewater Treatment Plant in Orange County, CA (21), and wastewater near the CNAC manufacturing plants in India; concentrations ranged from 0.97 to 1,800 mg/liter (42). Some of the studies pertaining to the assessment of the toxicity of CNACs revealed that 2C4NBA causes methemoglobinemia, reticulocytosis, and Heinz bodies in humans and reportedly is weakly mutagenic, teratogenic, and carcinogenic (38, 46). The U.S. Environmental Protection Agency and European Economic Community also have declared that 2C4NBA is one of the priority pollutants that is particularly harmful and persistent in the environment. Therefore, the waste management and detoxification of this compound is important for protecting the environment and human health.
Nitroaromatic compounds (NACs) and CNACs are structural analogs. The microbial degradation of NACs has been extensively investigated (15, 32, 40, 41). Although structurally related to the NACs, the CNACs are extremely resistant (12, 31, 49, 50, 51) to microbial degradation due to the simultaneous existence of two strong electron-withdrawing chlorine and nitro groups; thus, the knowledge of its microbial degradation is very limited. Most investigations of the degradation of CNACs have revealed cometabolic transformations. In one such study, the degradation of chloronitrobenzenes was carried out by the coculture of two microbial strains, Pseudomonas putida and Rhodococcus sp. (31). In another study, Bruhn et al. (5) constructed 4-chloro-2-nitrophenol (4C2NP) assimilatory bacteria by transferring the plasmid-carried 4-chlorocatechol-degrading genes from Ralstonia eutropha JMP134 into Pseudomonas sp. strain N31. Before the conjugation experiments, the recipient strain was able to remove nitrite from 4C2NP but failed to degrade 4-chlorocatechol. Some studies have reported CNAC degradation by mixed cultures. Beunink and Rehm (3) reported the degradation of 4C2NP by mixed culture in a coupled anaerobic-aerobic process. In contrast, very few reports are available for the microbial degradation of CNACs by a single bacterial isolate(s) (12, 25, 27, 28, 37). Based upon the intermediates identified from the reports cited above, it could be argued that the reductive dehalogenization (12, 31, 51) or partial reduction of nitro groups (29, 49, 50) is involved in the initial steps of CNAC degradation. However, the present study describes the involvement of the oxidative dehalogenation-initiated degradation of 2C4NBA by a newly isolated Acinetobacter strain. 2C4NBA subsequently is metabolized by strain RKJ12 into 2-hydroxy-4-nitrobenzoic acid (2H4NBA) and 2,4-dihydroxybenzoic acid (2,4-DHBA) by a mono-oxygenase, which is further degraded by an ortho ring cleavage dioxygenase, ultimately leading to tricarboxylic acid (TCA) cycle intermediates via the formation of salicylic acid and catechol. The present report also demonstrates that the genes for the whole 2C4NBA degradation pathway probably are located on the transmissible plasmid.
MATERIALS AND METHODS
Chemicals.
2C4NBA, 2H4NBA, 2,4-DHBA, salicylic acid, catechol, sodium succinate, and universal primers were purchased from Sigma. Labeled 2C4NBA and 2,4-DHBA-coenzyme A (CoA) were procured from American Radiolabeled Chemicals, St. Louis, MO. H218O (81% 18O) and 18O2 (98% 18O) were from Cambridge Isotope Laboratories. NAD+, NADH, and NADPH were obtained from Boehringer, Germany. All other chemicals used were of the highest purity available locally.
Isolation and characterization of the bacterial strain.
The test organism used in this study, RKJ12, was isolated from CNAC-contaminated soil in India by an enrichment culture technique described by Prakash et al. (34). The morphological features were determined and conventional biochemical tests performed using standard methods as described by Smibert and Krieg (39). The 16S rRNA gene was amplified using universal bacterium-specific primers 27F and 1492R as described by Goodwin et al. (13), and the reaction product was analyzed on an ABI PRISM 377 automated DNA sequencer (Perkin-Elmer Applied Biosystems). The 16S rRNA gene sequence of the new isolate was compared to those in the EMBL, GenBank, and DDBJ databases using BLAST version 2.2.12 from the National Center for Biotechnology Information (NCBI) (1).
Bacterial growth conditions.
Cells of the strain RKJ12 were grown aerobically in mineral salts medium (MSM) (34) that was supplemented with a 20 mM final concentration of 2C4NBA. Whenever needed, 10 mM sodium succinate (SS) was used to supplement the growth of the microorganism. Due to the relatively low water solubility of 2C4NBA, 4.5 g/liter (∼20 mM) of 2C4NBA was incubated with MSM (pH 7.2) at 30°C with shaking at 150 rpm for 24 h as described by Katsivela et al. (25). The mineral salts medium containing dissolved 2C4NBA (20 mM) was filtered before inoculation and used to determine the kinetics of 2C4NBA degradation at 30°C with aeration. Bacterial growth was determined by monitoring the optical density at 600 nm (OD600) spectrophotometrically (Perkin-Elmer). To investigate the ability of strain RKJ12 to utilize the pathway intermediates, cells were cultivated in MSM in the presence of various metabolites (10 to 20 mM) singly as the sole carbon source under identical conditions.
Isolation of metabolites.
After incubation, the spent broth of the cell culture was centrifuged (8,000 × g for 10 min), and the supernatant was extracted with ethyl acetate (acidic and neutral) as described by Ghosh et al. (12). The concentrated residue was resuspended in a small volume (approximately 100 μl) of methanol or ethyl acetate as needed prior to analysis.
Chloride release, nitrite release, and Rothera test.
The release of chloride ions was detected and quantified by following a colorimetric method as described by Bergmann and Sainik (2). Similarly, nitrite ions were detected spectrophotometrically in the culture supernatants using the method described previously by White et al. (47). For the detection of the ortho or meta cleavage product, a Rothera test was performed as described by Holding and Collee (20).
Chemical analyses.
Metabolites obtained from resting-cell studies were resolved by HPLC analysis using a Waters 600 model (Wein, Austria) equipped with a Waters 996 photodiode array detector operating at 215 to 600 nm and C18 reversed-phase (particle size, 5 μm; 4.6 by 250 mm; Waters Spherisorb ODS2) column. The mobile phase consisted of 1% acetic acid in methanol and 1% acetic acid in water (35:65) with an isocratic flow rate of 1.0 ml min−1. Quantitative analysis was performed by HPLC using a standard curve(s) constructed with an authentic standard(s).
The gas chromatography (GC) study was carried out using an AutoSystem-XL gas chromatograph equipped with a DB-1 (100% dimethyl polysiloxane) capillary column (30 m by 0.25 mm) and a flame ionization detector (Perkin Elmer, MA). Temperatures for injector, oven, and detector were kept constant at 280, 200, and 250°C, respectively.
GC-mass spectrometry (GC-MS) analysis was carried out using a GC-MS-QP5000 instrument (Shimadzu, Tokyo, Japan) equipped with a quadrupole mass filter and a DB-1 capillary column with ionization of 70 eV, scan interval of 1.5 s, and mass range of 40 to 700 Da.
Mass spectrometry-electrospray ionization (MS-ESI) analysis was performed on a Finnigan LTQ-FT mass spectrometer with a range of 50 to 4,000 Da (Thermo Electron Corporation, Waltham, MA) equipped with an online mass detection assembly from a linear ion trap and an ion cyclotron (7 T magnet) with Fourier-transformed ion cyclotron resonance mass spectrometry.
1H nuclear magnetic resonance (1H-NMR) and 13C-NMR spectroscopy were performed using a Bruker Avance DRX500 spectrometer (Bruker, Germany), operating at 500 and 125 MHz for 1H and 13C nuclei, respectively. Experiments were performed in deuterated dimethylsulfoxide at 20°C in 5-mm NMR tubes. Chemical shifts (δ) in parts per million (ppm) refer to tetramethylsilane [Si(CH3)4] as the internal standard.
Fourier transform infrared (FT-IR) spectra were obtained in the range of 4,000 to 450 cm−1 by using a Perkin-Elmer Paragon 500 FT-IR spectrophotometer. Solid samples dispersed in KBr pellets were used for FT-IR analysis.
Preparation of resting cells.
Resting-cell studies with strain RKJ12 were carried out as described by Takeo et al. (43) with minor modifications. Briefly, the cells were grown on 10 mM sodium succinate until mid-log phase (OD600 of approximately 1.2) and induced with 2C4NBA. After 2 h, induced cells were harvested, washed twice with ice-cold 10 mM phosphate buffer (pH 7.2), and resuspended in 10 ml of the same buffer. The test compound was added at a concentration of 2 mM, and 1.0 ml of samples was removed at different time intervals. The collected samples were immediately centrifuged at 15,000 rpm at 4°C for 2 min and analyzed by HPLC.
14C trapping studies.
14C trapping experiments were performed as described by Jain et al. (22) with little modification. Unlabeled 2H4NBA or 2-chloro-4-hydroxybenzoic acid (2C4HBA) (final concentration of each compound, 2 mM) was added to the separate samples of washed 2C4NBA-grown cells (A600 of 1.2) along with >99% pure 2C4NBA (0.2 mM) (specific activity, 0.30 μCi; American Radiolabeled Chemicals, St. Louis, MO). The samples were analyzed by HPLC, and radioactivities of solvent-extracted metabolites were determined with a scintillation counter (Beckman LS6800 scintillation counter).
Preparation of cell extract.
Cell extract was prepared by cell lysis using ultrasonication (Vibra-Cell VCX 130; Newtown, CT) as described by Hofmann et al. (19).
Partial purification of 2C4NBA mono-oxygenase.
The partial purification of 2C4NBA mono-oxygenase was carried out at 4°C as described previously by Kadiyala and Spain (24).
Enzyme assays.
To elucidate the biochemical characterization of 2C4NBA degradation, different enzymatic assays were performed with cell extracts of RKJ12. The protein estimation was carried out by the method of Bradford (4). The enzymatic transformations were carried out by recording cell extract-catalyzed changes in UV-visible spectra over a wavelength of 210 to 450 nm using a Lambda EZ 201 UV/Vis (Perkin Elmer, Waltham, MA).
The activity of 2C4NBA mono-oxygenase was determined by measuring the formation of 2H4NBA (maximum absorbance [Amax], 280 nm) and 2,4-DHBA (Amax, 317 nm) as described by Green and Shangguan (14). Further, the reaction products formed were quantified and identified by HPLC and GC-MS analysis. The stoichiometric O2 uptake with respect to NADPH oxidation was measured simultaneously on aliquots from the same reaction mixture placed in a Clark oxygen electrode cell, and quartz cuvettes were positioned in a Shimadzu Model 160 double beam spectrometer (Shimadzu, Kyoto, Japan) set to track NADPH oxidation at 340 nm.
The labeling experiments with H218O (81% 18O) or 18O2 (98% 18O) (Cambridge Isotope Laboratories) were carried out as described previously by Schenk et al. (36).
The activities of 2,4-dihydroxybenzoyl–CoA ligase and reductase were measured by the quantification of substrate and product by HPLC and electrospray ionization-mass spectrophotometry (ESI-MS) (resolution, 100,000; mass, 50 to 4,000 Da) as described previously by Geissler et al. (10). The radioactive reductive dehydroxylation assays were carried out as described by Laempe et al. (26) using labeled [U-14C]2,4-DHBA-CoA and [18O-4OH]2,4-DHBA-CoA. The enzymatic activity of salicylate hydroxylase was measured as described by White-Stevens and Kamin (48) by measuring the formation of catechol at an Amax of 276 nm. Finally, the activity of catechol-1,2-dioxygenase was assayed by measuring the rate of formation of cis,cis-muconate at an Amax of 260 nm as described by Hegeman (17).
Plasmid characterization.
Attempts were made to obtain derivatives which were incapable of utilizing 2C4NBA as the sole source of carbon by treatment with mitomycin C as described previously by Prakash et al. (34) to confirm the possible involvement of plasmid in 2C4NBA degradation. The plasmid DNA isolation was carried out by the method of O' Sullivan and Klaenhammer (30). The plasmid-curing experiment was carried out by growing the cells of RKJ12 for three cycles in the presence of mitomycin C (2.5 μg/ml). The dilutions were plated out on nutrient agar, and colonies were picked and patched on MSM agar containing 2C4NBA as the sole carbon source to score 2C4NBA− derivatives. The conjugation experiments were performed by triparental mating to transfer the 2C4NBA+ phenotype (2C4NBA+, Strs) to the recipient strain Pseudomonas putida PaW340 (2C4NBA− Strr, Trp−) using pRK2013 (in Escherichia coli HB 101) as a helper plasmid. Following the triparental plate mating, the transconjugants were obtained on the selective medium, which contained 2C4NBA, streptomycin (200 μg/ml), and tryptophan (20 μg/ml).
Nucleotide sequence accession number.
The DNA sequence reported here is available in GenBank under accession number HM568506.
RESULTS AND DISCUSSION
Using an enrichment culture technique, a 2C4NBA-degrading bacterium, strain RKJ12, was isolated from CNAC-contaminated soil from India containing residues of hexachlorocyclohexane (HCH) and hexachlorobenzene (HCB) pesticides. Results based on conventional biochemical tests and 16S rRNA sequencing showed that strain RKJ12 belongs to the genus Acinetobacter. Furthermore, based upon polyphasic taxonomic investigations, strain RKJ12 represents a new species of the genus Acinetobacter, designated Acinetobacter sp. RKJ12.
Growth profiles using 2C4NBA indicated that the cells of strain RKJ12 utilized this substrate as the sole source of carbon, nitrogen, and energy at a concentration of 20 mM; chloride ions were released initially into the culture medium with the concomitant degradation of 2C4NBA. However, the release of nitrite ions was observed with a time difference of about 8 h between the initiation of the release of chloride and nitrite ions. The sequential removal of both ions from the medium suggested that the pathway was initiated by mono-oxygenase mechanisms. Furthermore, the observation of the initial release of chloride ions in the medium clearly indicated that the degradation of 2C4NBA was initiated by an oxidative dechlorination reaction. The results from growth characteristics and kinetics of degradation revealed that 20 mM 2C4NBA was completely degraded within 28 to 32 h of incubation, wherein the consumption of substrate also was correlated with an increase in optical density (Fig. 1). RKJ12 also was capable of utilizing pathway intermediates such as 2H4NBA, 2,4-DHBA, salicylic acid, and catechol individually as sole sources of carbon and energy. However, the analogous substrates 2C4NBA, 2,3- dihydroxybenzoic acid, 2,5-dihydroxybenzoic acid, m-salicylic acid, and resorcinol (1,3-dihydroxybenzene) did not support growth, indicating that none of these compounds were involved in the 2C4NBA degradation pathway.
Fig. 1.
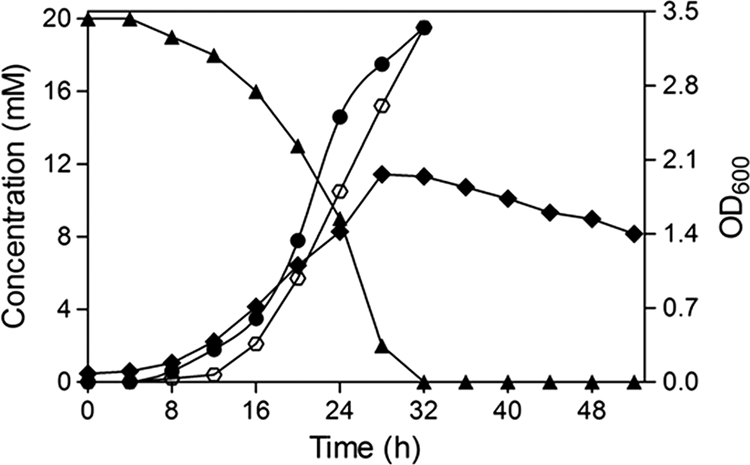
Growth profile and degradation of 2C4NBA by Acinetobacter sp. strain RKJ12 when grown on 2C4NBA as the sole source of carbon, nitrogen, and energy; strain RKJ12 metabolized 2C4NBA with the concomitant release of chloride and nitrite. ♦, Optical density of the culture; ▴, depletion of 2C4NBA; •, chloride release; ⎔, nitrite release. The values represented are averages from triplicates.
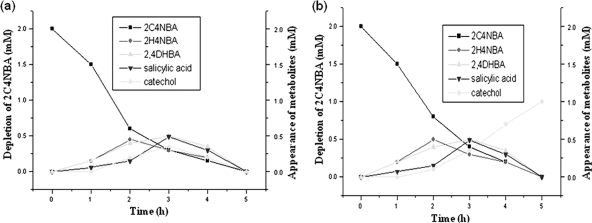
To characterize the metabolites of the 2C4NBA degradation pathway, time courses of resting cells were carried out on the ethyl acetate-extracted, different-time-interval samples using HPLC and GC-MS. Sample collected at 1 h demonstrated the appearance of metabolite I with an HPLC retention time (10.94 min), UV-visible (UV-vis) spectra (Amax, 281 nm), and molecular ion [M+] peak (at m/z 183) that corresponded well with those of authentic 2H4NBA. The appearance of 2H4NBA early in the induction period with the simultaneous release of chloride ions again suggested that the pathway was initiated by oxidative dehalogenation. Further, the samples collected at 3 and 4 h of incubation indicated the gradual depletion of metabolite I with the concomitant stoichiometric formation of three metabolites nearby. Metabolite II (HPLC retention time, 8.95 min; UV-vis spectra Amax, 315 nm; molecular ion [M+] peak, m/z 154) was identical to authentic 2,4-DHBA. Metabolites III and IV (detected by HPLC at 4.10 and 2.12 min, respectively, UV-vis spectra [Amax] of 295 and 257 nm, respectively, and with molecular ions at m/z 138 and 110, respectively) closely matched authentic salicylic acid and catechol, respectively (Fig. 2a). The mass fragmentation patterns of each of the metabolites also corresponded well with authentic standards. Furthermore, no intermediates were detected in samples collected at 5 h, suggesting the complete utilization of these compounds. These results therefore indicated that 2H4NBA, 2,4-DHBA, salicylic acid, and catechol were intermediates in 2C4NBA degradation.
Fig. 2.
(a) Quantification of 2C4NBA depletion and formation of intermediates by Acinetobacter sp. RKJ12 by HPLC analysis. 2C4NBA depletion (▪) and the formation of 2H4NBA (♦), 2,4 DHBA (▴), salicylic acid (▾), and (♦) catechol are shown. (b) Quantification of 2C4NBA and depletion and formation of intermediates by Acinetobacter sp. RKJ12 by HPLC analysis in the presence of 2,2-dipyridyl, an iron-chelating ring cleavage inhibitor. 2C4NBA depletion (▪) and the formation of 2H4NBA (•), 2,4-DHBA (▴), salicylic acid (▾), and catechol (♦) are shown.
The same time-course experiment was carried out with 2,2-dipyridyl, an iron chelator. It inhibits certain aromatic ring cleavage enzymes that require ferrous ion for their activities (4). As expected, the stoichiometric conversion of 2C4NBA into respective intermediates was not apparent, and the maximum accumulation of a single metabolite (catechol; retention time, 5.36 min; m/z 110) was exhibited even after 5 h of incubation (Fig. 2b) The accumulation of a single metabolite when 2,2-dipyridyl was incorporated into the medium indicated that catechol was the substrate for ring cleavage in the 2C4NBA degradation pathway.
To further ensure that 2H4NBA was produced from 2C4NBA by initial oxidative dechlorination reactions, two 14C trapping experiments were performed. When unlabeled 2H4NBA or 2C4HBA was included in such experiments as an isotope trap, the 14C label rapidly accumulated from 14C-labeled 2C4NBA in the 2H4NBA pool. After 15 min, 90% of the 14C was trapped into 2H4NBA. However, only 2% was recovered as 2C4HBA even after 20 min. The nearly stoichiometric accumulation of the radiolabeled 2H4NBA pool clearly shows that 2H4NBA is an oxidative dechlorinated metabolite in the 2C4NBA degradation pathway.
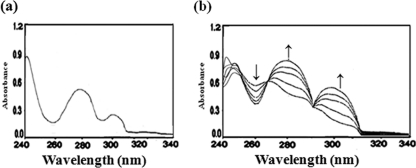
To further strengthen the results of the biochemical characterization of 2C4NBA degradation, different enzymatic assays were carried out with cell extracts of RKJ12. The positive activity of 2C4NBA mono-oxygenase (2.0 U mg−1) was measured by a decrease in the absorbance of 2C4NBA (Amax, 262 nm) with a concomitant increase in the absorption (Amax, 280 and 317 nm) corresponding to the absorbance of 2H4NBA and 2,4-DHBA, respectively (Fig. 3). Further, 2H4NBA (m/z 183) and 2,4-DHBA (m/z 154) were identified as reaction products. The formation of reaction products with stoichiometric release and chloride and nitrite ions clearly suggested that the mono-oxygenase initiated dechlorination and denitration reactions in 2C4NBA degradation.
Fig. 3.
Enzyme activity of 2C4NBA mono-oxygenase with the extract of 2C4NBA-induced cells of RKJ12. (a) Activity of 2C4NBA mono-oxygenase in negative-control sodium succinate-grown cells. (b) Depletion of 2C4NBA (at 262 nm; ↓) and formation of 2H4NBA and 2,4-DHBA (at 282 nm and 317 nm, respectively; ↑).
Furthermore, the stoichiometric NADPH and oxygen consumption analysis revealed that 2 mol NADPH and 1 mol O2 were consumed for every 1 mol 2C4NBA hydroxylation (conversion of 1 mol 2C4NBA into 2,4-DHBA). However, 1 mol NADPH and 1 mol O2 were required for the hydroxylation of 2H4NBA, indicating mono-oxygenase-catalyzed dechlorination and denitration reactions in 2C4NBA degradation with at least one oxygenolytic step.
The enzyme that catalyzed the two sequential mono-oxygenations of 2C4NBA was partially purified and separated into two components (A and B) by anion-exchange chromatography and size-exclusion chromatography. Fractions containing component A catalyze the NAD(P)H-dependent reduction of 2,6-dichlorophenolindophenol but not cytochrome c, and this component was identified as a reductase. The NAD(P)H-2,6-dichlorophenolindophenol reduction activity coeluted with component A using Sephacryl S-300 columns. The fraction with an elution volume of 70 ml showed maximum chloride and nitrite release activities (when combined with component B) and also maximum reductase activity (3.0 μmol/min/mg protein). Further, component A exhibits a flavoprotein spectrum, with a peak of 445 nm, and the addition of NADPH did not reduce the flavoprotein absorption at 445 nm. The results suggest that component A is a flavoprotein. However, component B showed no reductase activity with electron acceptors tested and appears to be an oxygenase. Cytochrome P-450 inhibitors at 0.5 mM had very little effect (<5 to 6% inhibition) on the chloride and nitrite release from 2C4NBA or 2H4NBA by reconstituted mixtures of components A and B. However, methimazole, an inhibitor of flavin mono-oxygenase at 0.5 mM, greatly inhibited (∼60% inhibition) the enzymatic release of chloride and nitrite ions from the substrates. This observation also suggests that the enzyme system contains a flavoprotein reductase. Both components are essential for two mono-oxygenation reactions that catalyze sequential NADPH-dependent dechlorination and denitration of 2C4NBA into 2,4-DHBA. Therefore, based upon this preliminary characterization, the enzyme is an oxidoreductase mono-oxygenase that catalyzes the transformation of 2C4NBA into 2,4-DHBA by mono-oxygenation. Further, the characterization of this enzyme currently is under investigation.
The incubation of enzyme extract with 2C4NBA in an 18O2-enriched atmosphere yielded a product whose retention time and mass spectrum were identical to those of 2H4NBA. However, when enzyme extract was incubated with H218O and 2C4NBA, the product yielded had the same retention time as 2H4NBA but a mass spectrum with major peaks that were 2 mass units larger than those of untreated 2H4NBA, which is consistent with the incorporation of one atom of 18O into 2H4NBA, indicating that mono-oxygenase catalyzed the initial hydrolytic dechlorination reaction (6, 36, 52, 53). Likewise, the incubation of enzyme extract with 2H4NBA in an atmosphere of 18O2 or H218O separately yielded a product with molecular ion peaks and fragments 2 mass units larger than those of untreated 2H4NBA, which is consistent with the incorporation of one atom of 18O into 2,4-DHBA under an atmosphere enriched with 18O2 but not under an atmosphere enriched with H218O, suggesting that the denitration was an oxygenolytic reaction (23, 52, 53).
The enzymatic activity of 2,4-dihydroxybenzoyl–CoA ligase (2.0 U mg−1) was measured by HPLC and ESI-MS analyses. The depletion of 2,4-dihydroxybenzoic acid (retention time, 9.14 min; m/z 154) with the concomitant formation of 2,4-dihydroxybenzoyl–CoA (retention time, 9.0 min; m/z 1045.151) indicated the positive activity of 2,4-dihydroxybenzoyl–CoA ligase. Furthermore, the formation of salicylic acid-CoA (retention time, 4.3 min; m/z 908.173) was detected immediately upon the addition of the electron donor (NADH plus H+), suggesting the possible activity of 2,4-dihydroxybenzoyl–CoA reductase.
To further examine the reductive dehydroxylation reaction, radioactive assays were carried out using labeled [U-14C]2,4-dihydroxybenoic acid–CoA (specific activity, 0.70 μCi) and [18O-4OH]2,4-dihydroxybenoic acid–CoA (specific activity, 0.85 μCi). The consumption of [U-14C]2,4-dihydroxybenzoic acid–CoA and the formation of [14C]salicylic acid-CoA (retention time, 4.6 min; m/z 910.175) clearly indicated the positive activity of 2,4-dihydroxybenzoic acid reductase in strain RKJ12. Similarly, in the [18O-4OH]2,4-dihydroxybenzoic acid–CoA assays, [18O-4OH]2,4-dihydroxybenzoic acid–CoA was reduced to salicylic acid-CoA (retention time, 4.3 min; m/z 908.173) by eliminating the labeled 4-hydroxyl group reductively. The observed reductive dehydroxylation reaction under aerobic conditions is in close agreement with earlier reports. Crawford (8) and Fairley et al. (9) investigated the unusual aerobic reductive dehydroxylation of 2HBA to BA. Reductive dehydroxylation is a common reaction in anaerobic bacteria, and the enzyme responsible for this reaction, benzoyl-CoA reductase, may be oxygen sensitive (16). However, Gescher et al. (11) and Poole (33) evaluated the effect of oxygen on purified benzoyl-CoA reductase and reported that some of its subunits partially work under aerobic conditions. The rigorous confirmation of this interesting reaction must await the results of enzyme purification experiments, which are currently in progress. To further support the reductive dehydroxylation reaction in RKJ12, experiments with FT-IR, 1H-NMR, and 13C-NMR were devised.
The activity of salicylate hydroxylase (1.6 U mg−1) was measured by the depletion of salicylic acid (Amax, 297 nm) with concomitant increase in the formation of 0.17 mM catechol (Amax, 276 nm; m/z 110), demonstrating the positive activity of salicylate hydroxylase. Finally, the activity of ring cleavage catechol 1,2-dioxygenase (1.8 U mg−1) was measured by the formation of cis,cis-muconate (Amax, 260 nm), indicating the mode of ortho ring cleavage in 2C4NBA degradation. The mode of ortho ring cleavage in catechol also was confirmed by a Rothera test. The 2C4NBA-grown whole-cell lysate of RKJ12 was incubated with catechol. The development of red-brown color with the formation of cis,cis-muconate in the reaction mixture suggested the mode of ortho ring cleavage in the 2C4NBA degradation pathway (18).
The results described above, particularly the reductive dehydroxylation of 2,4-DHBA into salicylic acid, were additionally supported by FT-IR, 1H-NMR, and 13C-NMR spectral studies. The cells of RKJ12 were grown on 2C4NBA for 20 h, and the compounds present in the supernatant fluids were extracted and analyzed by thin-layer chromatography (TLC). The compounds which had the same Rf values as those of 2,4-DHBA and salicylic acid were further purified from the silica gel preparative TLC plate 60F254 for spectral studies.
The FT-IR spectra of hydroxyl absorption bands spread from 3,514 to 2,138 cm−1, with the highest absorption intensity peak at 3,375 cm−1. However, the carbonyl absorption (COOH) band was reflected at 1,650 cm−1, corresponding to the FT-IR spectrum of authentic 2,4-DHBA. Thus, the shifting of the hydroxyl absorption band toward lower wave numbers at 3,248 cm−1 and an increase in the carbonyl (COOH) absorption band toward higher wave numbers at 1,665 cm−1 due to the dehydroxylation of C-4 OH indicated the formation of salicylic acid from 2,4-DHBA by reductive dehydroxylation.
The 1H-NMR and 13C-NMR studies also were carried out for the final confirmation of the dehydroxylation reaction by RKJ12. The chemical shifts of the carbonyl (COOH) and two hydroxyl groups were observed at 10.78 ppm. One H-3 singlet at 7.20 ppm and two doublets at 6.89 and 6.69 ppm also were detected at positions H-5 and H-6, respectively, corresponding to the 1H-NMR spectra of the authentic 2,4-DHBA. The formation of two broadened singlets at 13.44 and 11.52 ppm for one carbonyl and hydroxyl (C-2 OH), respectively, two H-3 and H-6 doublets at 6.96 and 7.82 ppm, respectively, and two H-4 and H-5 triplets at 7.52 and 6.92 ppm, respectively, suggested the transformation of 2,4-DHBA into salicylic acid by reductive (C-4 OH) dehydroxylation.
Furthermore, the formation of 13C-NMR carbon signal peaks at C-1 (δ 120.81), C-2 (δ 149.30), and C-4 (δ 157.50), respectively, represents the presence of one carbonyl group (COOH) at C-1 and two hydroxyl groups at C-2 and C-4, corresponding to 13C-NMR spectra of authentic 2,4-DHBA. The decrease in the carbon signal at C-4 (δ 157.50 to 136.80) and C-1 (δ 120.81 to 115.13) and the increase in the carbon signals at C-2 (δ 149.30 to 163.31), C-3 (δ 111.61 to 119.22), C-5 (δ 115.86 to 118.30), and C-6 (δ 125.92 to 132.41) indicated (C-4 OH) the dehydroxylation of 2,4-DHBA into salicylic acid. The 1H-NMR and 13C-NMR spectra of the compound also were comparable with those of authentic salicylic acid. Therefore, based upon these results, strain RKJ12 transformed 2,4-DHBA into salicylic acid by reductive dehydroxylation mechanisms.
Plasmid characterization.
The involvement of a catabolic plasmid(s) in the degradation of organic compounds has been shown previously by Prakash et al. and Rani et al. (34, 35). Recently, Wu et al. (49) reported the possible involvement of an 89-kb plasmid in 4-chloronitrobenzene degradation. Therefore, to check if a plasmid is present in strain RKJ12 that could be involved in 2C4NBA degradation, attempts were made to isolate the plasmid from this strain. A plasmid of ∼55 kb in size (Fig. 4) was found to be present in RKJ12 using E. coli strain V517 as a standard marker for plasmids (32). Attempts were made to obtain 2C4NBA− derivatives to determine the role of this plasmid in strain RKJ12. There was an approximately 3% loss of the 2C4NBA+ phenotype when mitomycin C was present during growth. When such mitomycin C-derived 2C4NBA− derivatives were checked for the presence of a plasmid, the plasmid of ∼55 kb was absent (Fig. 4). The plasmid-cured strains also were unable to grow on 2C4NBA as the sole carbon source, indicating the possible involvement of this plasmid in 2C4NBA degradation. Conjugation studies also were carried out to transfer the 2C4NBA+ phenotype (2C4NBA+, Strs) to the plasmid-free strain Pseudomonas putida PaW 340 (2C4NBA−, Strr, Trp−). Following triparental plate matings, transconjugants (2C4NBA+, Strr, Trp−) were obtained at a frequency of 10−6 on selective medium. Plasmid DNA was isolated from such transconjugants, and they carried the same plasmid (∼55 kb) that was present in the wild-type strain RKJ12 (Fig. 4). Such transconjugants also were capable of utilizing 2C4NBA and their metabolites as sole carbon sources and released chloride and nitrite ions from 2C4NBA, as in the case of wild-type strain RKJ12. The GC and HPLC analysis results were comparable to those obtained with RKJ12 (data not shown). These results demonstrate that the degradation of 2C4NBA may be plasmid encoded, and the genes for its degradation reside on the ∼55-kb transmissible plasmid.
Fig. 4.

Agarose gel electrophoresis of plasmid isolated from Acinetobacter sp. RKJ12. Lane M, E. coli V517 as a standard plasmid size marker; A and B, wild-type strain RKJ 200; C and D, mitomycin C-treated CNBA− derivatives; and E and F, CNBA+ transconjugants. Chr, chromosomal DNA.
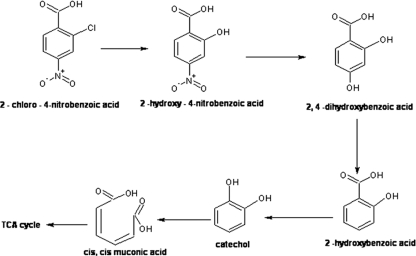
We believe that this account of the degradation of 2C4NBA by strain RKJ12 is the first comprehensive report involving both oxidative and reductive mechanisms. The organism initiates the metabolism of 2C4NBA by a mono-oxygenase-catalyzed oxidative dechlorination pathway, leading to the formation of 2H4NBA and 2,4-DHBA. The further degradation of 2C4NBA then proceeds via reductive dehydroxylation involving the formation of salicylic acid and catechol, ultimately forming TCA cycle intermediates by catechol-1,2-dioxygenase (Fig. 5). The organism RKJ12 metabolized 2C4NBA by an uncommon reductive dehydroxylation reaction, and the enzyme responsible for this conversion, benzoyl-CoA reductase, may be oxygen sensitive (16). However, using HPLC, MS-ESI, FT-IR, NMR, and radioactive assays, convincing evidence was obtained that 2,4-DHBA was transformed into salicylic acid by a reductive dehydroxylation mechanism. Considering that none of the Acinetobacter strains have been reported to possess the genetic makeup for ring-hydroxylating mono-oxygenase and reductive dehydroxylase, it is important to understand the genetic features of the organism RKJ12 in 2C4NBA degradation. Interestingly, strain RKJ12 harbors a catabolic plasmid probably containing genes for 2C4NBA assimilation. Therefore, such an organism(s) could prove useful for the decontamination of pollutants in the environment. Further investigations of the involvement of catabolic plasmid, cloning, and the characterization of novel catabolic genes and the purification of key enzymes will allow us to better understand this unique 2C4NBA degradation pathway in Acinetobacter strain RKJ12.
Fig. 5.
Proposed pathway for the degradation of 2C4NBA by Acinetobacter sp. RKJ12.
ACKNOWLEDGMENTS
The present study was supported by the Department of Biotechnology (DBT) and the Council of Scientific and Industrial Research (CSIR), India.
We thank Jim Spain for suggesting 14C trapping experiments. We also thank Avtar Singh, RSIC, Panjab University, Chandigarh, India, for carrying out the 1H-NMR, 13C-NMR, and IR analyses.
Footnotes
This article is dedicated to the memory of the late R. K. Jain.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Bergmann J. G., Sainik J. 1957. Determination of trace amounts of chlorine in naphtha. Anal. Chem. 29:241–243 [Google Scholar]
- 3. Beunink J., Rehm H. J. 1990. Coupled reductive and oxidative degradation of 4-chloro-2-nitrophenol by a co-immobilized mixed culture system. Appl. Microbiol. Biotechnol. 34:108–115 [DOI] [PubMed] [Google Scholar]
- 4. Bradford M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizating the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 5. Bruhn C., Bayly R. C., Knackmuss H. J. 1988. The in vivo construction of 4-chloro-2-nitrophenol assimilatory bacteria. Arch. Microbiol. 150:171–177 [Google Scholar]
- 6. Chae J. C., Kim Y., Kim Y. C., Zylstra G. J., Kim C. K. 2000. Genetics structure and functional implication of the fcb gene culture for hydrolytic dechlorination of 4-chlorobenzoate from Pseudomonas sp. DJ-12. Gene 258:109–116 [DOI] [PubMed] [Google Scholar]
- 7. Chemical Information Services 1994. Directory of world chemical producers 1995/96. Chemical Information Services, Oceanside, NY [Google Scholar]
- 8. Crawford R. L. 1976. Pathways of 4-hydroxybenzoate degrading among species of Bacillus. J. Bacteriol. 127:204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fairley D. J., et al. 2002. Aerobic metabolism of 4-hydroxybenzoic acid in Archaea via an unusual pathway involving an intramolecular migration (NIH shift). Appl. Environ. Microbiol. 68:6246–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geissler J. F., Harwood C. S., Gibson J. 1988. Purification and properties of benzoate-coenzyme A ligase: a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J. Bacteriol. 170:1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gescher J., Eisenreich W., Wörth J., Bacher A., Fuchs G. 2005. Aerobic benzoyl-CoA catabolic pathway in Azoarcus evansii: studies on the non-oxygenolytic ring cleavage enzyme. Mol. Microbiol. 56:1586–1600 [DOI] [PubMed] [Google Scholar]
- 12. Ghosh A., et al. 2010. Degradation of 4-nitrophenol, 2-chloro-4-nitrophenol, and 2,4-dinitrophenol by Rhodococcus imtechensis strain RKJ300. Environ. Sci. Technol. 44:1069–1077 [DOI] [PubMed] [Google Scholar]
- 13. Goodwin K. D., Tokaczyk R., Stephens F. C., Saltzman E. S. 2005. Degradation of toluene inhibition of methyl bromide biodegradation in seawater and isolation of a marine toluene oxidizer that degrades methyl bromide. Appl. Environ. Microbiol. 71:3495–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Green T. R., Shangguan X. 1993. Stoichiometry of O2 metabolism and NADPH oxidation of the cell-free latent oxidase reconstituted from cytosol and solubilized membrane from resting human neutrophils. J. Biol. Chem. 268:857–861 [PubMed] [Google Scholar]
- 15. Groenewegen P. E. J., et al. 1992. Novel degradative pathway of 4-nitrobenzoate in Comamonas acidovorans NBA-10. J. Gen. Microbiol. 138:1599–1605 [DOI] [PubMed] [Google Scholar]
- 16. Harwood C. S., Gibson J. 1997. Shedding light on anaerobic benzene ring degradation: a process unique to prokaryotes. J. Bacteriol. 179:301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hegeman G. D. 1966. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. 1. Synthesis of enzymes of the wild type. J. Bacteriol. 91:1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heinaru E., et al. 2001. Reversible accumulation of p-hydroxybenzoate and catechol determines the sequential decomposition of phenolic compounds in mixed substrate cultivations in pseudomonads. FEMS Microbiol. Ecol. 137:79–89 [Google Scholar]
- 19. Hofmann K. W., Knackmuss H. J., Heiss G. 2004. Nitrite elimination and hydrolytic ring cleavage in 2,4,6-trinitrophenol (picric acid) degradation. Appl. Environ. Microbiol. 70:2854–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holding A. J., Collee J. G. 1997. Routine biochemical tests. Methods Microbiol. 16:1–32 [Google Scholar]
- 21. Howard P. H., Santodonato J., Saxena J., Malling J., Greninger D. 1976. Investigation of selected environmental contaminants: nitroaromatics. US EPA report no. EP A-56012-76-010; US NTIS PB-275078 Environmental Protection Agency, Washington, DC [Google Scholar]
- 21a. International Agency for Research on Cancer 1997. Evaluation of carcinogenic risk of chlorinated aromatics to humans. IARC Monogr. Eval. Carcinog. Risks Hum. 69:33–6399336729 [Google Scholar]
- 22. Jain R. K., Dreisbach J. H., Spain J. C. 1994. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by Arthrobacter sp. Appl. Environ. Microbiol. 60:3030–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson G. R., Jain R. K., Spain J. C. 2002. Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol. 184:4219–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kadiyala V., Spain J. C. 1998. A two-component mono-oxygenase catalyzes both the hydroxylation of p-nitrophenol and oxidative release of nitrite from 4-nitrocatechol in Bacillus sphaericus JS905. Appl. Environ. Microbiol. 64:2479–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katsivela E., Wary V., Pipper D. H., Wittich R.-M. 1999. Initial reactions in the biodegradation of 1-chloro-4-nitrobenzene by a newly isolated bacterium, strain LW-1. Appl. Environ. Microbiol. 65:1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laempe D., Jahn M., Fuchs G. 1999. 6-Hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 263:420–429 [DOI] [PubMed] [Google Scholar]
- 27. Leisinger T., Bader R. 1993. Microbial dehalogenation of synthetic organohalogen compounds: hydrolytic dehalogenases. Chimia 47:116–121 [Google Scholar]
- 28. Lenke H., Knackmuss H.-J. 1996. Initial hydrogenation and extensive reduction of substituted 2,4-dinitrophenols. Appl. Environ. Microbiol. 62:784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Livingston A. G. 1993. A novel membrane bioreactor for detoxifying industrial waste-water. I. Biodegradation of 3-chloronitrobenzene in an industrially produced waste-water. Biotechnol. Bioeng. 41:927–936 [DOI] [PubMed] [Google Scholar]
- 30. O'Sullivan D. J., Klaenhammer T. R. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park H. S., Lim S. J., Chang Y. K., Linvingston A. G., Kim H. S. 1999. Degradation of chloronitrobenzenes by a coculture of Pseudomonas putida and a Rhodococcus sp. Appl. Environ. Microbiol. 65:1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park H. S., Kim H.-S. 2000. Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J. Bacteriol. 182:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poole S. T. 2003. Benzoyl-CoA reductase: a biological birch reduction. Ph.D. thesis University of Maryland, College Park, MD [Google Scholar]
- 34. Prakash D., Chauhan A., Jain R. K. 1996. Plasmid-encoded degradation of p-nitrophenol by Pseudomonas cepacia. Biochem. Biophys. Res. Commun. 224:375–381 [DOI] [PubMed] [Google Scholar]
- 35. Rani M., Prakash D., Sobti R. C., Jain R. K. 1996. Plasmid-mediated degradation of o-phthalate and salicylate by a Moraxella. Biochem. Biophys. Res. Commun. 220:377–381 [DOI] [PubMed] [Google Scholar]
- 36. Schenk T., Muller R., Lingens F. 1990. Mechanism of enzymatic dehalogenation of pentachlorophenol by Arthrobacter sp. strain ATCC 33790. J. Bacteriol. 172:7272–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schenzle A., Lenke H., Spain J. C., Knackmuss H-J. 1999. Chemoselective nitro group reduction and reductive dechlorination initiate degradation of 2-chloro-5-nitrophenol by Ralstonia eutropha JMP134. Appl. Environ. Microbiol. 65:2317–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shimizu M., Yasui T., Matsumoto N. 1983. Structural specificity of aromatic compounds with special reference to mutagenic activity in Salmonella typhimurium-a series of chloro- or fluro-nitrobenzene derivatives. Mutat. Res. 116:217–238 [DOI] [PubMed] [Google Scholar]
- 39. Smibert R. M., Krieg N. R. 1994. Phenotypic characterization, p. 611–654 In Gerhardt P., Murray R. G. E., Wood W. A., Krieg N. R. (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 40. Spain J. C., Gibson D. T. 1991. Pathway for biodegradation of para nitrophenol in a Moraxella sp. Appl. Environ. Microbiol. 57:812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Spanggord R. J., Spain J. C., Nishino S. F., Mortelmans K. E. 1991. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol. 57:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swaminathan K., Kondawar V. K., Chakrabarti T., Subrahmanyam P. V. R. 1987. Identification and quantification of organics in nitro aromatic manufacturing wastewaters. Indian Environ. Health 29:32–38 [Google Scholar]
- 43. Takeo M., et al. 2003. Cloning and characterization of a 4-nitrophenol hydroxylase gene cluster from Rhodococcus sp. PN1. J. Biosci. Bioeng. 95:139–145 [PubMed] [Google Scholar]
- 44. Van de Meent D., et al. 1986. Organic micropollutants in Dutch coastal waters. Water Sci. Tech. 18:73–81 [Google Scholar]
- 45. Van Zoest R., van Eck G. T. M. 1991. Occurrence and behaviour of several groups of organic micropollutants in the Scheldt estiary. Sci. Total Environ. 103:57–71 [Google Scholar]
- 46. Weisburger E. K., et al. 1978. Testing of twenty-one environmental aromatic amines or derivatives for long-term toxicity or carcinogenicity. Environ. Pathol. Toxicol. 2:325–356 [PubMed] [Google Scholar]
- 47. White G. F., Snape J. R., Nicklin S. 1996. Biodegradation of glycerol trinitrate and pentaerythritol tetranitrate by Agrobacterium radiobacter. Appl. Environ. Microbiol. 62:637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. White-Stevens R. H., Kamin H. 1972. Studies of a flavoprotein, salicylate hydroxylase. I. Preparation, properties and the uncoupling of oxygen reduction from hydroxylation. J. Biol. Chem. 247:2358–2370 [PubMed] [Google Scholar]
- 49. Wu J. F., Shen X. H., Zhou Y. G., Liu S. J. 2004. Characterization of p chloronitrobenzene-degrading Comamonas CNB-1 and its degradation of p-chloronitrobenzene. Acta Microbiol. Sin. 44:8–12 [Google Scholar]
- 50. Wu J. F., Sun C. W., Jiang C. Y., Liu Z. P., Liu S. J. 2005. A novel 2-aminophenol 1,6-dioxygenase involved in the degradation of p-chloronitrobenzene by Comamonas sp. strain CNB-1: purification, properties, genetic cloning and expression in Escherichia coli. Arch. Microbiol. 183:1–8 [DOI] [PubMed] [Google Scholar]
- 51. Wu J. F., et al. 2006. Novel partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1. Appl. Environ. Microbiol. 72:1759–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xiao Y., et al. 2006. Characterization of genes involved in the initial reactions of 4-chloronitrobenzene degradation in Pseudomonas putida ZWL73. Appl. Microbiol. Biotechnol. 73:166–171 [DOI] [PubMed] [Google Scholar]
- 53. Xun L., Webster C. M. 2004. A monooxygenase catalyzes sequential dechlorinations of 2,4,6-trichlorophenol by oxidative and hydrolytic reactions. J. Biol. Chem. 279:6696–6700 [DOI] [PubMed] [Google Scholar]
- 54. Yoshida T. 1993. Determination of p-chloronitrobenzene and its metabolites in urine by reversed-phase high-performance liquid chromatography. J. Chromatogr. 613:79–88 [DOI] [PubMed] [Google Scholar]