Abstract
The plant pathogen Ralstonia solanacearum, which causes bacterial wilt disease, is exposed to reactive oxygen species (ROS) during tomato infection and expresses diverse oxidative stress response (OSR) genes during midstage disease on tomato. The R. solanacearum genome predicts that the bacterium produces multiple and redundant ROS-scavenging enzymes but only one known oxidative stress response regulator, OxyR. An R. solanacearum oxyR mutant had no detectable catalase activity, did not grow in the presence of 250 μM hydrogen peroxide, and grew poorly in the oxidative environment of solid rich media. This phenotype was rescued by the addition of exogenous catalase, suggesting that oxyR is essential for the hydrogen peroxide stress response. Unexpectedly, the oxyR mutant strain grew better than the wild type in the presence of the superoxide generator paraquat. Gene expression studies indicated that katE, kaG, ahpC1, grxC, and oxyR itself were each differentially expressed in the oxyR mutant background and in response to hydrogen peroxide, suggesting that oxyR is necessary for hydrogen peroxide-inducible gene expression. Additional OSR genes were differentially regulated in response to hydrogen peroxide alone. The virulence of the oxyR mutant strain was significantly reduced in both tomato and tobacco host plants, demonstrating that R. solanacearum is exposed to inhibitory concentrations of ROS in planta and that OxyR-mediated responses to ROS during plant pathogenesis are important for R. solanacearum host adaptation and virulence.
INTRODUCTION
Plants produce reactive oxygen species (ROS) in response to invading pathogens, and an effective oxidative stress response (OSR) contributes to the fitness of phytopathogenic bacteria (18, 39, 40, 51). However, research on the role of regulators of the OSR in the virulence of plant-pathogenic bacteria is limited and contradictory. In Agrobacterium tumefaciens, oxyR is necessary for tumorigenesis (34), but Erwinia chrysanthemi does not need oxyR for soft-rot virulence (32). OxyR is a redox-sensing LysR family transcriptional regulator that has been well characterized in several bacteria (23, 43, 52). In the absence of oxidative stress, OxyR is reduced and acts as a repressor of several genes, including oxyR itself (47, 52). In the presence of hydrogen peroxide, the conserved cysteines of OxyR (C199 and C208, in Escherichia coli) form a disulfide bond that changes its conformation and converts OxyR into a transcriptional activator (52, 53). In E. coli, OxyR-regulated genes include catalase (kat), alkyl hydroperoxide reductase (ahp), glutaredoxin (grx), and glutathione reductase (gor) (53).
The plant-pathogenic bacterium Ralstonia solanacearum causes bacterial wilt disease on many economically important crops, including tomato (19). Multiple quantitative virulence factors contribute to the disease (11); however, little is known about how R. solanacearum adapts to its host environment, which is a critical prerequisite for pathogen success. An in vivo expression technology (IVET) screen performed on R. solanacearum during midphase tomato disease revealed that this bacterium encounters a stressful environment while in the host (7). Mutagenesis of stress response genes identified in this screen significantly decreased R. solanacearum's virulence on tomato (8).
Host plants generate ROS in the form of hydrogen peroxide in response to infection by R. solanacearum (13, 30), so this pathogen experiences oxidative stress during pathogenesis. The available R. solanacearum genomes predict multiple and redundant OSR genes (13), including an in planta-induced peroxidase, Bcp (7), and Dps (nonspecific DNA binding protein from starved cells), which is induced by tomato root exudates and is regulated by OxyR (9). However, mutants lacking either bcp or dps are only slightly reduced in virulence (9, 13). These findings suggest that either the concentrations of ROS in planta are sublethal or that the bacterium's remaining OSR genes compensate for the loss of Bcp and Dps.
We created an R. solanacearum mutant lacking oxyR, the only apparent OSR regulator in this bacterium (12, 13). We hypothesized that OSR genes would be differentially expressed in the oxyR mutant background in R. solanacearum, thereby contributing to host adaptation and virulence in host adaptation and virulence. We found that R. solanacearum oxyR is essential for survival of hydrogen peroxide stress, for hydrogen peroxide-inducible gene expression, and for full bacterial wilt virulence. Further, OxyR regulates at least five OSR-related genes, including itself.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains were grown in LB medium (33) at 37°C. R. solanacearum strains in this study were derived from a wild-type tomato isolate, the type strain K60 (phylotype II, sequevar 7) (25). R. solanacearum strains were grown in CPG broth (20) or CPG solid medium with 0.05% (wt/vol) tetrazolium chloride (25). Antibiotics were added to cultures at the following concentrations: 25 μg/ml kanamycin, 12.5 μg/ml gentamicin, and 15 μg/ml tetracycline; catalase (MP Biomedicals, Solon, OH) was added at 100 units/plate. Growth curve analyses were performed in minimal Boucher's minimal medium (BMM) (6) with 0.2% (wt/vol) glucose and supplemented with either 250 μM hydrogen peroxide or 5 μM paraquat (Ultra Scientific, North Kingstown, RI). Overnight cultures of R. solanacearum strain K60 and oxyR mutant K2690 grown in BMM with glucose to an optical density at 600 nm (OD600) of 0.1 were exposed to 150 μM hydrogen peroxide for 15 min before RNA extraction. In planta growth of R. solanacearum strains was assessed by quantifying multiplication of the organism in tobacco leaves (Nicotiana tabacum cv. Bottom Special) as previously described (46). The bacterial strains, selected plasmids, and primers used in this study are listed in Table S1 in the supplemental material.
DNA manipulations.
DNA isolation, PCR, cloning, and Southern hybridization were carried out by standard protocols (4). Competent cells of E. coli and R. solanacearum were created as described previously (1). Unless otherwise noted, chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO), and molecular biology reagents and kits were purchased from Promega (Madison, WI).
Oligonucleotides and sequence analysis.
DNA and protein sequences were analyzed using Biology Workbench (http://workbench.sdsc.edu/), NCBI BLAST (2), NEB cutter (http://tools.neb.com/NEBcutter2/index.php), and the genomic databases of R. solanacearum strains UW551 (http://vision.biotech.ufl.edu/mycap/jsp/project/description.jsp?projectID=1) and GMI1000 (http://bioinfo.genotoul.fr/annotation/iANT/bacteria/ralsto/). Primers were purchased from Integrated DNA Technologies (Coralville, IA). DNA sequencing was performed at the University of Wisconsin Biotechnology Center (Madison, WI).
Construction of K2690.
A 1,048-bp DNA fragment containing the oxyR gene was amplified by PCR using primers listed in Table S1 in the supplemental material. The resulting fragment was AT cloned into pSTBlue-1, creating pSToxyR, and sequenced. The aacCI gentamicin resistance gene cassette from pUCGM was introduced into a unique SacI site in oxyR, creating pSToxyR::aacCI. The resulting mutant allele was introduced into R. solanacearum through triparental mating using the helper plasmid pRK600 in E. coli HB101 and donor plasmid pSJYoT carrying the oxyR::aacC1 EcoRI fragment from pSToxyR::aacCI. Transconjugants were plated on media containing gentamicin and catalase, which was necessary to permit growth of the oxyR mutant strain. Allelic replacement of the oxyR gene in R. solanacearum was confirmed by PCR and Southern blot analysis (data not shown). The confirmed oxyR mutant was called K2690.
Complementation of K2690.
The oxyR and promoter primers (see Table S1 in the supplemental material) were used to amplify a 1,953-bp fragment containing the oxyR gene and its upstream region from R. solanacearum biovar 3 phylotype I strain GMI1000. The resulting fragment was AT cloned into pSTBlue-1 and sequenced. Using EcoRI sites, the 1,953-bp fragment was subcloned into the low-copy-number plasmid pUFJ10, which is stably maintained in R. solanacearum (15), creating pUFJ10::oxyR. For trans-complementation, pUFJ10 and pUFJ10::oxyR were introduced into K2690 and wild-type R. solanacearum by electroporation.
Virulence assays.
Tomato plants of susceptible cultivar Bonny Best and the wilt-tolerant line Hawaii 7996, and tobacco plants of the susceptible cultivar Petit Havana were inoculated using a naturalistic soil soak assay as previously described (45). Briefly, a suspension of bacteria was poured into the pots of unwounded 15-day-old tomato plants or 30-day-old tobacco plants. Plants were kept in a growth chamber with 12-h day/12-h night cycles at 28°C, and disease symptoms were evaluated daily for 14 days using a 0 to 4 disease index. The assay was performed in triplicate with 16 plants per treatment in each assay for tomato plants and in duplicate with 10 plants per treatment in each assay for the tobacco plants.
Plating efficiency and catalase activity.
Overnight cultures of R. solanacearum strains were collected by centrifugation and resuspended in water, and the OD600 was adjusted to 0.1. The bacterial suspensions were serially diluted and plated on CPG plates with and without catalase. Results presented are the average of three independent experiments, each done in duplicate. Catalase activity was assayed visually by adding 1 ml 880 mM hydrogen peroxide to 3 ml of R. solanacearum overnight cultures adjusted to an OD600 of 1.0 with fresh CPG. The experiment was repeated four times.
RNA extraction, cDNA synthesis, and quantitative RT-PCR.
After exposure to hydrogen peroxide for 15 min, R. solanacearum cells were treated with 1.25 ml of 5% ethanol (EtOH)-phenol solution per 10 ml of culture to preserve the RNA profile (26) and collected by centrifugation. Total nucleic acids were isolated with MasterPure complete DNA and RNA purification kit (Epicentre Biotechnologies, Madison, WI), followed by four Baseline-ZERO DNase treatments (Epicentre Biotechnologies, Madison, WI). RNA was quantified with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE), and RNA quality was assessed with a 2100 Bioanalyzer (Agilent Technologies Deutschland GmbH). cDNA was synthesized from 1 μg RNA with SuperScript III first-strand synthesis system for reverse transcriptase PCR (RT-PCR) (Invitrogen, Carlsbad, CA). Quantitative real-time PCR was performed in 25-μl volumes with 2 μl of cDNA, a 400 nM concentration of each primer, and 12.5 μl of Power SYBR green master PCR mix (Applied Biosystems, Warrington, United Kingdom) in an ABI 7300 real-time PCR system (Applied Biosystems). Primers were designed to amplify 100- to 200-bp fragments (see Table S1 in the supplemental material). Standard curves with known DNA concentrations were used to determine primer pair efficiency, and primer products were verified with melting-curve analysis according to the ABI real-time PCR system software. All samples were run in triplicate, and RNA with no reverse transcriptase was used as a control for genomic DNA contamination. Relative fold changes in expression of each target gene were calculated with the Pfaffl method (37), using the wild-type strain in the absence of hydrogen peroxide as the calibrator sample and three constitutively expressed normalization reference genes (glmS, gyrB, and pcrA) (48). Results presented are the average of two independent RNA extractions per tested condition. Significant values are those with a P value of ≤0.05 and at least a 1.5-fold difference from that of the wild type in the absence of hydrogen peroxide.
Statistical analysis.
Statistical analysis was performed with Minitab statistical software (Minitab, Inc.) and GraphPad Prism software (GraphPad Software, Inc.).
RESULTS
Cloning of R. solanacearum oxyR.
The R. solanacearum GMI1000 and UW551 genomes contain a predicted oxyR gene with 96% amino acid identity. We used the R. solanacearum phylotype II, sequevar 1 strain UW551 genome sequence (15) to design primers to clone oxyR from the related phylotype II strain K60. Sequence analysis revealed that K60's OxyR is 99% identical at the amino acid level to that of UW551. Multiple sequence alignments among four other characterized OxyR proteins demonstrate high conservation in the area surrounding the two essential hydrogen peroxide-responsive cysteines (Fig. 1).
Fig. 1.
Sequence of Ralstonia solanacearum OxyR. ClustalW multiple sequence alignment of OxyR sequences from R. solanacearum strain GMI1000 and related bacteria. Boldface letters indicate the conserved active cysteines in OxyR. An asterisk indicates ful conservation, a colon indicates conservation of strong groups, and a period indicates conservation of weak groups. GenBank accession numbers are as follows: Erwinia chrysanthemi, CAB40388; Escherichia coli K-12, AAC76943.1; Agrobacterium tumefaciens, AAK88806; Ralstonia solanacearum K60, JN382247; and Pseudomonas syringae DC3000, AA053620.1.
Mutagenesis of R. solanacearum oxyR.
To understand the role of oxyR in the OSR and virulence of R. solanacearum, we disrupted the strain K60 oxyR open reading frame (ORF) with a gentamicin resistance cassette. The resulting oxyR mutant strain, named K2690, grew as well as its wild-type parent in minimal medium with glucose as a sole carbon source (Fig. 2A) or in susceptible tobacco leaves (cultivar Bottom Special; data not shown), demonstrating that this mutant is not an auxotroph and suffers no general growth defects. However, K2690 had a significantly (P = 0.003) lower ability to form colonies on rich medium plates without catalase than the the wild type (3.17 × 106 CFU/ml versus 1.02 × 108 CFU/ml, respectively); this phenotype was rescued by adding exogenous catalase to the plates (data not shown). Consistent with this observation, broth cultures of K2690 had no detectable catalase activity, assessed visually as rapid formation and accumulation of bubbles after the addition of hydrogen peroxide. Both these phenotypes were returned to the wild type following introduction of a wild-type copy of oxyR in trans.
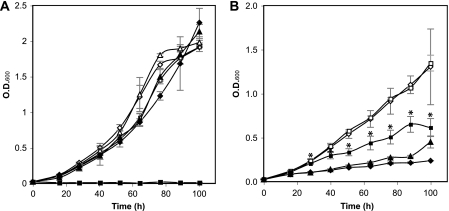
Fig. 2.
Effect of oxidative stress on R. solanacearum strain growth in minimal medium. Growth of wild-type K60 carrying empty vector pUFJ10 (diamonds), oxyR mutant K2690 carrying empty vector pUFJ10 (squares), and K2690 complemented with pUFJ10::oxyR (triangles) strains in the absence (empty symbols) and presence (filled symbols) of 250 μM H2O2 (A) or 5 μM paraquat (B) is shown. Results are the average of two independent experiments, with two replicates for each treatment in each experiment; bars represent standard errors of the means. An asterisk indicates a statistically significant difference according to Student's t test (P < 0.05).
R. solanacearum required oxyR for growth in the presence of hydrogen peroxide.
To determine the role of oxyR in the R. solanacearum hydrogen peroxide stress response, we cultured wild-type strain K60 and oxyR mutant K2690 in minimal medium with glucose as a sole carbon source in the absence or presence of 250 μM hydrogen peroxide. The wild-type strain suffered a slight growth delay in the presence of hydrogen peroxide but ultimately reached the same cell density as that in medium without hydrogen peroxide. However, strain K2690 did not grow in the presence of 250 μM hydrogen peroxide (Fig. 2A). The wild-type growth pattern was restored when a copy of oxyR was introduced into strain K2690 in trans (Fig. 2A). K2690 grew slowly in the presence of 150 μM hydrogen peroxide, reaching a final density well below that of the wild-type strain (data not shown).
R. solanacearum strain K2690 grew better than the wild type in the presence of paraquat.
To test the role of oxyR in the superoxide stress response of R. solanacearum, we cultured K2690 in minimal medium in the absence or presence of a 5 μM concentration of the superoxide-generating chemical paraquat. Growth of wild-type K60 was severely affected in the presence of paraquat. Unexpectedly, in the presence of paraquat, K2690 grew significantly better than the wild-type strain (P < 0.05) starting at 28 h postinoculation (Fig. 2B). It did not, however, reach densities comparable to those in the absence of paraquat. The growth of K2690 in the presence of paraquat was reduced to wild-type levels after a wild-type copy of oxyR was added in trans.
R. solanacearum needs oxyR for full virulence.
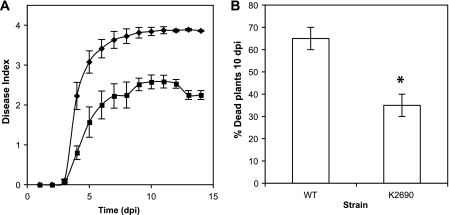
To determine the role of oxyR in bacterial wilt virulence, we performed a naturalistic disease assay on susceptible and tolerant tomato and susceptible tobacco plants. The tomato line Hawaii 7996 is horizontally resistant to bacterial wilt, but it readily forms latent infections (16). Under the conditions tested, K60 killed most Hawaii 7996 plants by 14 days postinoculation, but strain K2690 was significantly delayed in disease symptom onset and development, killing approximately half the infected plants by day 14 (Fig. 3A). A similar disease progress curve was observed in the highly susceptible tomato cultivar Bonny Best, where K2690 was also significantly less virulent than the wild type, although the difference between the R. solanacearum strains was smaller (data not shown). K2690 was also significantly reduced in virulence on another R. solanacearum host, tobacco (P = 0.05) (Fig. 3B).
Fig. 3.
Effect of oxyR mutation on R. solanacearum virulence. Disease progression curves of R. solanacearum strains on wilt-tolerant tomato line Hawaii 7996 (A) and on wilt-susceptible tobacco cultivar Petit Havana SRI (B) following soil soak inoculation of unwounded plants, with bacterial cell suspensions of strain K60 (diamonds) and oxyR mutant K2690 (squares). Disease symptoms were evaluated daily for 14 days using a 0 to 4 disease index. (A) Inoculum was ∼5 × 107 CFU/gram of soil. Results shown are the average of four independent experiments, with 16 plants per treatment per assay; bars indicate standard errors of the means. (B) Inoculum was ∼3 ×107 CFU/gram of soil. Results shown are the average of two independent assays, with 10 plants per treatment per assay; bars represent standard errors of the means. An asterisk indicates a statistically significant difference according to Student's t test (P = 0.05).
Oxidative stress response genes were differentially regulated in an R. solanacearum oxyR mutant.
To identify genes directly or indirectly regulated by OxyR and/or hydrogen peroxide in R. solanacearum, we performed quantitative RT-PCR (qRT-PCR) to measure expression of genes selected from homologs of known oxidative stress response and OxyR-regulated genes in other bacterial species. RNA was extracted from early-log-phase cells exposed, or not, to 150 μM hydrogen peroxide for 15 min. Relative fold changes in expression of tested genes are presented in Table 1. In the presence of hydrogen peroxide, R. solanacearum cells had increased expression of oxyR, catalase genes katG and katE, the alkyl hydroperoxide reductase gene ahpC1, and the glutaredoxin gene grxC. Induction of ahpC1 and both catalase genes was dependent on the presence of oxyR. However, expression of grxC and oxyR itself was enhanced in the absence of oxyR. In contrast, bcp, sodB, and sodC gene expression was reduced in the presence of hydrogen peroxide, independent of the presence of oxyR.
Table 1.
Relative fold change in expression of R. solanacearum genes in response to 150 μM hydrogen peroxide and oxyR
| Locusb | Gene | Fold change in expressiona |
Functionc | ||
|---|---|---|---|---|---|
| K2690 without H2O2 | With H2O2 |
||||
| K60 | K2690 | ||||
| RRSL03242 | oxyR | 15.49 | 5.79 | 24.55 | Transcriptional regulator |
| RRSL01521 | katE | −64.39 | 1.62 | −265.45 | Catalase |
| RRSL01521 | katG | −1.94 | 27.92 | −4.38 | Catalase |
| RRSL01699 | ahpC1 | 4.94 | 3.26 | −3.22 | Alkyl hydroperoxide reductase subunit |
| RRSL00135 | bcp | 1.54 | −3.14 | −4.55 | Peroxidase |
| RRSL03306 | grxC | 3.82 | 4.12 | 15.38 | Glutaredoxin |
| RRSL03640 | sodC | 1.27 | −2.19 | −3.05 | Cu,Zn superoxide dismutase |
| RRSL01451 | sodB | 1.23 | −6.14 | −9.29 | Fe superoxide dismutase |
Fold change in expression relative to the expression in parent strain K60 grown in the absence of hydrogen peroxide. Boldface numbers are statistically significantly different from gene expression in K60 growing without hydrogen peroxide (P ≤ 0.05), according to Student's t test, and are at least 1.5-fold different.
Gene locus tag in the R. solanacearum UW551 genome.
Putative function was assigned by similarity to proteins in the public databases.
DISCUSSION
A growing body of work demonstrates the importance of the oxidative stress response in eukaryote-bacterium interactions, including pathogenic ones (18, 36, 38–40, 42, 44, 51). However, there is scant and contradictory information about the role of the OSR regulator OxyR in the virulence of plant-pathogenic bacteria (33, 34). We previously found that the bacterial wilt pathogen R. solanacearum is exposed to ROS during host infection and that it possesses multiple and redundant ROS scavenging enzymes, some of which are induced in the host (13). However, the presence of multiple and redundant predicted OSR genes in the genome made it impossible to assess the importance of the OSR in R. solanacearum by deleting individual structural genes. Here we investigated the role of a predicted OSR regulator, oxyR, in this pathogen's virulence and ROS susceptibility.
We found that R. solanacearum possesses an oxyR homolog that was necessary for survival in the presence of hydrogen peroxide. The oxyR gene product was also required for wild-type virulence levels on susceptible and tolerant tomato plants and on susceptible tobacco plants, consistent with our previous observation of elevated hydrogen peroxide levels in plants infected with R. solanacearum (13). It is known that hydrogen peroxide generated in the plant's defensive oxidative burst triggers host defense expression at the site of infection and in distant tissues, leading to systemic acquired immunity (3). However, neither the concentrations of hydrogen peroxide that induce host defense activation nor the concentrations of ROS experienced by pathogens during infection are known. Our results indicate that during host infection, R. solanacearum encounters inhibitory concentrations of ROS and that it needs a functional OSR to overcome them. In addition, the strikingly poor ability of K2690 to form colonies on agar plates, and this mutant's lack of detectable catalase activity, suggests that oxyR is an essential part of the hydrogen peroxide stress response in R. solanacearum as in other bacteria.
The plating defect of oxyR mutants is well documented for several bacteria (29, 32, 35, 50). This defect is believed to result because bacteria beginning to grow on plates launch a global stress response that includes genes in the OxyR regulon (10). In Pseudomonas aeruginosa, the plating defect was attributed to autooxidizable components in the medium (17). This likely explains why supplementing the plates with catalase recovers the plating defect of K2690 to wild-type levels. Low-temperature-induced viable but nonculturable (VBNC) R. solanacearum cells are partially restored to growth on plates by the addition of catalase to the agar plates (49). Oxidative damage to cell components is hypothesized to induce the VBNC state (5). The recovery of K2690 colonies on plates with added catalase suggests that under these conditions, a proportion of the oxyR mutant cells could be in a VBNC state and that oxidative stress may trigger the VBNC state in R. solanacearum.
The increased growth rate of strain K2690 in the presence of the superoxide-generating agent paraquat was unexpected. Although paraquat severely reduced growth of wild-type strain K60, in the presence of paraquat, the oxyR mutant grew better than the wild type. This phenotype resulted in loss of oxyR, since the introduction of a wild-type copy of oxyR into K2690 reduced its growth in paraquat to wild-type levels. R. solanacearum genomes predict the presence of superoxide dismutases (Sod), which convert superoxide into hydrogen peroxide (14); however, the Sod genes were expressed at similar levels in wild-type and K2690 cells. We speculate that R. solanacearum has superoxide stress response genes other than sod that are induced in the oxyR mutant background.
OxyR, sublethal concentrations of hydrogen peroxide, or both, appear to regulate several putative R. solanacearum OSR genes. Expression of bcp, sodB, and sodC genes was reduced in the presence of hydrogen peroxide regardless of the presence of oxyR, suggesting that these genes are not under the control of OxyR.
Following hydrogen peroxide exposure, oxyR and grxC were induced in K2690 and in the wild-type strain, suggesting that OxyR acts as a repressor of these genes. grxC encodes a putative glutaredoxin that, accompanied by glutathione, helps maintain the redox status of the cell in E. coli (27) and reduces oxidized OxyR, creating a feedback loop regulation of OxyR activity (52, 53). Our results suggest that R. solanacearum uses a similar mechanism to regulate OxyR activity and gene expression. The bacterium's two catalase genes, katE and katG, and ahpC1 were induced by hydrogen peroxide in an OxyR-dependent manner. These results are consistent with the lack of detectable catalase activity in K2690 and with OxyR-dependent activation of kat and ahpC genes in other bacteria (22–24, 31, 35, 50, 53). ahpC1 is predicted to encode one of two predicted alkyl hydroperoxide reductase systems in R. solanacearum, which in E. coli is the primary scavenger of endogenous and/or low concentrations of hydrogen peroxide (41). We found that ahpC1 expression trended higher in K2690 in the absence of hydrogen peroxide than in its presence, although the difference was not statistically significant. This could indicate that R. solanacearum OxyR is both a repressor and an activator of ahpC1. OxyR has been reported to activate and repress transcription of a target gene in other bacterial systems (21, 23, 28). This putative ahpC1 expression pattern might explain the unexpectedly strong growth of K2690 in the presence of paraquat. Consistent with this hypothesis, overexpression of ahpC in an Xanthomonas campestris oxyR mutant resulted in increased superoxide resistance compared to that in an oxyR mutant alone (50).
In summary, the diverse phenotypes of an R. solanacearum oxyR mutant indicate that this regulator plays a central role in the oxidative stress response of this plant pathogen. In particular, an OxyR-mediated OSR is needed for full bacterial wilt virulence on several different plant hosts. Our in vitro gene expression data suggest that much of the OxyR regulon is conserved among several bacterial species. However, global gene expression studies of an oxyR mutant during disease development are needed to fully decipher the biological importance of this regulon. Once the full set of OxyR-regulated and oxidative stress-responsive genes are identified, it will be possible to more conclusively determine the specific mechanisms used by R. solanacearum to adapt to the oxidative host environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Shah (Louisiana State University) for valuable technical assistance.
This research was supported by USDA-CSREES Plant Biosecurity Award 07-55605-17843 and by the University of Wisconsin—Madison College of Agricultural and Life Sciences.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Allen C., Huang Y., Sequeira L. 1991. Cloning of genes affecting polygalacturonase production in Pseudomonas solanacearum. Mol. Plant Microbe Interact. 4:147–154 [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alvarez M. E., et al. 1998. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92:773–784 [DOI] [PubMed] [Google Scholar]
- 4. Ausubel F. M., et al. 1998. Current protocols in molecular biology, vol. 1-4 John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 5. Barer M. R., Hardwood C. R. 1999. Bacterial viability and culturability. Adv. Microb. Physiol. 41:94–137 [DOI] [PubMed] [Google Scholar]
- 6. Boucher C., Barberis P., Trigalet A., Demery D. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131:2449–2457 [Google Scholar]
- 7. Brown D. G., Allen C. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol. Microbiol. 53:1641–1660 [DOI] [PubMed] [Google Scholar]
- 8. Brown D. G., Swanson J. K., Allen C. 2007. Two host-induced Ralstonia solanacearum genes, acrA and dinF, encode multidrug efflux pumps and contribute to bacterial wilt virulence. Appl. Environ. Microbiol. 73:2777–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Colburn-Clifford J. M., Scherf J. M., Allen C. 2010. Ralstonia solanacearum Dps contributes to oxidative stress tolerance and to colonization of and virulence on tomato plants. Appl. Environ. Microbiol. 76:7392–7399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cuny C., Lesbats M., Dukan S. 2007. Induction of a global stress response during the first step of Escherichia coli plate growth. Appl. Environ. Microbiol. 73:885–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Denny T. P. 2006. Plant pathogenic Ralstonia species, p. 573–644In Gnanamanickam S. S. (ed.), Plant-associated bacteria. Springer Publishing, Dordrecht, The Netherlands [Google Scholar]
- 12. Flavier A. B., Schell M. A., Denny T. P. 1998. An RpoS (sigma S) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol. Microbiol. 28:475–486 [DOI] [PubMed] [Google Scholar]
- 13. Flores-Cruz Z., Allen C. 2009. Ralstonia solanacearum encounters an oxidative environment during tomato infection. Mol. Plant Microbe Interact. 22:773–782 [DOI] [PubMed] [Google Scholar]
- 14. Fridovich I. 1995. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64:97–112 [DOI] [PubMed] [Google Scholar]
- 15. Gabriel D., et al. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant Microbe Interact. 19:69–79 [DOI] [PubMed] [Google Scholar]
- 16. Grimault V., Anais G., Prior P. 1994. Distribution of Pseudomonas solanacearum in the stem tissues of tomato plants with different levels of resistance to bacterial wilt. Plant Pathol. 43:663–668 [Google Scholar]
- 17. Hassett D. J., et al. 2000. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J. Bacteriol. 182:4557–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hassouni M. E., Chambost J. P., Expert D., Van Gijsegem F., Barras F. 1999. The minimal gene set member msrA encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc. Natl. Acad. Sci. U. S. A. 96:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayward A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65–87 [DOI] [PubMed] [Google Scholar]
- 20. Hendrick C. A., Sequeira L. 1984. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heo Y.-J., et al. 2010. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J. Bacteriol. 192:381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hishinuma S., Yuki M., Fujimura M., Fukumori F. 2006. OxyR regulated the expression of two major catalases, KatA and KatB, along with peroxiredoxin, AhpC in Pseudomonas putida. Environ. Microbiol. 8:2115–2124 [DOI] [PubMed] [Google Scholar]
- 23. Ieva R., et al. 2008. OxyR tightly regulates catalase expression in Neisseria meningitidis through both repression and activation mechanisms. Mol. Microbiol. 70:1152–1165 [DOI] [PubMed] [Google Scholar]
- 24. Imlay J. A. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelman A. 1954. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in tetrazolium medium. Phytopathology 44:693–695 [Google Scholar]
- 26. Khodursky A., et al. 2003. Escherichia coli spotted double-strand DNA microarrays: RNA extraction, labeling, hybridization, quality control, and data management. Methods Mol. Biol. 224:61–78 [DOI] [PubMed] [Google Scholar]
- 27. Lillig C. H., Berndt C., Holmgren A. 2008. Glutaredoxin systems. Biochim. Biophys. Acta 1780:1304–1317 [DOI] [PubMed] [Google Scholar]
- 28. Loprasert S., Fuangthong M., Whangsuk W., Atichartpongkul S., Mongkolsuk S. 2000. Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 37:1504–1514 [DOI] [PubMed] [Google Scholar]
- 29. Maciver I., Hansen E. J. 1996. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect. Immun. 64:4618–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mandal S., Das R. K., Mishra S. 2011. Differential occurrence of oxidative burst and antioxidative mechanism in compatible and incompatible interactions of Solanum lycopersicum and Ralstonia solanacearum. Plant Physiol. Biochem. 49:117–123 [DOI] [PubMed] [Google Scholar]
- 31. Michán C., Manchado M., Dorado G., Pueyo C. 1999. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J. Bacteriol. 181:2759–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miguel E., et al. 2000. Evidence against a direct antimicrobial role of H2O2 in the infection of plants by Erwinia chrysanthemi. Mol. Plant Microbe Interact. 13:421–429 [DOI] [PubMed] [Google Scholar]
- 33. Miller J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Nakjarung K., Mongkolsuk S., Vattanaviboon P. 2003. The oxyR from Agrobacterium tumefaciens: evaluation of its role in the regulation of catalase and peroxide responses. Biochem. Biophys. Res. Commun. 304:41–47 [DOI] [PubMed] [Google Scholar]
- 35. Ochsner U. A., Vasil M. L., Alsabbagh E., Parvatiyar K., Hassett D. J. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okinaka Y., Yang C.-H., Perna N. T., Keen N. T. 2002. Microarray profiling of Erwinia chrysanthemi 3937 genes that are regulated during plant infection. Mol. Plant Microbe Interact. 15:619–629 [DOI] [PubMed] [Google Scholar]
- 37. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rediers H., Rainey P. B., Vanderleyden J., De Mot R. 2005. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69:217–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saenkham P., Eiamphungporn W., Farrand S., Vattanaviboon P., Mongkolsuk S. 2007. Multiple superoxide dismutases in Agrobacterium tumefaciens: functional analysis, gene regulation, and influence on tumorigenesis. J. Bacteriol. 189:8807–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santos R., et al. 2001. Essential role of superoxide dismutase on the pathogenicity of Erwinia chrysanthemi strain 3937. Mol. Plant Microbe Interact. 14:758–767 [DOI] [PubMed] [Google Scholar]
- 41. Seaver L. C., Imlay J. A. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith S. G., Wilson T. J., Dow J. M., Daniels M. J. 1996. A gene for superoxide dismutase from Xanthomonas campestris pv. campestris and its expression during bacterial-plant interactions. Mol. Plant Microbe Interact. 9:584–593 [DOI] [PubMed] [Google Scholar]
- 43. Sund C. J., et al. 2008. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol. Microbiol. 67:129–142 [DOI] [PubMed] [Google Scholar]
- 44. Tamir-Ariel D., Navon N., Burdman S. 2007. Identification of genes in Xanthomonas campestris pv. vesicatoria induced during its interaction with tomato. J. Bacteriol. 189:6359–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tans-Kersten J., Guan Y., Allen C. 1998. Ralstonia solanacearum pectin methylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl. Environ. Microbiol. 64:4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tans-Kersten J., Huang H., Allen C. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183:3597–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toledano M., et al. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897–909 [DOI] [PubMed] [Google Scholar]
- 48. Vandesompele J., et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Elsas J. D., van Oberbeek L. S., Trigalet A. 2005. Viable but non-culturable state in Ralstonia solanacearum: is there a realistic threat to our strategic concepts? p. 103–119In Allen C., Prior P., Hayward A. C. (ed.), Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, St. Paul, MN [Google Scholar]
- 50. Vattanaviboon P., Whangsuk W., Mongkolsuk S. 2003. A suppressor of the menadione-hypersensitive phenotype of a Xanthomonas campestris pv. phaseoli oxyR mutant reveals a novel mechanism of toxicity and the protective role of alkyl hydroperoxide reductase. J. Bacteriol. 185:1734–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu X. Q., Pan S. Q. 2000. An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 35:407–414 [DOI] [PubMed] [Google Scholar]
- 52. Zheng M., Aring, slund F. A., Storz G. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718–1722 [DOI] [PubMed] [Google Scholar]
- 53. Zheng M., et al. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.