Abstract
Campylobacter jejuni and C. coli were quantified and typed, using multilocus sequence typing (MLST), from fecal samples collected from a mixed cattle and sheep farm during summer. Cattle had a significantly higher prevalence than sheep (21.9% [74/338] and 14.0% [30/214], respectively), but both decreased over time. There were no differences in the average Campylobacter concentrations shed by cattle (600 CFU g−1) and sheep (820 CFU g−1), although sheep did show a significant temporal reduction in the number of Campylobacter organisms shed in their feces. A total of 21 different sequence types (STs) (97.7% C. jejuni, 2.3% C. coli) were isolated from cattle, and 9 different STs were isolated from sheep (40.6% C. jejuni, 59.4% C. coli). The Campylobacter population in cattle was relatively stable, and the frequencies of genotypes isolated showed little temporal variation. However, the composition of subtypes isolated from sheep did show significant temporal differences. The cattle and sheep consistently showed significant differences in their carriage of Campylobacter species, STs, and CCs despite the fact that both were exposed to the same farming environment. This work has highlighted the patterns of a Campylobacter population on a ruminant farm by identifying the existence of both temporal and between-host variations.
INTRODUCTION
Campylobacter is the most commonly recognized etiological agent of bacterial gastroenteritis in many developed countries (5, 7), with incidence rates of 123.4 and 13.02 cases per 100,000 in Scotland and the United States, respectively, during 2009 (8, 33). Campylobacter jejuni accounts for approximately 90% of human Campylobacter infections, and approximately 10% are caused by C. coli (36). The sporadic nature of Campylobacter infection is partially responsible for underreporting and for sources of infection rarely being identified (13, 32).
The expanding use of multilocus sequence typing (MLST) (10, 28) has enabled the accumulation of transferable data that can aid in the study of Campylobacter population structure. There is increasing evidence of host-associated lineages (12, 26, 27) that can ultimately be used to attribute human infection to its origin (36, 37, 42). In Scotland, the majority (58 to 78%) of clinical Campylobacter cases were related to chickens, in contrast to 18 to 38% of cases associated with ruminants (37). The role of ruminants as a major reservoir of Campylobacter is further highlighted by the rural-urban association of strains. While 42% of infections in children less than 5 years old who reside in rural locations are associated with cattle strains, in contrast, 43% of infections in children within the same age bracket who reside in urban locations are associated with chickens (42). Ruminants constantly shed Campylobacter organisms into the environment, where they act as a reservoir for human infection via ingestion from contaminated food or via contact with ruminants or their feces, either directly or indirectly. Individual Campylobacter prevalence in cattle (the percentage of positive animals within a single herd) has been reported to range from 1.6% to 89%, with C. jejuni accounting for 55.4% to 100% and C. coli for between 0% and 30% of the Campylobacter organisms isolated (3, 4, 6, 14, 17, 18, 20, 25, 29, 30, 31, 35, 40, 46). Sheep have been reported to show a prevalence of approximately 25 to 30% (22, 23, 35, 41) and tend to carry higher proportions of C. coli than cattle, typically ranging from 30 to 47% (1, 6, 17, 35, 47), with the remainder being C. jejuni. Cattle and sheep appear to shed similar average concentrations, ranging from <102 CFU g feces−1 to 107 CFU g feces−1 (3, 6, 30, 35, 40, 41).
The Campylobacter subtypes tend to show poor discrimination between cattle and sheep (27, 35), especially when either is compared to other, less closely related hosts, such as poultry; this is likely due to both their similarity in physiology and mixing of livestock (36). This work aims to identify temporal variation in the prevalence, shedding rates, and diversity of C. jejuni and C. coli shed by cattle and sheep on a single farm over a 19-week period and to identify any between-host variation despite similar exposures to the Campylobacter load, strains, and environmental conditions.
MATERIALS AND METHODS
Collection of field samples.
Freshly voided feces were collected weekly from a single farm in Aberdeenshire, Scotland, over a 19-week period (i.e., weeks 20 to 38 of the year) between May and September 2006. The sampled flock contained approximately 200 sheep and a herd of 48 beef cattle. In order to establish the full genetic diversity of Campylobacter strains present, intensive sampling was performed near the beginning (61 cattle and 20 sheep at week 23) and at the end (75 cattle and 25 sheep at week 38) of the study. During the intervening 16 weeks (week 24 was excluded due to the large number of samples collected the previous week), 13 cattle and 11 sheep fecal samples were collected each week. To overcome any sampling limitations due to the relatively low numbers of samples taken on a weekly basis and to aid in providing more easily observable temporal trends, the data were combined into 3 sampling periods, each consisting of 6 weeks, as follows: sampling period 1, weeks 20 to 26 (excluding week 24); sampling period 2, weeks 27 to 32; sampling period 3, weeks 33 to 38.
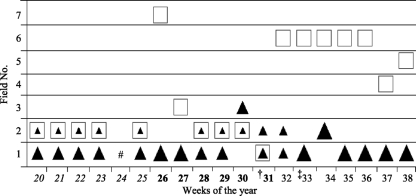
During the sampling, the herd of cattle and flock of sheep were intermittently split, mixed, or separated and moved between different fields by the farmer for husbandry and commercial reasons. Figure 1 shows the weekly locations of cattle and sheep with the numbers present at each sampling point. Overall, the cattle and sheep were mixed for the majority of time during the 1st and 2nd sampling periods but remained completely separate during the final sampling period (Fig. 1).
Fig. 1.
Weekly locations of livestock during the 19-week sampling showing when cattle and sheep were mixed, separated, or adjacent to each other. #, no sampling was carried out; †, introduction of grain into cattle diet; ‡, 30 new cattle replaced 30 of the previous herd, field 6 (approximately 1.5 km offsite) and field 7 (approximately 0.5 km offsite). The filled triangles represent cattle, where the large size represents the whole cattle herd (48 cattle), the middle size represents 30 to 33 cattle, and the small size represents 15 to 18 cattle. The squares represent 200 sheep. The different text styles in the weeks of the year (italics, boldface, and regular) indicate 6-week sampling periods 1 to 3.
Isolation of Campylobacter.
Campylobacter was isolated from 10-g aliquots of freshly collected feces (35, 37, 39). Briefly, the fecal samples were individually diluted (10:90 [wt/vol]) in a supplemented nutrient broth (DM180D; Mast, United Kingdom) containing 5 antibiotics and growth supplement (SRO232E; Oxoid, United Kingdom), further serially diluted, and plated onto supplemented (SR155E; Oxoid, United Kingdom) charcoal cefoperazone deoxycholate agar (CCDA) (CM0739; Oxoid, United Kingdom) for enumeration. Both the plates and the broth were incubated for 2 days under microaerophilic conditions, after which presumptive Campylobacter cells were counted and a further 100 μl of enrichment broth was plated and incubated for 2 days to confirm presence or absence. The presence of thermophilic Campylobacter species was initially confirmed using a latex agglutination test (M46CE; Microgen, United Kingdom). Following isolation of a single colony, DNA was extracted using Chelex 100 resin (142-1253; Bio-Rad Laboratories). All positive isolates were stored in nutrient broth plus 15% glycerol at −80°C. During the weeks in which large numbers of fecal samples were analyzed (weeks 23 and 38), multipicks (5) were purposely performed from positive plates to increase the chances of detecting >1 sequence type (ST) from multiple shedders. During all other sampling times, multipicks were performed only when the Campylobacter colonies appeared to differ in morphology or color.
Molecular typing.
C. jejuni and C. coli species were initially confirmed by PCR using the Pgm primer set, which is specific for C. jejuni and C. coli (28), and further typed by MLST as previously described (15, 39). Sequences were assembled and alleles were assigned using the STARS software available on the PubMLST website (http://pubmlst.org/campylobacter/), and sequence types and clonal complexes (CCs) were assigned to the array of seven alleles (21). The successful assignment of an allele number to each of the seven loci and subsequent allocation of a sequence type further confirmed the species as C. jejuni or C. coli.
Campylobacter output load.
The total number of Campylobacter CFU shed each day by cattle or sheep was calculated using the following formula: ROL = (CF)N, where ROL is the reservoir output load, C is the average shedding concentration (CFU g−1, including nonshedders), F is the mass of feces (g) excreted per day per animal (26 kg per day per animal for beef cattle and 4.1 kg per day for each ewe) (38, 43), and N is the number of animals present (48 cattle and 200 sheep) (43).
Statistical analysis.
The prevalence values of cattle and sheep with the binominal 95% confidence intervals (CI) were calculated in Excel (39). The average counts of Campylobacter CFU shed, with the corresponding 95th percentiles, were calculated using a Poptools (http://www.cse.csiro.au/poptools/) bootstrap method with 10,000 iterations of sampling with replacement. For samples detected only by enrichment (i.e., <100 CFU g−1), a random array of numbers between 1 and 100 is inserted for each iteration. Fisher's exact test of differentiation was used to statistically identify the presence of differences between cattle and sheep or between sampling periods for a single ST, species, or prevalence (44; http://www.quantitativeskills.com/sisa/statistics/twoby2.htm).
The exact test of differentiation (16, 34) involving 100,000 iterations using the Markov chain method was used to compare the whole range of STs/CCs and was performed with Arlequin software (11).
Finally a randomization test was performed using pairwise analysis to determine if Campylobacter STs became more dissimilar with increasing time differences between collection points.
RESULTS
Cattle.
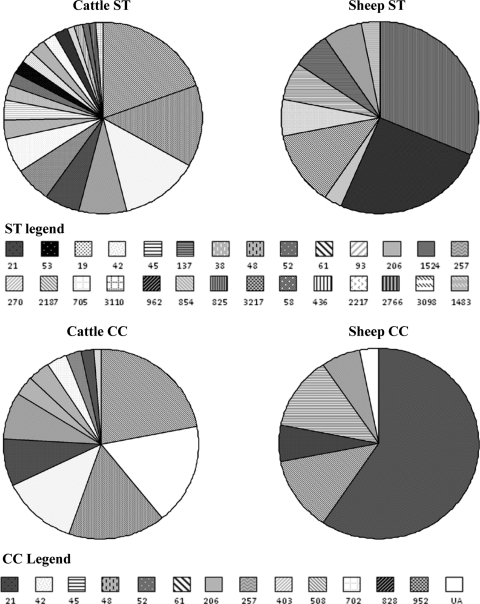
C. jejuni and C. coli were isolated from 74/338 (21.9% [95% CI, 17.5 to 26.3%]) cattle fecal samples, 62.2% by direct plating, with the remainder detected by enrichment alone. The majority of Campylobacter organisms isolated from cattle were C. jejuni (97.7% [95% CI, 94.6 to 100.9%]), with the remainder (2.3% [95% CI, 0.85 to 5.45%]) being C. coli. The average concentration of Campylobacter shed by cattle (including nonshedders) was 600 CFU g−1 (95% CI, 280 to 980 CFU g−1), accounting for 7.5 × 108 CFU of Campylobacter excreted per day by the herd of cattle. Twenty-one different STs were isolated from cattle, 16 of which belonged to 11 CCs, with 5 STs not assigned to a CC (see Fig. 5). The 3 most common STs isolated from cattle were ST-61 (19.5% [95% CI, 11.2 to 27.9%]), ST-48 (13.8% [95% CI, 6.6 to 21.0%]), and ST-42 (12.6% [95% CI, 5.7 to 19.6%]). Excluding the unassigned CC, the 3 most common CCs isolated from cattle were CC-61 at 21.8% (95% CI, 13.2 to 30.5%), CC-48 at 16.1% (95% CI, 8.4 to 23.8%), and CC-42 at 12.6% (95% CI, 5.7 to 19.6%). Campylobacter coli was isolated from only two cattle fecal samples (2.3%) and belonged to ST-962 (CC-828). From the 74 positive pats, 87 different isolates were obtained due to multiple shedders. Two different STs were isolated from 7 pats (9.46% [95% CI, 2.8 to 16.1%]), and 3 different STs were isolated from 3 pats (4.1% [95% CI, −0.4 to 8.5%]).
Fig. 5.
Prevalence of each Campylobacter sequence type and clonal complex isolated from cattle and sheep during the 18 weeks of sampling.
Temporal changes in Campylobacter shedding by cattle.
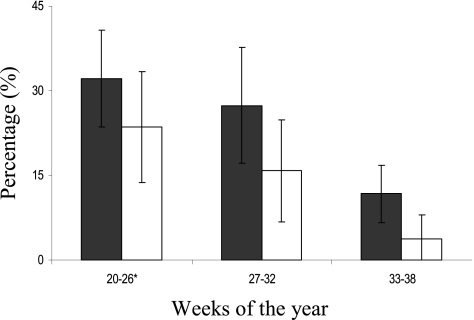
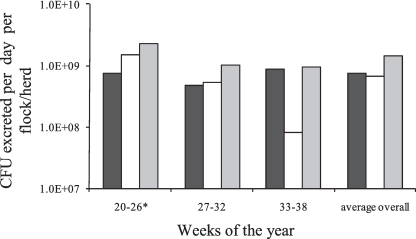
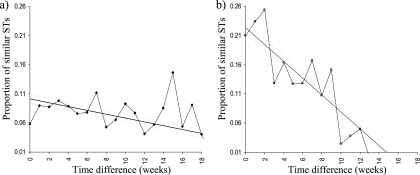
Cattle showed a highly significant temporal decline in Campylobacter prevalence between the 1st and final sampling times, where the percentage of positive cattle fell from 32.1% (95% CI, 23.5 to 40.8%) to 11.8% (95% CI, 6.7 to 16.9%) (P <0.00005) (Fig. 2). The rate of decline was more pronounced between the middle and late summer, since no significant changes were observed between the 1st and 2nd sampling periods. The concentrations of Campylobacter shed in cattle feces showed no significant differences over the whole sampling period (Fig. 3), and similarly, any decline in ROL was absent in cattle (Fig. 4). It was observed that the highest Campylobacter counts from cattle (750 CFU g−1), although not significantly different from the other periods, occurred at the time of lowest prevalence (11.8%) during the final sampling period (Fig. 2 and 3). The compositions of both STs and CCs isolated from cattle (see Fig. 6) showed no significant difference between the 3 sampling times, using the exact test of differentiation, and was further confirmed using the pairwise comparison test, although a slight decreasing trend in the proportion of similar STs with an increasing time difference was observed (see Fig. 7).
Fig. 2.
Campylobacter prevalence in cattle (filled bars) and sheep (open bars), including the 95% binomial confidence intervals (error bars), during each sampling period. *, excluding week 24.
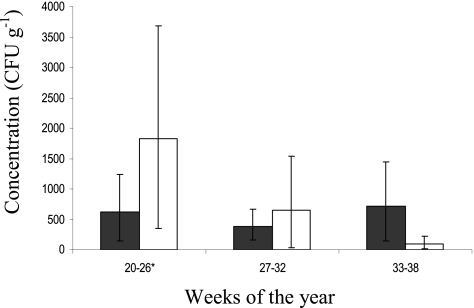
Fig. 3.
Average Campylobacter shedding concentrations in cattle (filled bars) and sheep (open bars) during each sampling period. The error bars represent the 95% CI. *, excluding week 24.
Fig. 4.
Campylobacter reservoir output loads (CFU per day) of cattle (n = 48; black bars), sheep (n = 200; white bars), and all the ruminants included in the study (248; gray bars) calculated from the average shedding and prevalence for each of the 3 sampling times and overall. *, excluding week 24.
Fig. 6.
Temporal prevalence of Campylobacter sequence types in cattle (a) and sheep (b) and the clonal complexes isolated from cattle (c) and sheep (d) during each of the three sampling periods (see the legend to Fig. 5 for symbols). *, excluding week 24.
Fig. 7.
Pairwise clustering randomization test to show temporal genetic variation in Campylobacter sequence types isolated from cattle (a) and sheep (b). The straight line shows the linear average and overall trend. The 2 flanking gray lines are the 2.5% and 97.5% confidence levels of the linear average. A diagonal line from left to right shows that the STs become more dissimilar with increasing time difference between collection times; the steeper the gradient, the more dissimilar. A horizontal line for the linear average would show no dissimilarities over time.
Sheep.
Campylobacter was isolated from 30/214 (14.0% [95% CI, 9.4 to 18.7%]) fecal samples from sheep, and similar to cattle, 60.0% were detected by direct plating with the remainder via enrichment. The predominant species was C. coli at 59.4% (95% CI, 76.4 to 42.4%), with the remainder being C. jejuni. The average concentration of Campylobacter shed by sheep (including nonshedders) was 820 CFU g−1 (95% CI, 271 to 1,455 CFU g−1) with an average ROL of 6.7 × 108 CFU day−1. Nine different STs were identified from sheep, which belonged to 5 different CCs plus 1 unassigned (Fig. 5). The 3 most common STs isolated from sheep were C. coli ST-825 at 31.3% (95% CI, 15.2 to 47.3%), C. coli ST-962 at 25.0% (95% CI, 10.0 to 40.0%), and C. jejuni ST-61 at 12.5% (95% CI, 1.0 to 24.0%). The 3 most common CCs were CC-828 at 59.4% (95% CI, 42.4 to 76.4%), CC-61 at 12.5% (95% CI, 1.0 to 24.0%), and CC-45 at 12.5% (95% CI, 1.0 to 24.0%). Two individual fecal samples from sheep each had 2 STs isolated (6.67% [95% CI, −2.3 to 15.6%]).
Temporal changes in Campylobacter shedding by sheep.
Sheep showed a highly significant temporal decline in both Campylobacter prevalence and the concentrations shed. The percentage of positive sheep declined from 23.6% (95% CI, 13.8 to 33.4%) to 3.8% (95% CI, −0.4 to 8.0%) (P < 0.0005) (Fig. 2), and the concentrations shed (including nonshedders) fell from an average of 1,800 CFU g−1 (95% CI, 347 to 3,679 CFU g−1) to 100 CFU g−1 (95% CI, 13 to 228 CFU g−1) (Fig. 3) (P <0.05) between the 1st and final sampling times. Since the values for both prevalence and concentration declined, the ROL from sheep also showed a similar pattern, declining from 1.48 × 109 to 8.30 × 107 CFU day−1 (P< 0.05). No temporal difference was observed between the 1st and 2nd sampling periods (Fig. 4).
Using the exact test of differentiation, a significant difference in the compositions of STs and CCs isolated from sheep was identified between sampling periods (Table 1). For STs, this was apparent only when comparing either the 1st or 2nd sampling period to the final sampling period. However, for CCs, the significant difference remained between the 3 sampling periods (P<0.05) (Fig. 6). This temporal difference was further enforced using a pairwise analysis, where STs became less similar with increasing time differences between sample collections (Fig. 7).
Table 1.
P values for the exact test of differentiation comparing the frequencies of STs and CCs from cattle and sheep for each sampling period
| CC/ST and host | Sampling perioda |
P valueb |
||||
|---|---|---|---|---|---|---|
| Sheep |
Cattle |
|||||
| T1 | T2 | T3 | T1 | T2 | ||
| STs | ||||||
| Sheep | T1 | |||||
| T2 | 0.120 | |||||
| T3 | 0.003 | 0.030 | ||||
| Cattle | T1 | 0.000 | 0.000 | 0.190 | ||
| T2 | 0.000 | 0.000 | 0.397 | 0.545 | ||
| T3 | 0.000 | 0.000 | 0.032 | 0.342 | 0.570 | |
| CCs | ||||||
| Sheep | T1 | |||||
| T2 | 0.043 | |||||
| T3 | 0.009 | 0.014 | ||||
| Cattle | T1 | 0.000 | 0.000 | 0.024 | ||
| T2 | 0.000 | 0.000 | 0.091 | 0.371 | ||
| T3 | 0.000 | 0.001 | 0.014 | 0.341 | 0.423 | |
T1 to T3, sampling periods 1 to 3.
Boldface indicates significant differences (<0.05) between sampling times or hosts.
Comparison between hosts.
During the 1st sampling period, the prevalences in cattle and sheep were not significantly different. However, during the 2nd and 3rd sampling periods and overall, the cattle had a significantly greater prevalence than sheep using Fisher's exact test (P < 0.05 and P < 0.01) (Fig. 2). Throughout the study, the proportions of species isolated from cattle and sheep remained significantly different, with C. coli predominating in sheep (59.4%) compared to cattle (2.3%) (P < 0.00001). Both the concentration of Campylobacter shed per gram of feces and the overall number excreted per day per group of animals were not significantly different between cattle and sheep overall or at any point in the study. Of the 27 different STs isolated from ruminants, only 3 were identified in both cattle and sheep (ST-962, ST-61, and ST-257). Most importantly, the STs and CCs were consistently and statistically significantly different throughout the duration of the study between the 2 hosts, which is evident in Fig. 5 and 6.
The Campylobacter population was relatively stable in cattle, where the only temporal difference was observed in a reduced prevalence from early to late summer. In contrast, sheep showed significant changes, with both the prevalence and the concentration shed decreasing with time, along with changes in the composition of Campylobacter STs and CCs carried.
DISCUSSION
The Campylobacter prevalence in cattle was significantly higher than that in sheep at the 2nd and 3rd sampling times and overall (P < 0.05), but they showed similar significant declines between May and September, from 32.8% to 11.8% and from 23.6% to 3.8% for cattle and sheep, respectively (Fig. 2). Any seasonal trends must be interpreted cautiously to determine whether they are the result of true seasonal variation or of husbandry factors, such as housing, diet, age, and stress. For example, feedlot cattle have previously been reported to show a higher prevalence than those on pasture (14), which is likely a result of increased exposure between animals in housing (4, 29). However, contradicting this, dairy cattle had the highest prevalence during summer when grazing as opposed to when they were housed during the winter months (17). The presence of seasonal variation in dairy cattle is further supported by the work of Kwan et al. (25). The isolation of C. coli from cattle was relatively low (2.3%) and was similar to the values reported elsewhere. For example, 2.0% of beef cattle sampled at abattoirs in eastern Canada were positive for C. coli (14), and C. coli failed to be isolated from dairy cattle in both Denmark and New Zealand (2, 30). However, it was calculated that 10% of isolates from Scottish cattle were C. coli (35), much greater than reported here.
The Campylobacter prevalence in sheep (14.0%) was generally lower than reported elsewhere (range, 17 to 29.3%) (17, 35, 41, 47). This study appears to agree with the true seasonal variation reported to exist in sheep, where the highest prevalence was recorded during the summer months (17). This variation has also been reported to occur in lambs (41) but strongly contradicts the suggestion that seasonality is absent in adult grazing ewes (41). However, the percentage of C. coli present in sheep during the current work (59.4%) is greater than reported elsewhere. The proportion of C. coli isolated from sheep ranged from 8.7% to a maximum of 47% (17, 23, 35, 41, 47). Neither the difference in Campylobacter prevalence between the cattle and sheep nor the seasonal variation was apparent during a study comparing spatiotemporal homogeneity of cattle and sheep in Scotland (35), but this may be partially due to the variation in the ruminants sampled and to farm practices.
Cattle and sheep showed similar rates of Campylobacter shedding. The shedding rates for Campylobacter from positive cattle was 2,700 CFU g−1 (95% CI, 1,305 to 4,405 CFU g−1), greater than the 920 CFU g−1 recorded for positive cows over 4 months old (30) but considerably lower than the average of 2.7 × 104 CFU g−1 (35). The consistent shedding rates and ROL recorded for cattle throughout the sampling season (Fig. 3 and 4) agrees with the absence of temporal variation in the shedding concentrations of beef and mixed cattle (19, 35, 40). However, temporal variation in the concentrations shed by dairy cattle has been suggested to exist (40). The number of Campylobacter shed in the feces of positive sheep was 5,800 CFU g−1 (95% CI, 2,252 to 10,112 CFU g−1). In contrast to the cattle counts, a significant temporal reduction in the concentration of Campylobacter shed in sheep feces was present, and it continued to decline between early and late summer (Fig. 3 and 4). Lambs have also been reported to show a general decline in numbers of Campylobacter shed (most probable number [MPN]) between May and September (41). This may provide further information on the presence and absence of the much debated seasonal trends of both Campylobacter prevalence and shedding in ruminants. Very few studies have compared cattle and sheep prevalences in conjunction with shedding rates within the same farming environment over time.
The overall Campylobacter population in cattle was diverse, with 21 different STs (Fig. 5). More than a single sequence type was isolated from 13.5% (10/74) of individual fecal pats, similar to the reported 12.5% of cattle feces that had more than 1 different serotype isolated (31). However, this is likely an underestimate, since due to material, time, and space limitations, multipicks were purposely performed (5 colonies from each positive sample) only during the two large sampling periods at the beginning and end of the study. It is therefore likely that some of the fecal samples had more than one ST present but failed to be selected. The diversity of STs and CCs was lower in sheep than in cattle, where 9 different STs were identified. In sheep, only two fecal samples had 2 STs isolated (6.67% [95% CI, 2.3 to 15.6%]), generally showing fewer multiple shedders than in cattle.
The 3 most common CCs isolated from cattle (CC-61, 21.8%; CC-48, 16.1%; and CC-42, 12.6%) contributed half of all isolates from cattle, were persistently dominant, and were isolated at each of the 3 sampling times. All of these CCs have been previously reported to be among the most common subtypes isolated from cattle (9, 12, 26). Clonal complex 61 has been found in similar percentages in cattle, ranging from 21.8 to 24.2% (21, 25, 26, 45); CC-48 in cattle has ranged from 7.69 to 26.1% (9, 12, 25, 26). The third most common CC (CC-42) has also been isolated from cattle in proportions ranging from 0% to 20.5% (9, 24–26). The three most common CCs identified from sheep were C. coli CC-828 (59.4%) and C. jejuni CC-61 (12.5%), and CC-45 (12.5%), and they contributed 84.4% of all isolates from sheep. The high proportion of C. coli CC-828 (ST-962, ST-825, and ST-854) (59.4%) identified from sheep in the current study contrasts with only 3.9% isolated from sheep in the United Kingdom (21). Many studies screen for only C. jejuni (rather than C. coli), which may partially explain why the proportion of C. coli isolated from sheep in the United Kingdom is low. An interesting observation is that both Scotland and Grampian also appear to show a greater predominance of C. coli CC-828 in sheep (33.0% and 36.4%, respectively) (45), but whether this is due to a sampling bias (i.e., testing for both species) or a true geographic variance is unknown. Clonal complex 61 has already been associated with bovine (12, 26) and ovine (9) origins. In sheep, the percentage of CC-61 (ST-61) was identical to the value recorded by Manning et al. (26) at 12.5%. Here, the prevalence of CC-45 in sheep (12.5%) (ST-45 and ST-137) was higher than in other reports, where it ranged from 0.0 to 5.0% (9, 26). Clonal complex 45 has previously been associated with a range of other hosts, as well as with “other” environmental sources, such as cats and dogs (24), poultry, humans (9, 26), wildlife, and water (12), and therefore, this CC may have the ability to adapt to a number of different hosts.
In the current work, no temporal variation in the STs or CCs was observed in cattle, and this is supported by other reports, in which amplified fragment length polymorphism (AFLP) and MLST also failed to detect any time-related changes in the composition of the Campylobacter population in cattle (20, 35). Only CC-61 showed changes in its isolation rates from 5 dairy farms between spring and summer (25), which is the only known occurrence of significant temporal differences in the genotypic makeup of the Campylobacter population in cattle. This study supports the notion that the Campylobacter population in cattle can sufficiently maintain itself via transmission between individuals within the herd (20) and that the influx of less well-adapted Campylobacter types from other environmental sources (30) is resisted by the presence and persistence of dominant and host-adapted genotypes (9). This is further emphasized by the fact that no changes were observed in the genotypic composition during the replacement of 30 cattle within the herd with new cattle. From the three STs isolated from sheep that belonged to CC-828, only ST-962 (2.3%) was present in cattle. This may be due to low bovine specificity restricting any extensive transfer from sheep, which is further supported by the fact that ST-962, ST-825, and ST-854 have not yet been recorded from cattle (21, 45). In strong contrast to the cattle, the CCs and STs isolated from sheep here showed statistically significant temporal changes in their compositions. The proportions of STs present in sheep significantly differed between the start and end of the study, and the CCs consistently showed significant differences between the 3 sampling periods.
Although some degree of overlap is apparent between Campylobacter isolates from bovine and ovine sources, it is evident from this study that there are large differences in the frequencies of particular genotypes. Only 3 of the 27 STs and 4 of the 14 CCs isolated in this study were shared between cattle and sheep. It has been suggested that despite significant differences in the subtypes identified in cattle and sheep, the common STs are the same for both hosts (35). Here, only 1 CC (CC-61) was among the top 3 isolated from both cattle and sheep. The present study is valuable because comparisons between cattle and sheep are often made from isolates acquired from different locations or farms. In a spatiotemporal analysis of Campylobacter in both cattle and sheep, it was suggested that an increase in similarities of genotype composition occurred only when the collections were made from the same farm at the same time (35). Here, the two hosts were regularly mixed on the same farm, thus having similar environmental and Campylobacter exposures, yet they continued to show highly significant differences in their Campylobacter CC and ST carriage.
Conclusion.
Cattle appear to have a higher prevalence and a more diverse but stable population of Campylobacter than sheep, where the only temporal variation was in the percentage of positive animals. Sheep in this study were more subject to temporal change; Campylobacter prevalence, shedding rates, and the frequencies of subtypes in the Campylobacter population evidently differed with time. This provides further evidence that a strong host association exists between ovine and bovine sources for some sequence types and contributes to the currently limited knowledge on the temporal patterns of the Campylobacter population in ovine sources. These differences between cattle and sheep need to be further researched and confirmed, especially when attempting to assess the importance of ruminants as a source of human infection.
ACKNOWLEDGMENTS
We thank Oxford University for performing the sequencing for use in MLST. We also thank those who reviewed the paper for their input and constructive criticism.
We thank the Biotechnology and Biological Sciences Research Council for funding the Ph.D. studentship and the FSA for funding the sequencing.
Footnotes
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Açik M. N., Cetinkaya B. 2006. Heterogeneity of Campylobacter jejuni and Campylobacter coli strains from healthy sheep. Vet. Microbiol. 115: 370–375 [DOI] [PubMed] [Google Scholar]
- 2. Adhikari B., Connolly J. H., Madie P., Davies P. R. 2004. Prevalence and clonal diversity of Campylobacter jejuni from dairy farms and urban sources. N Z. Vet. J. 52: 378–383 [DOI] [PubMed] [Google Scholar]
- 3. Adhikari B., et al. 2002. Wild birds, flies, and rodents as reservoirs of Campylobacter spp. on dairy farm. MAF technical paper 2002/18. Ministry of Agriculture and Forestry, Wellington, New Zealand [Google Scholar]
- 4. Besser T. E., et al. 2005. Increasing prevalence of Campylobacter jejuni in feedlot cattle through the feeding period. Appl. Environ. Microbiol. 71: 5752–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blaser M. J. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176: S103–S105 [DOI] [PubMed] [Google Scholar]
- 6. Brown P. E., et al. 2004. Frequency and spatial distribution of environmental Campylobacter spp. Appl. Environ. Microbiol. 70: 6501–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buzby J. C., Allos B. M., Roberts T. 1997. The economic burden of Campylobacter-associated Guillain-Barre syndrome. J. Infect. Dis. 176: S192–S197 [DOI] [PubMed] [Google Scholar]
- 8. Centers for Disease Control and Prevention 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb. Mortal. Wkly. Rep. 59: 418–422 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5914a2.htm Accessed February 2011 [PubMed] [Google Scholar]
- 9. Colles F. M., Jones K., Harding R. M., Maiden M. C. J. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69: 7409–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dingle K. E., et al. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39: 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Excoffier L., Laval G., Schneider S. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1: 47–50 http://www.la-press.com/article.php?article_id=188 Accessed February 2011 [PMC free article] [PubMed] [Google Scholar]
- 12. French N., et al. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7: 1116–1126 [DOI] [PubMed] [Google Scholar]
- 13. Frost J. A., Gillespie I. A., O'Brien S. J. 2002. Public health implications of Campylobacter outbreaks in England and Wales, 1995–9: epidemiological and microbiological investigations. Epidemiol. Infect. 128: 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia M. M., et al. 1985. Isolation, characterization, and serotyping of Campylobacter jejuni and Campylobacter coli from slaughter cattle. Appl. Environ. Microbiol. 49: 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gormley F. J., et al. 2008. Has retail chicken played a role in the decline of human campylobacteriosis? Appl. Environ. Microbiol. 74: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goudet J., Raymond M., deMeeus T., Rousset F. 1996. Testing differentiation in diploid populations. Genetics 144: 1933–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grove-White D. H., Leatherbarrow A. J. H., Cripps P. J., Diggle P. J., French N. P. 2010. Temporal and farm-management-associated variation in the faecal-pat prevalence of Campylobacter jejuni in ruminants. Epidemiol. Infect. 138: 549–558 [DOI] [PubMed] [Google Scholar]
- 18. Hoar B. R., Atwill E. R., Elmi C., Utterback W. W., Edmondson A. J. 1999. Comparison of fecal samples collected per rectum and off the ground for estimation of environmental contamination attributable to beef cattle. Am. J. Vet. Res. 60: 1352–1356 [PubMed] [Google Scholar]
- 19. Inglis G. D., Kalischuk L. D., Busz H. W. 2004. Chronic shedding of Campylobacter species in beef cattle. J. Appl. Microbiol. 97: 410–420 [DOI] [PubMed] [Google Scholar]
- 20. Johnsen G., et al. 2006. Intestinal carriage of Campylobacter jejuni and Campylobacter coli among cattle from South-western Norway and comparative genotyping of bovine and human isolates by amplified-fragment length polymorphism. Acta Vet. Scand. 48: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jolley K., Chan M.-S. February 2011, accession date PubMLST: Campylobacter jejuni multilocus sequence typing website. Oxford University, Oxford, United Kingdom: http://pubmlst.org/campylobacter/ [Google Scholar]
- 22. Jones K. 2001. Campylobacters in water, sewage and the environment. J. Appl. Microbiol. 90: 68S–79S [DOI] [PubMed] [Google Scholar]
- 23. Jones K., Howard S., Wallace J. S. 1999. Intermittent shedding of thermophilic Campylobacters by sheep at pasture. J. Appl. Microbiol. 86: 531–536 [DOI] [PubMed] [Google Scholar]
- 24. Kärenlampi R., Rautelin H., Schonberg-Norio D., Paulin L., Hanninen M. L. 2007. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwan P. S. L., et al. 2008. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74: 3626–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manning G., et al. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69: 6370–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCarthy N. D., et al. 2007. Host-associated genetic import in Campylobacter jejuni. Emerg. Infect. Dis. 13: 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller W. G., et al. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43: 2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minihan D., et al. 2004. Campylobacter spp. in Irish feedlot cattle: a longitudinal study involving pre-harvest and harvest phases of the food chain. J. Vet. Med. B Infect. Dis. Vet. Public Health 51: 28–33 [DOI] [PubMed] [Google Scholar]
- 30. Nielsen E. M. 2002. Occurrence and strain diversity of thermophilic Campylobacters in cattle of different age groups in dairy herds. Lett. Appl. Microbiol. 35: 85–89 [DOI] [PubMed] [Google Scholar]
- 31. Nielsen E. M., Engberg J., Madsen M. 1997. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol. Med. Microbiol. 19: 47–56 [DOI] [PubMed] [Google Scholar]
- 32. Pebody R. G., Ryan M. J., Wall P. G. 1997. Outbreaks of Campylobacter infection: rare events for a common pathogen. Commun. Dis. Rep. CDR Rev. 7: R33–R37 [PubMed] [Google Scholar]
- 33. Pollock K., Locking M., Browning L., Smith-Palmer A., Brownlie S. 2010. Gastro-intestinal and foodborne infections; laboratory reports for common bacterial, protozoal and viral Infections 2009. HPS Weekly 44: 38–41 http://www.documents.hps.scot.nhs.uk/ewr/pdf2010/1005.pdf Accessed February 2011 [Google Scholar]
- 34. Raymond M., Rousset F. 1995. An exact test for population differentiation. Evolution 49: 1280–1283 [DOI] [PubMed] [Google Scholar]
- 35. Rotariu O., et al. 2009. Spatiotemporal homogeneity of Campylobacter subtypes from cattle and sheep across northeastern and southwestern Scotland. Appl. Environ. Microbiol. 75: 6275–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheppard S. K., et al. 2009. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 134: 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sheppard S., et al. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48: 1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith K. A., Frost J. P. 2000. Nitrogen excretion by farm livestock with respect to land spreading requirements and controlling nitrogen losses to ground and surface waters. Part 1: cattle and sheep. Bioresour. Technol. 71: 173–181 [Google Scholar]
- 39. Sproston E. L., et al. 2010. Multi-locus sequence types of Campylobacter carried by flies and slugs acquired from local ruminant faeces. J. Appl. Microbiol. 109: 829–838 [DOI] [PubMed] [Google Scholar]
- 40. Stanley K. N., Wallace J. S., Currie J. E., Diggle P. J., Jones K. 1998. The seasonal variation of thermophilic Campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85: 472–480 [DOI] [PubMed] [Google Scholar]
- 41. Stanley K. N., Wallace J. S., Currie J. E., Diggle P. J., Jones K. 1998. Seasonal variation of thermophilic Campylobacters in lambs at slaughter. J. Appl. Microbiol. 84: 1111–1116 [DOI] [PubMed] [Google Scholar]
- 42. Strachan N., et al. 2009. Attribution of Campylobacter infections in Northeast Scotland to specific sources by use of multilocus sequence typing. J. Infect. Dis. 199: 1205–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strachan N. J. C., MacRae M., Ogden I. D. 2005. Quantification of the Escherichia coli O157 reservoir in Grampian, Scotland. Vet. Rec. 156: 282–283 [DOI] [PubMed] [Google Scholar]
- 44. Uitenbroek D. G. 1997. SISA-binomial. http://www.quantitativeskills.com/sisa/statistics/twoby2.htm Accessed February 2011
- 45. University of Aberdeen 2008. Database for CaMPS—the campylobacter MLST project in Scotland. http://www.food.gov.uk/multimedia/pdfs/publication/fullreportcamps.pdf Accessed February 2011
- 46. Wesley I. V., et al. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl. Environ. Microbiol. 66: 1994–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zweifel C., Zychowska M. A., Stephan R. 2004. Prevalence and characteristics of shiga toxin-producing Escherichia coli, Salmonella spp. and Campylobacter spp. isolated from slaughtered sheep in Switzerland. Int. J. Food Microbiol. 92: 45–53 [DOI] [PubMed] [Google Scholar]