Abstract
A new pathway of molinate mineralization has recently been described. Among the five members of the mixed culture able to promote such a process, Gulosibacter molinativorax ON4T has been observed to promote the initial breakdown of the herbicide into ethanethiol and azepane-1-carboxylate. In the current study, the gene encoding the enzyme responsible for molinate hydrolysis was identified and heterologously expressed, and the resultant active protein was purified and characterized. Nucleotide sequence analysis revealed that the gene encodes a 465-amino-acid protein of the metal-dependent hydrolase A subfamily of the amidohydrolase superfamily with a predicted molecular mass of 50.9 kDa. Molinate hydrolase shares the highest amino acid sequence identity (48 to 50%) with phenylurea hydrolases of Arthrobacter globiformis and Mycobacterium brisbanense. However, in contrast to previously described members of the metal-dependent hydrolase A subfamily, molinate hydrolase contains cobalt as the only active-site metal.
INTRODUCTION
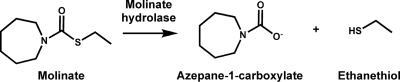
Molinate is a herbicide which is extensively applied to rice fields worldwide. When this thiocarbamate herbicide is applied to the flooded paddies, it dissipates into the environment largely through volatilization. However, (photo)chemical and microbiological molinate transformation also occurs (28), resulting in accumulation of oxidized metabolites, such as oxomolinate and molinate sulfoxide (11, 13), which have increased toxicity (5). The only biological system described so far as being able to mineralize molinate and use the herbicide as the sole source of carbon, energy, and nitrogen is a five-membered bacterial mixed culture (2, 6). Among the five community members, Gulosibacter molinativorax ON4T (gen. nov., sp. nov.), the only representative of this genus thus far (21), is responsible for the initial breakdown of molinate. In contrast to previously described molinate transformation reactions (13), G. molinativorax ON4T cleaves the thioester bond of molinate, releasing ethanethiol and azepane-1-carboxylate (ACA) (Fig. 1) (1). While this hydrolysis proceeds in the absence of oxygen, the further metabolism and mineralization of ACA by G. molinativorax ON4T necessitate oxygen. Ethanethiol is not transformed by this organism but is spontaneously oxidized to diethyl disulfide. At molinate concentrations of >2 mM, the accumulating sulfur compounds are toxic for G. molinativorax ON4T and molinate mineralization is achieved only when other mixed-culture members able to degrade the sulfur-containing metabolites are present (1, 2).
Fig. 1.
Transformation of molinate by molinate hydrolase in G. molinativorax ON4T.
In the present study, the molinate hydrolase (MolA) from G. molinativorax ON4T responsible for the key step of molinate breakdown was purified and characterized. The encoding gene was identified and expressed in Escherichia coli. This enzyme belongs to the metal-dependent hydrolase A subfamily of the amidohydrolase superfamily; however, in contrast to previously characterized members, it contains cobalt as a cofactor. To the best of our knowledge, this is the first description of an enzyme involved in molinate breakdown.
MATERIALS AND METHODS
Chemicals.
Molinate (S-ethyl azepane-1-carbothioate [C9H17NOS] of 97% purity) was obtained from Herbex, Produtos Químicos (Portugal). Analytical-standard grade S-ethyl dipropyl(thiocarbamate) (EPTC; C9H19NOS), vernolate [S-propyl dipropyl(thiocarbamate) (C10H21NOS)], cycloate [S-ethyl cyclohexyl(ethyl)thiocarbamate (C11H21NOS)], thiobencarb [S-4-chlorobenzyl diethyl(thiocarbamate) (C12H16ClNOS)], and 1,10-phenanthroline (C12H8N2·H2O) were obtained from Sigma-Aldrich. All other reagents were analytical grade and from commercial sources.
Bacterial strain, growth conditions, and preparation of cell extracts.
G. molinativorax ON4T was grown in Luria broth medium with 1 mM molinate at 30°C and 120 rpm until late exponential growth phase. Cell extracts were obtained by resuspending the harvested cells in 50 mM phosphate buffer (pH 7.4), followed by disruption with a French press (Aminco, Silver Spring, MD) and centrifugation at 66,000 × g for 40 min at 4°C.
Purification of native enzyme.
Native molinate hydrolase was purified using fast protein liquid chromatography (FPLC; Amersham Biosciences, United Kingdom). All columns were purchased from GE Healthcare (United Kingdom) unless otherwise specified. Cell extracts were loaded onto a MonoQ 5/50GL column, and elution was carried out with a 0 to 0.5 M NaCl linear gradient over 33 ml at a flow rate of 0.3 ml min−1. Active fractions were pooled, mixed with 4 M (NH4)2SO4 to obtain a final concentration of 1 M (NH4)2SO4, and applied to a Source 15PHE PE 4.6/100 column. Proteins were eluted with a 1 to 0 M (NH4)2SO4 linear gradient over 33 ml at a flow rate of 0.3 ml min−1. Active fractions were pooled and concentrated using a Centricon YM-30 filter (Millipore).
Electrophoretic methods.
The purification efficiency was evaluated by electrophoresis on 12.5% SDS-polyacrylamide gels, essentially as previously described (20). The proteins were stained with Page Blue protein staining solution according to the manufacturer's instructions (Fermentas, Lithuania), and Precision Plus protein standards (Bio-Rad) were used as markers. Gel images were analyzed using Quantity One one-dimensional analysis software (Bio-Rad).
Two-dimensional (2D) gel electrophoreses were performed with 100 to 200 mg of protein as previously described (12). In brief, isoelectric focusing was performed on ReadyStrip IPG strips (17 cm; pH 3 to 10 or 4 to 7; Bio-Rad), and second-dimension separation was performed on 10 to 15% SDS-polyacrylamide gels (20 by 20 cm2). The quantification of molinate hydrolase was performed as previously described (16).
Amino acid sequencing.
N-terminal amino acid and inner protein sequences were determined by Edman degradation and quadrupole time-of-flight mass spectrometric (Q-TOF) sequencing, respectively, as previously described (12).
Determination of molecular mass.
The molecular mass of native molinate hydrolase was determined by gel filtration using a Superose 12 HR 10/10 column (Amersham Pharmacia Biotech). The proteins were eluted with 16 ml of 50 mM phosphate buffer (pH 7.4) supplemented with 150 mM NaCl at a flow rate of 0.2 ml min−1. The column was calibrated for molecular mass determinations using chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), aldolase (158 kDa), and ferritin (440 kDa), obtained from Bio-Rad.
Transmission electron microscopy (TEM) analysis.
The purified enzyme was negatively stained for energy-filtered transmission electron microscopy (EFTEM) and analyzed in the elastic bright-field mode (energy slit width, 10 eV; objective aperture, 60 μm), as previously described in detail (30), using a Libra120 Plus electron microscope (Zeiss, Germany).
Identification of the gene encoding molinate hydrolase.
Genomic DNA of G. molinativorax ON4T as the template was extracted as described previously (18). Part of the molinate hydrolase-encoding gene was amplified by PCR at an annealing temperature of 50°C using the degenerate primers 5′-ACNATHGCNATHGTNGGNGG-3′ and 5′-CCNGCNARNARNGCNGCCATNGG-3′, designed on the basis of the determined N-terminal sequence and the internal peptide sequence (underlined) MGETIAIVGGTLIDGN and MGDPMAALLAGTANPAK. The single approximately 1.2-kb fragment obtained was cloned into the pGEM-T Easy vector (Promega) and sequenced. The regions flanking the ∼1.2-kb fragment were obtained by arbitrary PCR (ARB-PCR) (4). The specific primers targeting the molinate hydrolase-encoding gene to be used in ARB-PCR were designed on the basis of regions of this fragment that had not been used for degenerated primer design. The specific primers 5′-TCGGATGTACCTCGATCTGG-3′ and 5′-CTGCCGTCTTCAATGAACACG-3′ were used in the first and second cycles of arbitrary PCR, respectively, to confirm the N terminus, and the specific primers 5′-TATGTCCCAGGGTGAACTCC-3′ and 5′-AGGACCACTTCGTGTGGATG-3′ were used to reach the stop codon (see the figure in the supplemental material). The ARB-PCR products were separated by electrophoresis in agarose gels, and bands were excised and cloned into pGEM-T Easy (Promega). The obtained gene sequence was confirmed by PCR amplification using genomic DNA of G. molinativorax ON4T and sequencing with 16 specific primers.
Recombinant production of molinate hydrolase.
The molinate hydrolase-encoding gene (molA) was amplified from genomic DNA of G. molinativorax ON4T using primers 5′-GGGGTACCGAGAATCTTTATTTTCAGGGAGAAACGATCGCGATTGT-3′ and 5′-CCCAAGCTTCTAGTCGAGGACGTTCGCA-3′ (in the two sequences, the KpnI and HindIII restriction sites, respectively, are underlined), Pfu DNA polymerase (Fermentas), and an annealing temperature of 60°C. The approximately 1.4-kb fragment obtained was ligated into pGEM-T Easy (Promega), and ligation products were used to transform the ultracompetent strain E. coli JM109 (Promega). The transformants were grown on Luria broth-agar plates supplemented with 100 μg ml−1 ampicillin and 1% glucose and screened for the proper insert size by PCR amplification with M13 primers. The insert integrity was verified by sequencing. For preparing a strep-tagged fusion, the molinate hydrolase-encoding sequence was subcloned via the KpnI and HindIII restriction sites into plasmid pASK-IBA7plus (IBA, Germany). The ligation product was transformed into E. coli JM109 (Promega). For protein production, the strain was grown at 30°C in Luria broth medium containing 100 μg ml−1 ampicillin. Cells were induced with 0.2 μg ml−1 anhydrotetracycline at an A600 of 1.0 and harvested after 18 h of incubation at 15°C. The strep-tagged recombinant protein was purified from E. coli JM109(pASKmolA) cell extracts using a Strep-Tactin Sepharose gravity flow column (IBA) with a 1-ml bed volume. The column was washed using Tris-HCl (100 mM, with 150 mM NaCl, pH 8.0), and strep-tagged molinate hydrolase was eluted with Tris-HCl (100 mM, with 150 mM NaCl and 2.5 mM desthiobiotin, pH 8.0) at a flow rate of 0.1 ml min−1. Active fractions were pooled, concentrated (30,000-molecular-weight cutoff [MWCO]; Vivaspin), and further purified by size-exclusion chromatography using a Superose 12 PC 3.2/30 column (Amersham Pharmacia Biotech). Proteins were eluted with 24 ml of Tris-HCl (100 mM, pH 8.0, supplemented with 150 mM NaCl) at a flow rate of 0.3 ml min−1.
DNA extraction, quantification, and sequencing.
All PCR products were separated on 1% (wt/vol) agarose gels, and bands were visualized by ethidium bromide staining (25). Prior to sequencing, PCR products were purified with a QIAquick PCR purification kit (Qiagen, Germany), and plasmid DNA was extracted with a GeneJET plasmid miniprep kit (Fermentas). DNA quantification was performed using a Quant-iT double-stranded DNA HS assay kit with a Qubit fluorometer (Invitrogen) or through direct quantification using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific). All purified products were sequenced on both strands using a BigDye (version 1.1) ready reaction cycle sequencing kit (Applied Biosystems, Foster City, CA) on an ABI 373A automatic DNA sequencer (Perkin-Elmer, Applied Biosystems).
Sequence analysis.
The nucleotide sequences were translated into protein sequences (http://www.expasy.ch/tools/dna.html), and protein similarity searches were performed by PSI-BLASTN using default parameters (http://www.ncbi.nlm.nih.gov/). The sequences were aligned using the MUSCLE program (8). Phylogenetic analyses were performed with MEGA4 software (29), using the neighbor-joining method (24) with p-distance correction and pairwise deletion of gaps. A consensus tree was inferred from a total of 100 bootstrap trees generated.
Biochemical studies.
Molinate hydrolase activity was assayed by following substrate depletion through high-performance liquid chromatography as previously described (2). Appropriate amounts of cell extracts or 0.1 μM molinate hydrolase were incubated with 0.5 mM molinate in 50 mM phosphate buffer (pH 7.4 at 25°C) for up to 1 h. One unit is defined as the activity transforming 1 μmol of molinate per min. Recombinant molinate hydrolase stability was tested by measuring residual activity after incubation at −80, −20, and 4°C and at environmental temperature for up to 24 h. The substrate range was assessed using 0.1 to 0.8 μM molinate hydrolase incubated with 0.1 to 1.0 mM EPTC, vernolate, cycloate, or thiobencarb as substrates in 50 mM phosphate buffer (pH 7.4 at 25°C) for up to 24 h. The pH profile was determined over the pH range of 5.0 to 9.0 at 30°C using 50 mM sodium acetate (pH 5.0 to 6.0), 50 mM phosphate (pH 7.0 to 7.4), and 50 mM Tris-HCl (pH 8.0 to 9.0). The temperature profile was determined over the range of 10 to 50°C at pH 7.4. The pH and temperature profiles were determined using 0.1 μM recombinant protein incubated with 0.5 mM molinate. Kinetic data were assessed using 0.1 μM protein incubated with 0.05 to 2.0 mM molinate at 25°C and pH 7.4 for native and recombinant molinate hydrolase or at 30°C at pH 5.0 to 9.0 for recombinant protein and calculated from the initial velocities using the Michaelis-Menten equation by nonlinear regression (KaleidaGraph, version 3.6, software; Synergy Software). Standard protein quantification was performed using the method of Bradford (3).
Metal-free native enzyme was prepared by incubation of 0.3 μM molinate hydrolase with 4 mM 1,10-phenanthroline for 8 h at room temperature. 1,10-Phenanthroline was removed using a PD-10 desalting column (Amersham). For reactivation, the apoenzyme (0.6 μM) was incubated with divalent metals (10 μM to 10 mM) for 30 min at 25°C in the presence or absence of 100 mM sodium bicarbonate (26), followed by activity determination.
Metal analysis.
To determine the metal content, purified proteins were concentrated (30,000-MWCO; Vivaspin), washed using metal-free phosphate buffer (50 mM, pH 7.4), and quantified by measuring the absorbance at 280 nm, using the theoretical extinction coefficients 43,555 and 50,545 M−1 cm−1 for native and recombinant proteins, respectively (http://expasy.org/tools/protparam.html). Protein samples (400 μg) and the eluates from the final concentration step were digested in 4 ml of concentrated nitric acid (suprapure) by heating on a water bath at 95°C until dryness (3 h), resuspended in 4 ml of high-purity water, and analyzed by induction-coupled plasma-atomic emission spectroscopy (ICP-AES) and induction-coupled plasma-mass spectroscopy (ICP-MS) at the analytical laboratories of Requimte (New University of Lisbon) and the University of Aveiro, respectively.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was deposited in the DDBJ/EMBL/GenBank databases under the accession no. FN985594.
RESULTS
Purification of native molinate hydrolase.
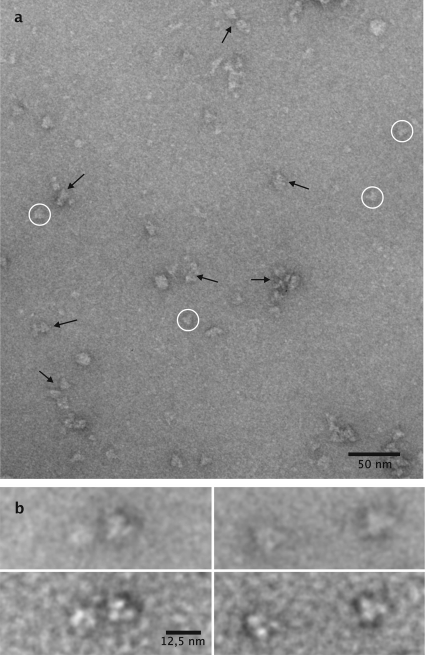
Molinate hydrolase was purified 90-fold from cell extracts by subsequent ion-exchange and hydrophobic interaction chromatography, giving a preparation with a specific activity of 505 ± 19 U g−1 protein. Molinate transformation into ethanethiol and ACA was confirmed by proton nuclear magnetic resonance (NMR) (data not shown). SDS-PAGE revealed a dominant band accounting for 95% of total protein, corresponding to a molecular mass of ∼50 kDa. The molecular mass of the native molinate hydrolase was estimated by gel filtration to be 310 ± 20 kDa. Purified and negatively stained enzyme particles of molinate hydrolase were analyzed by EFTEM (Fig. 2a). The sample is morphologically diverse; however, a characteristic triangular molecule (circled) appears to be the main constituent. The triangular molecules have morphologies comprising an edge length of 11.1 nm (mean, 11.1 ± 1.2 nm; n = 112; minimum, 8.0 nm; maximum, 15.5 nm; median, 11.2 nm; variance, 1.4 nm). Close-up views of individual particles (Fig. 2b) show the molecule to be composed of three protein masses at the vertices of the triangle, where the half-edge triangle length has a diameter of approximately 5.5 nm, roughly the equivalent of 60 to 70 kDa for a spheric protein (31), which is approximated to encompass two subunit mass equivalents. Characteristic bipartite edge-on views could be observed and are indicated by black arrows (Fig. 2a). The morphological data and size estimations based on gel-exclusion chromatography suggest that the molinate hydrolase might be composed of three planar dimers, which is a composition consistent with a homohexameric structure. However, from a strict morphological point of view, it has to be kept in mind that the triangular top-view projection is an isomorph with a distorted tetrahedral conformation of the enzyme molecule. This molecular arrangement would show two subunit masses in top-view projection in the plane, with two subunits oriented perpendicularly and visible only as a third single center of mass of 2 times 50 kDa (see Fig. 2b for close inspection). In this case, the data suggest a homotetramer structure with a lower molecular mass. Given the low accuracy of size-exclusion chromatography in the determination of the molecular mass of a protein, further studies are needed to confirm the quaternary structure of molinate hydrolase.
Fig. 2.
Electron microscopic view of purified molinate hydrolase negatively stained with uranyl acetate. (a) An overview, which shows triangular protein masses (circled) to be the dominant molecules. Morphologies indicated by arrows represent biparticulate protein masses of the triangular molecules seen as edge-on views. (b) Detailed view of individual triangular protein masses either focused close to the Gaussian focus (upper row) or at 2.0 μm underfocused (lower row) for maximum contrast and shape discrimination.
Gene identification and sequence analysis.
The N-terminal sequence of the major protein spot observed after 2-D gel electrophoresis (GETIAIVGGTLIDGN) showed significant similarity (11 out of 15 amino acids) to a putative amidohydrolase family protein from the gammaproteobacterium NOR51-B (ZP_04958230). In addition, three of the nine inner sequences (VTTVFDTWNALEPVTIAR, VPLLTHTTSLEGLNTAIER, and VVVLDQDPLADITNMR) identified showed significant similarity (up to 78% identity) to phenylurea hydrolases (PUHs) of Arthrobacter globiformis (PuhA; GenBank accession no. ACL11849) or Mycobacterium brisbanense (PuhB; GenBank accession no. ACL11830) (19). On the basis of the N-terminal and inner sequences of the native protein, the gene encoding molinate hydrolase was localized as described in Materials and Methods. The gene (molA) comprises 1,398 bp and encodes a protein of 465 amino acids with a predicted molecular mass of 50.9 kDa. All nine inner peptide sequences predicted by Q-TOF sequencing and the N-terminal sequence could be localized in the deduced protein sequence (see the figure in the supplemental material).
molA encodes a protein of the metal-dependent hydrolase A subfamily (cd01299) of the amidohydrolase superfamily. The highest similarity (48 to 50% amino acid sequence identity) was observed with phenylurea hydrolases from A. globiformis D47 and Mycobacterium brisbanense JK1 (19).
Recombinant protein production and purification.
To confirm that the identified gene encodes an active molinate hydrolase, a fragment comprising the gene was inserted into the expression vector pASKIBA7plus, giving pASKmolA. The highest activity in cell extracts of E. coli JM109(pASKmolA) was obtained from cells induced at 15°C. Under these induction conditions, the specific activity observed in cell extracts (4 ± 0.1 U g−1 protein) was similar to that of cell extracts of G. molinativorax ON4T (6 ± 0.2 U g−1 protein). Lower activity was found in E. coli cells induced at higher temperatures. The recombinant protein was purified by subsequent affinity binding and size-exclusion chromatography, giving a preparation of >97% purity with a specific activity of 496 ± 28 U g−1, similar to that of the native protein.
Substrate specificity and kinetic properties.
Molinate hydrolase was highly stable at pH 7.4, and >98% of activity could be recovered after incubation for 24 h at temperatures between −80°C and room temperature. The kinetic constants, determined at 25°C and pH 7.4, of both native and recombinant proteins were very similar, with kcat and kcat/Km values of approximately 40 min−1 and 0.14 μM−1 min−1, respectively (Table 1).
Table 1.
pH dependence of kinetic parameters kcat, Km, and kcat/Km for molinate hydrolase
| Protein | pH | Temp (°C) | kcat (s−1) | Km (M) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|---|
| Recombinant | 5.0 | 30 | 1.8 × 10−4 ± 2.2 × 10−5 | 2.5 × 10−4 ± 2.5 × 10−4 | 0.71 ± 0.30 |
| Recombinant | 7.4 | 25 | 6.7 × 10−4 ± 2.8 × 10−5 | 2.7 × 10−4 ± 3.3 × 10−5 | 2.54 ± 0.33 |
| Native | 7.4 | 25 | 6.6 × 10−4 ± 1.0 × 10−5 | 2.8 × 10−4 ± 1.3 × 10−5 | 2.40 ± 0.12 |
| Recombinant | 7.4 | 30 | 6.9 × 10−4 ± 2.2 × 10−5 | 2.1 × 10−4 ± 2.3 × 10−5 | 3.30 ± 0.38 |
| Recombinant | 9.0 | 30 | 3.2 × 10−4 ± 5.4 × 10−5 | 5.2 × 10−4 ± 2.0 × 10−4 | 0.62 ± 0.26 |
Except for molinate, none of the other thiocarbamate herbicides tested were transformed at detectable rates (<0.002% that of molinate), irrespective of the substrate and/or protein concentration, suggesting that molinate hydrolase is highly specific for molinate. The fact that molinate hydrolase does not transform EPTC (which differs from molinate only by the presence of two propyl substituents on the central nitrogen, whereas in molinate a cyclic azepane ring is formed) suggests that the bulkiness of the substrate prevents access to the active site.
Molinate hydrolase showed a temperature and pH optimum for kcat of approximately 30°C and 7.5, respectively, with a significant decrease toward higher and lower temperatures and pH values; approximately 20 and 30% of maximum activity were observed at 10°C, 50°C, pH 5 and at pH 9.0, respectively. Similarly, the kcat/Km value decreased significantly toward both higher and lower pH values, whereas a rise in the Km value was observed only at higher pH (Table 1).
Evaluation of molinate hydrolase metal dependence.
As molinate hydrolase showed similarity with metal-dependent hydrolases, the presence of metal ions in both native and recombinant proteins was determined by ICP-MS. For both proteins, of the metals analyzed (Zn2+, Mg2+, Mn2+, Fe2+, Co2+, and Ni2+), only Co2+ was observed in amounts exceeding those for the control. The cobalt ion content of recombinant protein determined by both ICP-AES and ICP-MS was 1.1 ± 0.2 mol per mol of subunit.
To evaluate the metal dependence and assess if Co2+ is required for activity, metal-free native enzyme was prepared by incubation of purified protein with the chelating agent 1,10-phenanthroline. The obtained enzyme showed a residual specific activity of 1.5% ± 0.5% that of the control, which had been incubated under the same conditions but in the absence of 1,10-phenanthroline. According to the loss of activity, the enzyme was shown by ICP-MS to be almost free of Co2+ (∼0.02 mol per mol subunit).
Reactivation of metal-free native enzyme was attempted with Co2+. The activity of the protein increased as a function of the cobalt concentration, reaching almost complete reactivation (72% ± 3%) when incubated in the presence of 100 μM Co2+. Higher concentrations of Co2+ (0.5 to 10 mM) were shown to be inhibitory, and only 40 to 29% of specific activity was recovered. Incubation with 25 μM Zn2+ resulted in a recovery of 58% ± 3% of specific activity. Lower recovery values (∼32%) were obtained with 25 μM Mn2+, indicating that zinc and, to a lesser extent, manganese are capable of replacing cobalt in the active enzyme.
The presence of 100 mM sodium bicarbonate in reactivation experiments did not affect enzymatic activity.
DISCUSSION
In this study, molinate hydrolase from G. molinativorax ON4T catalyzing the hydrolysis of molinate to form ACA and ethanethiol was identified and characterized. High homology (>40% of amino acid identity) was observed only with the phenylurea hydrolases PuhA and PuhB. These enzymes, which are involved in the degradation of several phenylurea herbicides such as diuron and linuron (19), were found to oligomerize as homohexamers. Analysis of proteins of subfamily cd1292 identified after a PSI BLAST search to be related to molinate hydrolase identified a set of 853 proteins to be the most closely related. This set included proteins where either a structure or a function has been described and which exhibited amino acid sequence identities with molinate hydrolase of 21 to 25% and similarities of 36 to 40% (a representative subset of 172 related proteins is shown in Fig. 3). Proteins for which a function is described comprise prolidases, such as those from Microbacterium (formerly Aureobacterium) esteraromaticum (GenBank accession no. BAA77794) (17), organophosphorus insecticide hydrolase from Arthrobacter sp. strain B-5 (GenBank accession no. BAA85881), (23), or aryldialkylphosphatase from Nocardia sp. B-1 (GenBank accession no. JC1378) (22). Crystal structures for enzymes of validated function are available for two prolidases (the Sgx9260b environmental sequence from the Sargasso Sea, Protein Data Bank [PDB] accession number 3MKV, and Sgx9260c, PDB accession numbers 3FEQ and 3N2C) (33), two enzymes that catalyze the hydrolysis of l-Xaa–l-Arg/Lys dipeptides (Sgx9359b, PDB accession code 3BE7 and 3DUG; Caulobacter crescentus CB-15 Cc2672, PDB accession number 3MTW) (34), and a peptidase specific for the hydrolysis of l-Xaa–l-hydrophobic dipeptides (Sgx9355e, PDB accession number 2QS8) (32). These proteins contain the common metal-binding motif of amidohydrolase members (Hisa-X-Hisb-X-Lys-X-Hisc-X-Hisd-X-Asp) (27, 32–34). Even though these proteins were reported to contain variable amounts of Zn (up to 2 mol per mol of subunit), all of them are supposed to exhibit the typical subtype I binuclear metal center of proteins of the amidohydrolase family (27, 32–34). In these proteins, Znα is coordinated by Hisa, Hisb, and Asp and Znβ is coordinated by Hisc and Hisd. The carboxylated Lys residue bridges the two metal ions (27, 32–34). However, in the substrate-free Sgx9260c structure, the Lys residue is not carboxylated, and thus, the metal center is partially occupied with Zn+ and partially filled with water (33), whereas Sgx9355e crystallized without zinc (32). The same motif (Hisa-X-Hisb-X83-140-Lys-X25-70-Hisc-X18-25-Hisd-X52-143-Asp) was the most common among the ∼850 proteins closely related to molinate hydrolase. However, some variations were observed. Like in molinate hydrolase, Hisa is replaced by asparagine in previously described phenylurea hydrolases (19). This replacement, however, might not affect metal coordination, since asparagine is known to be involved in transition metal ion coordination in other metalloenzymes (15). However, further studies are needed to identify the metal-binding residues in both molinate and phenylurea hydrolases. Interestingly, in a group of 30 proteins from Mycobacterium spp. and Frankia spp., represented by an amidohydrolase from Frankia sp. EAN1pec (GenBank accession no. YP_0010505758), the Hisa-X-Hisb motif is replaced by Glu-X-Asn, and in a group of 59 proteins, represented by an amidohydrolase from Shewanella benthica KT99 (GenBank accession no. ZP_02158917), Hisc is replaced by Glu and Asp is replaced by His (Fig. 4). However, the functions and metal-binding characteristics of both groups are so far unknown.
Fig. 3.
Evolutionary relationships of members of the metal-dependent hydrolase A subfamily (cd1292) of the amidohydrolase superfamily. The evolutionary histories were inferred using the neighbor-joining method and the p-distance model. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons. Phylogenetic analyses were conducted in MEGA4 (17). Proteins are indicated by their GenBank or PDB accession number and the host strain. The bar represents 0.05 amino acid difference per site. Proteins of validated function are indicated by filled circles, proteins not sharing the His-His-Lys-His-His-Asp metal-binding motif are indicated by filled diamonds, and the protein of unknown function where a crystal structure is available is indicated by an open diamond.
Fig. 4.
Multiple-sequence alignment of protein regions of molinate hydrolase and selected members of the metal-dependent hydrolase A subfamily (cd1292) of the amidohydrolase superfamily. Putative metal-binding residues, typically of a His-His-Lys-His-His-Asp motif, are boxed. C., Caulobacter; M., Microbacterium; So., Sorangium; Sh., Shewanella.
The majority of the members described in the amidohydrolase superfamily are zinc dependent (divalent zinc ion or dinuclear zinc pair) (27). Also, PuhA and PuhB are described as containing a mononuclear zinc ion per subunit (19). Dinuclear Ni2+ and mono Fe2+ centers are present in some other metal-dependent hydrolases (27). Less common is the presence of heterodinuclear metal centers, as described for the phosphotriesterases responsible for the degradation of organophosphate insecticides in Agrobacterium radiobacter (OpdA) (14) and Sulfolobus solfataricus (SsoPox) (9). SsoPox contains a heterodinuclear Fe2+-Co2+ center (9). A similar heterodinuclear center was found in OpdA heterologously expressed in E. coli, whereas the native protein contains a heterodinuclear Fe2+-Zn2+ center (14).
For molinate hydrolase, the loss of activity in the presence of 1,10-phenanthroline and its reactivation by cobalt suggest that the protein is a Co-dependent hydrolase. Additionally, the observed cobalt stoichiometry and the absence of significant amounts of other metals, together with the fact that metal reconstitution of enzyme activity does not require lysine carbamylation, suggest that a mononuclear Co2+ is sufficient to have activity. This is a unique characteristic among the metal-dependent hydrolases of the amidohydrolase family.
It was shown that approximately 50% of molinate hydrolase activity could be achieved by addition of zinc to metal-free protein. The fact that cobalt ion can often substitute for zinc in vitro, giving comparable or higher levels of enzyme activity, is known (14, 15) and suggests that cobalt and zinc can bind to similar motifs. In fact, Glu, Asp, and His were shown to coordinate Co2+ in the cobalt-dependent prolidase of Pyrococcus furiosus (7, 10).
Metal-bound water molecules are known to be good nucleophiles from many well-studied hydrolases, e.g., the zinc l-X–l-Arg/Lys (34) and l-X–l-hydrophobic dipeptidases (32) or cobalt prolidases (7), supporting the hypothesis that the nucleophile used by molinate hydrolase is a cobalt-bound hydroxide ion. The metal-binding sites and bound metals that contribute to the structure and activity of molinate hydrolase are under investigation. A crystal structure will be necessary to confirm the active-site architecture.
Microbial molinate transformation was first reported in the literature as occurring through cometabolism and resulting in the accumulation of partially oxidized products such as the acid, alcohol, hydroxy, oxo, sulfone, and sulfoxide molinate derivatives (11, 13). To the best of our knowledge, G. molinativorax ON4T is the only organism so far known to be able to hydrolyze molinate into ACA and ethanethiol through the activity of a molinate hydrolase. Except for molinate, molinate hydrolase has no activity on other thiocarbamates, suggesting that the protein evolved in G. molinativorax ON4T to permit the utilization of this herbicide as a nutrient. However, it is difficult at the moment to hypothesize about possible ancestors of this enzyme, given the low similarity of molinate hydrolase with the majority of known proteins. G. molinativorax ON4T was isolated from an enrichment of soil and water collected at a site where the molinate-containing effluent of a pesticide-producing industry was discharged for several years. This fact supports specialization to use this herbicide as a nutrient. Several attempts were made to isolate other strains from pesticide-contaminated soils and water. However, G. molinativorax ON4T is still the only strain so far isolated and characterized, suggesting that it may be present in low numbers or be a slow grower in such habitats.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Acções Integradas Luso-Alemãs (A4/07 and A5/10) and the Fundação para a Ciência e a Tecnologia from the Ministério da Ciência e do Ensino Superior, Portugal (project PTDC/66653/AMB/2006).
We gratefully acknowledge Herbex, Produtos Químicos SA, for supplying molinate, as well as Luísa Barreiros and Ivone Moreira from LEPAE, Beatriz Cámara for experimental support, and Agatha Bielecka and Iris Plumeier from HZI for technical assistance. We are grateful to Victor Wray from HZI and Manuel Ferrer from CSIC, Madrid, Spain, for help with NMR and ICP-MS analyses, respectively. We are indebted to Melissa Wos-Oxley for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Barreiros L., et al. 2008. New insights into a bacterial metabolic and detoxifying association responsible for the mineralization of the thiocarbamate herbicide molinate. Microbiology 154:1038–1046 [DOI] [PubMed] [Google Scholar]
- 2. Barreiros L., et al. 2003. A novel pathway for mineralization of the thiocarbamate herbicide molinate by a defined bacterial mixed culture. Environ. Microbiol. 5:944–953 [DOI] [PubMed] [Google Scholar]
- 3. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 4. Caetano-Anollés G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85–94 [DOI] [PubMed] [Google Scholar]
- 5. Cochran R. C., Formoli T. A., Pfeifer K. F., Aldous C. N. 1997. Characterization of risks associated with the use of molinate. Regul. Toxicol. Pharmacol. 25:146–157 [DOI] [PubMed] [Google Scholar]
- 6. Correia P., Boaventura R. A., Reis M. A., Nunes O. C. 2006. Effect of operating parameters on molinate biodegradation. Water Res. 40:331–340 [DOI] [PubMed] [Google Scholar]
- 7. Du X., Tove S., Kast-Hutcheson K., Grunden A. M. 2005. Characterization of the dinuclear metal center of Pyrococcus furiosus prolidase by analysis of targeted mutants. FEBS Lett. 579:6140–6146 [DOI] [PubMed] [Google Scholar]
- 8. Edgar R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elias M., et al. 2008. Structural basis for natural lactonase and promiscuous phosphotriesterase activities. J. Mol. Biol. 379:1017–1028 [DOI] [PubMed] [Google Scholar]
- 10. Ghosh M., Grunden A. M., Dunn D. M., Weiss R., Adams M. W. 1998. Characterization of native and recombinant forms of an unusual cobalt-dependent proline dipeptidase (prolidase) from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 180:4781–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Golovleva L. A., Popovich N. A., Skryabin G. K. 1981. Transformation of ordram by microorganisms. Izv. Akad. Nauk. SSSR Ser. Biol. 3:348–358 [PubMed] [Google Scholar]
- 12. Heim S., Lleo M. M., Bonato B., Guzman C. A., Canepari P. 2002. The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. J. Bacteriol. 184:6739–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imai Y., Kuwatsuka S. 1986. Metabolic pathways of the herbicide molinate in four strains of isolated soil microorganisms. J. Pesticide Sci. 11:245–251 [Google Scholar]
- 14. Jackson C. J., et al. 2006. Anomalous scattering analysis of Agrobacterium radiobacter phosphotriesterase: the prominent role of iron in the heterobinuclear active site. Biochem. J. 397:501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jackson C. J., et al. 2007. The structure and function of a novel glycerophosphodiesterase from Enterobacter aerogenes. J. Mol. Biol. 367:1047–1062 [DOI] [PubMed] [Google Scholar]
- 16. Junca H., Plumeier I., Hecht H. J., Pieper D. H. 2004. Difference in kinetic behaviour of catechol 2,3-dioxygenase variants from a polluted environment. Microbiology 150:4181–4187 [DOI] [PubMed] [Google Scholar]
- 17. Kabashima T., Fujii M., Hamasaki Y., Ito K., Yoshimoto T. 1999. Cloning of a novel prolidase gene from Aureobacterium esteraromaticum. Biochim. Biophys. Acta 1429:516–520 [DOI] [PubMed] [Google Scholar]
- 18. Kanakaraj R., Harris D. L., Songer J. G., Bosworth B. 1998. Multiplex PCR assay for detection of Clostridium perfringens in feces and intestinal contents of pigs and in swine feed. Vet. Microbiol. 63:29–38 [DOI] [PubMed] [Google Scholar]
- 19. Khurana J. L., et al. 2009. Characterization of the phenylurea hydrolases A and B: founding members of a novel amidohydrolase subgroup. Biochem. J. 418:431–441 [DOI] [PubMed] [Google Scholar]
- 20. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 21. Manaia C. M., Nogales B., Weiss N., Nunes O. C. 2004. Gulosibacter molinativorax gen. nov., sp. nov., a molinate degrading bacterium, and classification of Brevibacterium helvolum DSM 20419 as Pseudoclavibacter helvolus gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 54:783–789 [DOI] [PubMed] [Google Scholar]
- 22. Mulbry W. W. 1992. The aryldialkylphosphatase-encoding gene adpB from Nocardia sp. strain B-1: cloning, sequencing and expression in Escherichia coli. Gene 121:149–153 [DOI] [PubMed] [Google Scholar]
- 23. Oshiro K., Kakuta T., Nikaidou N., Watanabe T., Uchiyama T. 1999. Molecular cloning and nucleotide sequencing of organophosphorus insecticide hydrolase gene from Arthrobacter sp. strain B-5. J. Biosci. Bioeng. 87:531–534 [DOI] [PubMed] [Google Scholar]
- 24. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 25. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Seffernick J. L., et al. 2002. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP is a metalloenzyme. Biochemistry 41:14430–14437 [DOI] [PubMed] [Google Scholar]
- 27. Seibert C. M., Rauschel F. M. 2005. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44:6383–6391 [DOI] [PubMed] [Google Scholar]
- 28. Soderquist C. J., Bowers J. B., Crosby D. G. 1977. Dissipation of molinate in a rice field. J. Agric. Food Chem. 25:940–945 [Google Scholar]
- 29. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary 405 genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 30. Winkler J., Lünsdorf H., Jokusch B. M. 1996. The ultrastructure of chicken gizzard vinculin as visualized by high-resolution electron microscopy. J. Struct. Biol. 116:270–277 [DOI] [PubMed] [Google Scholar]
- 31. Wrigley N. G. 1988. Electron micrographs of protein molecules: interconversion of size, shape and molecular weight made easy. Proc. R. Microsc. Soc. 23:299–302 [Google Scholar]
- 32. Xiang D. F., et al. 2009. Functional annotation of two new carboxypeptidases from the amidohydrolase superfamily of enzymes. Biochemistry 48:4567–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiang D. F., et al. 2010. Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily. Biochemistry 49:6791–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiang D. F., et al. 2009. Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes. Biochemistry 48:3730–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.