Abstract
The path of substrates in the multidrug efflux pump AcrB of Escherichia coli was examined by using labeling with a lipophilic substrate mimic, Bodipy FL maleimide. Four (out of eight) residues in the vestibule bound the dye, suggesting its role in substrate transport, whereas only one (out of nine) residue in the central cavity tested positive.
TEXT
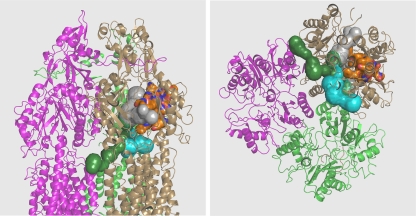
AcrB, a homotrimeric transporter protein, when complexed with TolC and AcrA forms an efficient multidrug efflux pump that confers much of the intrinsic resistance of Escherichia coli against a wide variety of antibacterial compounds (7, 9). Its asymmetric trimer structure was solved in 2006-2007 (6, 11, 12). Importantly, a large binding pocket deep in its periplasmic domain (Fig. 1, orange surface) was previously identified with substrates (6), and this site is thought to play a predominant role in the selection of drugs for export. This binding pocket is connected to the large cleft on the surface of the periplasmic domain by a large tunnel (Fig. 1, gray tunnel), whose branches also lead to the large central cavity (Fig. 1, cyan tunnel) as well as to the “vestibule” between protomers, located close to the membrane surface (Fig. 1, green tunnel) (6). This network of tunnels may represent the path substrates take before finally reaching the deep binding pocket for export. We have recently tested this idea by changing many residues facing the putative drug pathway into Cys and examining the covalent labeling of these residues with a low concentration of Bodipy FL N-(2-aminoethyl) maleimide (here called Bodipy FL maleimide) (3). This rather lipophilic reagent apparently behaves as an AcrB substrate, and in traveling through the normal substrate path(s) labels only the residues facing the tunnel and the binding site. Remarkably, Cys residues away from the path remained unlabeled, even when they were in the middle of a hydrophobic surface. However, our previous study was limited mostly to the residues lining the large, short tunnel connecting the cleft to the binding pocket. We now examine, in this study, the residues facing the central cavity as well as the vestibule.
Fig. 1.
Possible substrate pathways leading to the deep binding pocket. The structure of the AcrB asymmetric trimer 2DRD (6) is shown in cartoon representations. The binding protomer is shown in the sand color, the extrusion protomer in green, and the access protomer in mauve. The deep binding pocket (identified by the binding of minocycline [6]) is shown as an orange surface. A wide tunnel (gray) leads from the external cleft of the periplasmic domain to this pocket. This tunnel is also connected to the branches that lead to the vestibule (green) and central cavity (cyan). Left, side view; right, view from the bottom. Portions of the protein close to the viewer have been clipped off to better reveal the tunnels. The tunnels were visualized with the program CAVER (10), and the drawing was made with PyMol (http://www.pymol.org).
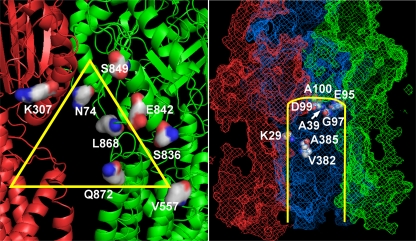
We selected K29, A39, E95, G97, D99, A100, V382, A385, and S462 from the central cavity and N74, K307, V557, S836, E842, S849, L868, and Q872 from the vestibule (Fig. 2). These selections were guided by the observation that their side chains faced the tunnels in the crystal structure 2DRD (6). Each of these residues was changed into Cys by using the QuikChange (Stratagene) approach in the plasmid pSPORT1-CLBH, expressing cysteine-less AcrB with a hexahistidine tag at its C terminus (13). RAM-1334 (MC4100 Δara ΔacrB) was the host strain, and the whole cells were treated with 6 μM Bodipy FL maleimide (Invitrogen) for 1 h at room temperature exactly as described earlier (3). (That Bodipy FL maleimide is an AcrB substrate was confirmed by the observation that the labeling of endogenous proteins under similar conditions increased significantly in RAM1334, in comparison with its isogenic acrB+ parent RAM1336 [Hong-Suk Kim, personal communication]). Cells were then lysed, and AcrB was isolated (3). The protein was resolved by SDS-PAGE, and the labeling was quantified by fluorescence as described earlier (3). The same gel was treated with Coomassie stain to quantify the protein bands. In every experiment, N274C, which is in the deep binding pocket and is strongly labeled by the dye (3), and K708C, which faces the periplasm just outside the cleft entrance and was shown not to be labeled by the dye (3), were included as positive and negative controls. Every assay was repeated at least twice.
Fig. 2.
Residues examined. (Left) Residues in the vestibule. Shown is the structure of the vestibule (yellow triangle), between two protomers of model 2DRD (6), in red and green. (Right) Residues in the central cavity (S462 is behind the G97 residue and is not visible). The protein is shown in a mesh representation and corresponds to access (red), binding (blue), and extrusion (green) protomers of model 2DRD. The yellow lines show the approximate position of the central cavity, with the proximal part of the model cut off to reveal the examined residues (in space-filling models).
We first tested for the functionality of the Cys mutant transporters by determining the susceptibility for novobiocin and erythromycin, both good substrates for AcrB efflux, using a disc inhibition assay. In this assay, most Cys mutants showed inhibition zones of the same size as the unaltered CLBHpump (7.0 mm for novobiocin and 9.5 mm for erythromycin), except G97C and R307C, which showed somewhat larger zones of inhibition (10.0 and 11.0 mm, respectively, for novobiocin and 12.0 and 14.5 mm for erythromycin). However, the zones were still significantly smaller than with the strain expressing no AcrB (15.0 mm for novobiocin and 17.5 mm for erythromycin), suggesting only a partial defect in efflux.
In the Bodipy FL maleimide modification assay of the residues in the vestibule area, four out of the eight residues tested, S836C, E842C, L868C, and Q872C, stained significantly, with S836C staining most strongly (Fig. 3). The positive residues were further tested by modification with a bulky substituent, pyrene maleimide, followed by the assay for the possible blocking of the efflux path, using Nile Red as the substrate (3). However, none of the residues gave positive results with this assay, perhaps due to the wide dimensions of the vestibule, as would be expected (Fig. 2, left).
Fig. 3.
Bodipy FL maleimide labeling of AcrB residues in the vestibule and the central cavity. N274C and K708C are positive and negative controls, respectively. The residues K29C, A39C, E95C, G97C, D99C, A100C, V382C, A385C, and S462C are in the central cavity. The residues N74C, R307C, V557C, S836C, E842C, S849C, L868C, and Q872 are in the vestibule. Each lane represents the labeling of a single cysteine residue by Bodipy FL maleimide (top) and a Coomassie stain (bottom) of the same gel. The labeling intensities (numbers below each lane) have been normalized (corrected for the amount of AcrB protein, determined with Coomassie stain), based on two independent experiments. The normalized labeling intensity in the positive control, N274C, was set as 100. Those residues showing intensities of <30 were considered negative.
Vestibules between protomer units were originally proposed as the sites of substrate entry by Murakami and coworkers in 2002 (5), in part because the units were very close to the outer surface of the cytoplasmic membrane and thus could accommodate amphipathic substrates that were hypothesized to partition partially into the membrane bilayer (7). The connection of the vestibule to the deep binding pocket (Fig. 1) was emphasized again by Murakami and coworkers in 2006 (6). Although the fact that AcrB substrates must contain a substantial lipophilic surface may be the outcome of the hydrophobic nature of the binding pocket and may not necessarily indicate their partial insertion into the membrane, it was gratifying to see that many of the residues facing the vestibule became labeled with Bodipy FL maleimide, suggesting the importance of the vestibule (in addition to the cleft, which is in the middle of the periplasm, far away from the membrane surface) in substrate capture.
When nine residues in the central cavity were tested in the Bodipy FL maleimide modification assay, only G97C gave a significant positive result. The neighboring residues T98C (from the previous study [3]), E95C, D99C, and A100C were negative. Again, as expected from the wide dimension of this large cavity, the efflux blocking assay with pyrene maleimide gave negative results with G97C.
The role of the central cavity in substrate capture is difficult to assess, especially because only one residue (G97C) tested gave a positive result. A further complication is that this Gly residue is located at the tip of a turn, and changing this residue is expected to alter the conformation of the surrounding residues. This result also demands close attention because the central cavity capture means that the substrate will enter into the export pathway not only from the periplasm but also from the cytosol. Some substrates were found in the central cavity in an early attempt at cocrystallization (17). However, the site-directed mutagenesis of nearby residues was later shown to have no discernible effect on activity (16), although their modification was initially thought to inactivate efflux (17). Furthermore, the observation (4, 14) that for the efficient efflux of drugs from cytosol, tripartite pumps, such as AcrB-AcrA-TolC, require help from singlet efflux pumps that extrude drugs into the periplasm suggests that the cytosolic capture by AcrB is at least inefficient. Although proteoliposome reconstitution of an AcrB homolog, AcrD, seemed to suggest that aminoglycosides are captured both from the periplasm and cytosol (1), streptomycin showed no evidence of cytosolic capture, and a recent reexamination of the data suggested that cytosolic capture of other aminoglycosides is marginal at best (8). However, the recent demonstration that the AcrB homolog MexB was active when expressed in a Gram-positive host (15) suggests that such capture does occur. Furthermore, in the AcrB homolog EmhB of Pseudomonas fluorescens, site-directed mutagenesis of residues at the top of the central cavity significantly affected the efflux of substrates (2). A more thorough investigation of this possible pathway is needed.
Acknowledgments
We thank Attilio V. Vargiu for his help in identifying candidate residues, especially in the vestibule area, and Hong-Suk Kim for carrying out the fluorescent labeling experiments of endogenous proteins.
This study was supported in part by a grant from the U.S. Public Health Service (AI-09644).
Footnotes
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Aires J. R., Nikaido H. 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 187:1923–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hearn E. M., Gray M. R., Foght J. M. 2006. Mutations in the central cavity and periplasmic domain affect efflux activity of the resistance-nodulation-division pump EmhB from Pseudomonas fluorescens cLP6a. J. Bacteriol. 188:115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Husain F., Nikaido H. 2010. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol. Microbiol. 78:320–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee A., et al. 2000. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J. Bacteriol. 182:3142–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murakami S., Nakashima R., Yamashita E., Yamaguchi A. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–593 [DOI] [PubMed] [Google Scholar]
- 6. Murakami S., Nakashima R., Yamashita E., Matsumoto T., Yamaguchi A. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179 [DOI] [PubMed] [Google Scholar]
- 7. Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nikaido H. 2011. Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77:1–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nikaido H., Takatsuka Y. 2009. Mechanisms of RND efflux pumps. Biochim. Biophys. Acta 1794:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Petrek M., et al. 2006. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics 7:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seeger M. A., et al. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295–1298 [DOI] [PubMed] [Google Scholar]
- 12. Sennhauser G., Amstutz P., Briand C., Storchenegger O., Grütter M. G. 2007. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 5:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takatsuka Y., Nikaido H. 2007. Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J. Bacteriol. 189:8677–8684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tal N., Schuldiner S. 2009. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc. Natl. Acad. Sci. U. S. A. 106:9051–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Welch A., Awah C. U., Jing S., van Veen H. W., Venter H. 2010. Promiscuous partnering and independent activity of MexB, the multidrug transporter protein from Pseudomonas aeruginosa. Biochem. J. 430:355–364 [DOI] [PubMed] [Google Scholar]
- 16. Yu E. W., Aires J. R., McDermott G., Nikaido H. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 187:6804–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu E. W., McDermott G., Zgurskaya H. I., Nikaido H., Koshland D. E., Jr 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976–980 [DOI] [PubMed] [Google Scholar]