Abstract
We used high-resolution tiling microarrays and 5′ RNA sequencing to identify transcripts in Desulfovibrio vulgaris Hildenborough, a model sulfate-reducing bacterium. We identified the first nucleotide position for 1,124 transcripts, including 54 proteins with leaderless transcripts and another 72 genes for which a major transcript initiates within the upstream protein-coding gene, which confounds measurements of the upstream gene's expression. Sequence analysis of these promoters showed that D. vulgaris prefers −10 and −35 boxes different from those preferred by Escherichia coli. A total of 549 transcripts ended at intrinsic (rho-independent) terminators, but most of the other transcripts seemed to have variable ends. We found low-level antisense expression of most genes, and the 5′ ends of these transcripts mapped to promoter-like sequences. Because antisense expression was reduced for highly expressed genes, we suspect that elongation of nonspecific antisense transcripts is suppressed by transcription of the sense strand. Finally, we combined the transcript results with comparative analysis and proteomics data to make 505 revisions to the original annotation of 3,531 proteins: we removed 255 (7.5%) proteins, changed 123 (3.6%) start codons, and added 127 (3.7%) proteins that had been missed. Tiling data had higher coverage than shotgun proteomics and hence led to most of the corrections, but many errors probably remain. Our data are available at http://genomics.lbl.gov/supplemental/DvHtranscripts2011/.
INTRODUCTION
Desulfovibrio vulgaris Hildenborough can obtain energy by reducing sulfate to sulfide while oxidizing organic material such as lactate or pyruvate. Such sulfate-reducing bacteria play a major role in the global sulfur and carbon cycles and are key drivers of biocorrosion (32). Sulfate-reducing bacteria are also important in the bioremediation of heavy metal ions such as uranyl, chromate, or zinc, which they can reduce to insoluble forms (28, 32, 48). D. vulgaris Hildenborough has become a model for the study of sulfate-reducing bacteria, as it was the first sulfate-reducing bacterium sequenced (21), and there have been many studies of the expression patterns of its mRNAs and proteins, as well as computational efforts to identify regulatory motifs (reviewed in reference 52). We are continuing to analyze the response of D. vulgaris Hildenborough to environmental stresses as part of ENIGMA—Ecosystems and Networks Integrated with Genes and Molecular Assemblies—which seeks to understand how environmental conditions affect the bioremediation of heavy metals.
As D. vulgaris Hildenborough is quite distantly related to well-studied bacteria such as Escherichia coli or Bacillus subtilis, relatively little is known about gene regulation in this organism. Tiling arrays and next-generation sequencing have been used successfully to map transcripts in other prokaryotes (47), so we undertook to characterize the transcripts of D. vulgaris Hildenborough. This should reveal how genes are expressed and should help to infer their regulation. We used two genome-wide methods to analyze D. vulgaris Hildenborough transcripts: high-resolution tiling microarrays and 5′ RNA-Seq analysis. Whereas most microarray studies aim to quantify gene expression, the goal of our tiling array experiments was to identify transcripts and their 5′ and 3′ boundaries. We used an array with 60-nucleotide (60-nt) probes spaced every 2 to 4 nucleotides on each strand, but even with such closely spaced probes, we were able to identify transcript boundaries only to within about 30 nucleotides. Thus, we used 5′ RNA-Seq as well. In 5′ RNA-Seq, an RNA ligase tags the 5′ ends of RNAs followed by reverse transcription, amplification, and sequencing; thus, 5′ RNA-Seq identifies 5′ ends to the precise nucleotide (7, 49). To classify these 5′ ends as transcript starts or RNA degradation products, we used the tiling data and the locations of promoter-like sequences. Finally, we did not determine the precise 3′ ends of the transcripts experimentally, but we were able to infer many of them, because most of the transcripts with clear 3′ ends had putative rho-independent terminators (25).
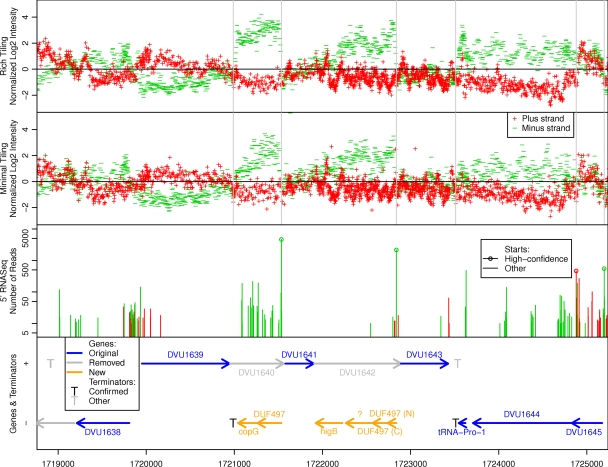
Preliminary analysis of our transcript data suggested that there were many errors in the genome annotation (the list of proteins predicted to be encoded by the genome). Although D. vulgaris Hildenborough has been the subject of many proteomics studies, we are not aware of any efforts to use proteomics data to correct its genome annotation. Thus, we combined the transcript data with shotgun proteomics data and homology evidence to revise the genome annotation. To illustrate our approach, Fig. 1 shows the tiling data and the 5′ RNA-Seq data for a six-kilobase region of the genome, along with transcript starts, rho-independent terminators, and revisions to the genome annotation.
Fig. 1.
Data for a region of the genome. We show the tiling and 5′ RNA-Seq data for kb 1719 to 1725 on the main chromosome, along with gene annotations, transcript starts, and terminators. The top two panels show normalized log levels from tiling data, with each probe plotted at its center. The genome-wide median value of 0 is shown as a horizontal black line, and vertical gray lines highlight the locations of key features from other panels, namely, high-confidence starts and confirmed terminators. The third panel shows the number of reads starting at each location across two 5′ RNA-Seq libraries from minimal media; note the log y axis. The bottom panel shows annotated genes (arrows) and predicted intrinsic terminators (25). For newly annotated genes we show which gene family they belong too, if any (DUF, domain of unknown function). Two of the newly annotated DUF497 genes have leaderless promoters, the data for the start of the transcripts for DVU1638 and for DVU1639 are ambiguous, DVU1645's transcript starts 24 nucleotides upstream of its start codon, and there is an antisense transcript for DVU1645 (an arsR-like regulator). The tiling data confirm the terminators for DUF497-copG and for tRNA-Pro-1.
MATERIALS AND METHODS
Strains and growth conditions.
Experiments were conducted within a Coy anaerobic chamber with an atmosphere of about 2% H and 5% CO, with the remainder being N. Desulfovibrio vulgaris Hildenborough (ATCC 29579; a gift from Terry Hazen's group), which was inoculated from 10% glycerol stock and grown in glass bottles with lactate-sulfate media at 30°C. Cells were collected at an optical density of around 0.3. Tiling data were collected from cells grown under two sets of conditions: one set used defined LS4D medium (30), and the other set used LS4 medium, which is LS4D medium supplemented with 0.1% (wt/vol) yeast extract. 5′ RNA-Seq data were collected using the defined LS4D medium.
RNA collection.
Bacterial pellets were collected by centrifuging cultures for 10 min at 10,000 × g and 4°C in RNase-free 50-ml polypropylene tubes. Supernatant was immediately poured off, and pellets were stored at −80°C. After thawing, RNA was extracted using RNeasy miniprep columns (Qiagen) with the optional on-column DNase treatment. RNA quality was confirmed with an Agilent Bioanalyzer; only samples with an RNA integrity number of around 9 or better were used. Ribosomal RNA (rRNA) was depleted using a MICROBExpress kit (Ambion), which uses magnetic beads coated with oligonucleotides that hybridize to rRNA. Those mRNA-enriched samples were analyzed using tiling arrays or 5′ RNA-Seq.
Tiling experiments.
First-strand cDNA was synthesized using random hexamer primers and a SuperScript indirect cDNA labeling system (Invitrogen); the reaction buffer was supplemented with actinomycin D to inhibit second-strand synthesis (36). First-strand cDNA was labeled with Alexa 555. About 2 mg of labeled first-strand cDNA was hybridized to a Nimblegen array. Nimblegen slides were scanned on an Axon Gene Pix 4200A scanner with 100% gain and analyzed with Nimblescan, with no local alignment and a border value of −1. For rich media, we used the average of the log intensities from two independent experiments, while for minimal media and the genomic control we did just one experiment.
As a control, we also hybridized genomic DNA to the tiling array. We used DNA from cells in the stationary phase to minimize copy number variations across the chromosome. Genomic DNA was extracted using a DNeasy blood tissue kit (Qiagen) and labeled using the Nimblegen comparative genomic hybridization protocol. Briefly, genomic DNA was sonicated to a level of 200 to 1,000 bp and amplified using a Klenow fragment and Cy3-labeled random nonamer primers.
To remove probes that might cross-hybridize, we mapped the probes to the genome (NC_002937 and NC_005863) with BLAT (24) and we ignored any probes whose second-best hit matched at a level of 50 or more nucleotides. We computed normalized log levels by using the genomic control and by using each probe's nucleotide content, followed by setting the median value to 0. First, we used linear regression to model the log intensity as a function of the log intensity in the genomic control and the probe's nucleotide content. To compute this model, we used only probes within the sense strands of genes because of differences in nucleotide composition between coding and noncoding regions and even between the coding and antisense strands of genes. The prediction of this model reflects the expected bias of the probe, so we subtracted this from the (raw) log intensity. We also removed the data for the 1% of probes with the lowest intensities in the genomic control, as these probes gave poor discrimination between coding and noncoding regions. Finally, we adjusted the normalized values so that their median was 0.
5′ RNA-Seq experiments.
Given an mRNA-enriched sample, we converted 5′-triphosphate ends to 5′-monophosphate with tobacco acid pyrophophatase, we blocked the 3′ ends with sodium periodate, and we added a sequencing adaptor (5′-ACACUCUUUCCCUACACGACGCUCUUCCGAUCU-3′) to the 5′ end with Ambion T4 RNA ligase (49). We used random hexamer primers with a sequencing adaptor on the 5′ end (5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTNNNNNN-3′) to obtain first-strand cDNA. We size-selected products of 150 to 500 bases from an agarose gel. We subjected the library to PCR amplification to enrich for products that contained both adaptors and to complete the 5′ adaptor by the use of primers 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ and 5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT-3′. We purified the PCR products and removed unincorporated nucleotides, primers, and adaptor-only products with AMPure XP beads (Agencourt). We also made a second library in which we used terminator 5′-phosphate-dependent exonuclease (Epicentre) to try to remove 5′-monophosphate (degraded) transcripts and then converted the 5′-triphosphate ends to 5′-monophosphate ends with RNA 5′ polyphosphatase (Epicentre) (7). Ligation and cDNA and PCR amplification conditions for the two libraries were similar. After each enzymatic reaction was performed using the exonuclease library, RNA was purified using Agencourt RNAClean XP beads. Molecules smaller than 100 nucleotides and unligated adaptors were mostly lost in these cleanup reactions.
For each library, the 32 nucleotides at the 5′ end were sequenced with a lane of Solexa by the University of California at Davis sequencing center and the reads were mapped to the genome with Eland software. Using the first library, 7.5 million reads mapped uniquely to the genome; using the second library, 15.5 million reads mapped uniquely to the genome. (Reads from ribosomal RNAs would not map uniquely, as D. vulgaris Hildenborough contains 5 to 6 nearly identical copies of each rRNA.) The two libraries gave similar results (the Spearman rank correlation coefficient of their counts was 0.67), and manual examination suggested that the exonuclease treatment had little effect. So we analyzed these libraries together and identified local peaks (within 50 nucleotides) in the combined numbers of reads in the libraries. Peaks with at least 10 reads in each library were considered potential transcript starts. (For analysis of nonspecific transcript starts, peaks with a total of 10 reads were considered.)
Identifying features in the tiling data.
Transcribed regions were defined by smoothing over 40 adjacent probes (roughly 150 nucleotides), using the moving average, and requiring a smoothed value above 0.
We identified rises or drops, corresponding to potential transcript starts and ends, based on “local correlation” to a step function (17). We used the data from 50 probes on either side of a potential rise or drop, and we investigated how similar this pattern was to a step function by measuring the absolute value of the correlation between this subset of the data and a series of −1 values followed by an equal number of +1 values. We measured the local correlation around every probe; to identify the center of the rise or drop, we used the local maximum of the local correlation within 21 probes. For transcript ends, we required a local correlation of at least 0.8 and also a 2-fold drop in intensity.
To identify a break in transcription within a potential operon or between a transcript boundary and a gene, we smoothed the normalized log level over five adjacent probes. If the minimum of the smoothed values was below 0, we identified a break in the transcript under those conditions. To identify breaks in putative operons, we also required a difference of at least 1 between the expression level of the upstream gene and that minimum.
Promoter sequence analysis.
We began with a preliminary set of 1,618 moderate-confidence transcription starts, based on a rise in rich media tiling data (a local correlation of at least 0.6 and association with a transcribed region) occurring within 30 nucleotides of a local peak in the 5′ RNA-Seq data. We extracted positions −40 to +1 relative to these putative promoters and analyzed only the strand in the orientation of transcription. We used BioProspector (27) to search for a bipartite motif with blocks of widths 10 and 8 separated by 10 to 18 nucleotides and kept the best of 12 runs of its Gibbs sampler. We used MEME software (4) to search for ungapped motifs of 30 to 35 nucleotides under the zero-or-one-occurrence-per-site model and found four significant motifs. We used patser software (23) to scan the entire genome for hits to any of the four MEME motifs and to correct for the high GC content of the D. vulgaris Hildenborough genome. For most analyses, we used only hits of 7 bits or above, which across the four motifs gave a hit every 111 nucleotides on each strand of the genome. We associated a motif hit with a 5′ RNA-Seq peak in cases in which the peak was within one nucleotide of the expected location.
Distinguishing transcription starts from RNA degradation products.
We used a semisupervised machine learning approach to classify local peaks in the 5′ RNA-Seq data as transcription starts or as “other.” For each local peak, we computed four features: (i) the total number of 5′ RNA-Seq reads mapped as starting at that location; (ii) whether the 5′ RNA-Seq peak was associated with a transcribed region and with a rise in the rich media tiling data with a local correlation of 0.6 or above, and if so, what the local correlation was; (iii) the corresponding value for minimal media tiling data, but with a threshold of 0.7 and without consideration of whether it was associated with a transcribed region; and (iv) b, the bit score of the best hit to any of the four MEME promoter motifs that occurred within 1 nucleotide of the putative transcription start, if any (weak hits of under 7 bits were ignored). For each feature, we inferred a model (a log-odds score for any given value) by comparing the distributions of the feature for transcription starts that represented results that were high or low confidence according to the other features. Specifically, we inferred an appropriate model by comparing starts with both a and b values to starts with neither. We inferred a model for rmin using an analogous method. We inferred a model for b by comparing starts with both rrich and rmin values to starts with neither. Finally, we inferred a model for rrich and rmin values by comparing starts with a total log odds score (from the other features) of above 4 to starts with a log odds score of under −4 (e4 ≈ 55, so these are about 55 times more likely to be genuine or false starts).
Models were inferred using binned subsets of the data, pseudocounts, and smoothing [see BinnedBinaryFit2() and BBFPredict() in utils.R at our website http://genomics.lbl.gov/supplemental/DvHtranscripts2011/]. We summed the log odds for each feature to get a final log odds score. This calculation is based on the assumption that the features are independently distributed among the false positives and among the true transcription starts, as in a naive Baysian classifier. Values above 0 indicate that the transcription start resembles the high-confidence transcription starts, and the magnitude of the log odds indicates the level of confidence. We considered starts above a log odds value of 4 to represent high-confidence starts. The distributions of the features for the high-confidence starts and the other starts are shown in Fig. S1 in the supplemental material. To estimate the false-positive rate, we used a randomized data set; we replaced the locations of all 5′ RNA-Seq peaks with random locations, we recomputed all features, we shuffled the resulting values to eliminate the (biological) agreement between them, and we applied the model.
Shotgun proteomics.
Mass spectra were collected for peptides derived from a variety of protein fractions from the ENIGMA project. We also used spectra from previously published whole-cell proteomics experiments performed with D. vulgaris Hildenborough grown under several sets of stress conditions (31, 42, 51). All spectra were analyzed against the six-frame translation of the genome. For protein fractions, spectra were analyzed with the Paragon algorithm in ProteinPilot 3.0 software (46), and peptides were considered confidently identified at a posterior probability of 0.95, resulting in 22,503 different peptides for 1,866 reading frames. For complete-proteome experiments, reading frames were considered confidently identified when they had a Mascot search engine score of 32 or greater, resulting in 1,556 reading frames detected. To estimate the rate of false-positive identification, we used proteins from the original annotation that were unlikely to be genuine (i.e., those that were expressed at a level at least two times higher on the antisense than on the sense strand and that lacked homology support). Of the 106 proteins identified, just five were detected, each with one peptide. Manual examination of the spectra suggested that these were false positives (H. Liu and A. M. Redding-Johanson, personal communication). Thus, the automated protein identification had a false-positive rate of around 5%. To prevent the possibility of annotation errors, spectra from proteins with a single identified peptide were checked manually as necessary.
Correcting gene annotations.
Annotated proteins that seemed inconsistent with the transcript data were checked manually before being removed or changed. We ignored genes not transcribed on either strand, as these could be expressed under some other set of conditions. Similarly, we cannot rule out the possibility that a transcript expressing the entire gene would have been seen under some other set of growth conditions; thus, a few genes with clear homology or proteomics support (using data presented above or from reference 33) were retained even though transcription in those regions was not consistent with expression of the predicted protein.
Data were viewed using Artemis software (43). Homology evidence for a given protein was examined using the domains and homologs tools on the MicrobesOnline website (13); the results showed HMMer 3 (http://hmmer.janelia.org/) or PSI-BLAST (44) hits to known families and FastBLAST (38) hits to other proteins. Potentially missed proteins were identified by examining proteomics data, by using CRITICA and RAST software, by using PSI-BLAST to search for homology to conserved domains (29) in the six-frame translation of the genome, by using blastx (16) to compare the six-frame translation of unannotated transcribed regions to annotated proteins in other organisms, and by checking candidate open reading frames (ORFs) with a MicrobesOnline sequence search and with the PFam website (http://pfam.janelia.org/). After initial changes to the annotation, we examined genes with significant overlaps. In particular, in cases in which a gene with homology or experimental support overlapped a gene without support, we removed the unsupported protein.
We began our analysis with the RefSeq database annotation from 2007. As of December 2010, the RefSeq annotation had changed (presumably based on comparative genomics analyses). The RefSeq update included 95 of the 505 changes that we made and another 23 changes (mostly changes to start codons) that are consistent with our data and with homology.
Revising operon structures.
We classified adjacent pairs of genes on the same strand based on whether there was a high-confidence “internal” transcript start (that is, between the upstream gene's start codon and the downstream gene's start codon) and whether there was a break in expression between the genes. Simple operon pairs had neither an internal transcript start nor a 2-fold drop in expression in the intergenic region. Nonoperon pairs showed a drop in expression of at least 2-fold to below a log level of 0 or had both an internal transcript start and a confirmed terminator. However, we classified pairs as having attenuators when they had a confirmed terminator but a log level of at least 0.25 throughout the intergenic region. If a pair had an internal promoter and no drop, then it was classified as a complex operon pair. The various thresholds were validated by manually examining the results.
Statistics.
All statistical tests and regressions were conducted using R software (http://www.r-project.org/).
RESULTS
We first discuss the reliability of the tiling data and the correspondence between 3′ ends of transcripts and rho-independent terminators. Then, we use 5′ RNA-Seq to identify exact transcript starts and to identify the sequence motifs for the sigma factors of D. vulgaris Hildenborough. Given the transcript boundaries, we revise the gene models (while also considering the proteomics data and homology evidence). Given the transcript boundaries and corrected gene annotations, we discuss the lengths of the 5′ and 3′ untranslated transcribed regions (UTRs), and we revise the operon structures. Finally, we show that nonspecific transcription occurs across much of the D. vulgaris Hildenborough genome and discuss potential mechanisms that might control it.
Reliability of tiling data.
We obtained tiling data for mRNA from cells grown with lactate as the carbon source and sulfate as the electron acceptor. We used both a defined minimal medium (LS4D) and a rich medium supplemented with yeast extract (LS4). We also hybridized an array to genomic DNA to measure the strength of each probe. We used this genomic control and the nucleotide content of the probes to normalize the tiling data and to estimate the level of expression at each probe. Log levels for rich and minimal media were quite similar, with a linear correlation of 0.93 across 2.004 million probes.
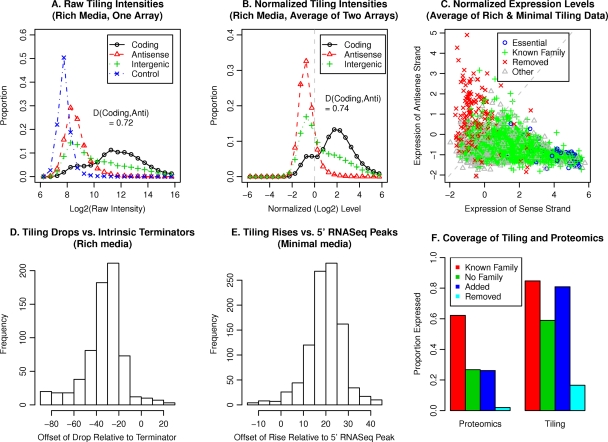
Probes for the coding regions of genes usually had higher raw intensity than did antisense probes for the opposite strand (Fig. 2A). To quantify the difference between the two distributions, we used the Kolmogorov-Smirnov D statistic, a nonparametric measure that ranges from 0 for two distributions that are identical to 1 for those that do not overlap. The D statistic improved upon normalization; for rich media, it improved from 0.72 to 0.74. The overlap between the distributions is primarily due to poorly expressed genes rather than to noise in individual probe measurements. For example, when we use only the most highly expressed two-thirds of the genes, then D improves to 0.95 for rich media. The poorly expressed genes can be seen in the left shoulder of the coding distribution (Fig. 2B) and at the left of Fig. 2C. Hundreds of genes exhibited little expression or were expressed primarily on the antisense (noncoding) strand, but genes that were expected to be essential were well expressed. As discussed below, the annotation of many of the poorly expressed regions as proteins seems questionable.
Fig. 2.
Quality and coverage of data. (A) The distribution of raw log intensities, as a function of probe type, for a single array hybridized to cDNA from rich LS4 media. Probes were classified as coding, antisense, or intergenic according to the original genome annotation; control probes have random sequences that do not match the D. vulgaris Hildenborough genome but have about the same GC content (63%). (B) The distribution of normalized log intensity for rich media (data represent averages of the results from two replicate experiments). The median value for the probes (excluding the random control probes) is 0 and is shown with a dashed vertical line. (C) The median normalized expression level for the sense and antisense strands of each protein-coding gene from the original annotation. The dashed line shows x = y. (D) The distribution of offsets between drops in the tiling data and the end of the intrinsic terminator's stem-loop. (E) The distribution of offsets between rises in the tiling data and peaks in 5′ RNA-Seq. (F) The proportions of different types of protein-coding genes that were detected by tiling or by shotgun proteomics. Genes were considered detected by tiling when the corresponding smoothed intensity value was above 0 throughout. Genes with a single high-confidence peptide were considered detected by proteomics. (A few removed “proteins” were detected, but manual examination showed that these were false positives.)
Figure 2A also shows that expression of most antisense probes was above that of control probes that did not match the genome sequence. This might reflect nonspecific transcription across the genome, as has been reported in studies of Escherichia coli (15, 45). We discuss nonspecific transcription in more detail below. The presence of nonspecific transcripts complicates the determination of a region as “expressed” or not. However, if it is assumed that the entire genome is transcribed at physiologically relevant levels on one strand or the other, the median across both strands then represents the boundary between expression and the absence of expression. As our tiling data are normalized to a median of 0, we use 0 as the threshold for expression (per the method described in reference 19).
Transcript ends.
If a transcript has a specific end, then the log level should drop sharply. By examining the “local correlation” to a step function (17) and how far the expression level dropped, we identified 771 sharp drops in tiling data from rich media and 483 sharp drops in tiling data from minimal media. For comparison, on the basis of our updated operon predictions (see below), we estimate that D. vulgaris Hildenborough has about 1,200 transcript ends.
When we compared these drops to predictions for intrinsic (rho-independent) terminators from TransTermHP (25), we found that the majority of sharp drops were located at intrinsic terminators (61% in rich media and 75% in minimal media). As shown in Fig. 2D, the drop tends to be at about −30 relative to the end of the terminator's stem-loop. Because the probes are 60 nucleotides long, this implies that the drop usually occurs around a probe that ends near the termination site. Overall, we confirmed 771 of 2,978 predicted terminators, but the predicted terminators often overlapped. Combining the overlapping predictions, we confirmed 549 distinct terminators.
There were just 25 sharp drops that were found in both rich and minimal media but were not predicted by TransTermHP. We examined these manually and removed three questionable ones, leaving 22 unexplained terminators. There was a terminator prediction on the corresponding strand for 13 of these, and it appears that these terminators are bidirectional, leaving just 9 unexplained sharp drops. To understand the termination of the remaining transcripts that lacked sharp drops, we examined a random sample of 10 genes from 342 that were well expressed under both sets of conditions (median log level of 1 or higher), were expected to be at the end of their operon (based on revised predictions as described below), and lacked confirmed terminators. For 6 of the 10 genes, transcription downstream of the gene dropped gradually, without any specific end being apparent; 3 of the remaining 4 had weak drops (below our threshold values) at putative intrinsic terminators. Overall, intrinsic terminators account for virtually all of the specific transcript ends, but a significant fraction of transcripts had heterogeneous 3′ ends.
The other major mechanism for terminating transcription in bacteria involves the rho protein (reviewed in reference 8). Although rho is not well understood, it could account for the heterogeneous ends, and it is estimated to account for about 20% of termination in E. coli (37). However, we suspect that rho activity is weaker in D. vulgaris Hildenborough than in E. coli. First, we observed an operon which contains the antisense strand of an entire protein-coding gene (gidB [DVU1250]; see Fig. S2 in the supplemental material). Similar cases of operons extending through the antisense portion of an entire gene have been observed in B. subtilis, which has weak rho activity, but not, as far as we know, in E. coli (14). Second, a transposon mutagenesis project studying Desulfovibrio alaskensis G20 (formerly D. desulfuricans G20) found several insertions in rho, which suggests that rho is not required for growth in Desulfovibrios (A. Arkin laboratory, unpublished data). In contrast, rho is essential in E. coli (5, 50). Finally, large numbers of transcripts with heterogeneous 3′ ends have been reported in the archaeon Halobacterium salinarium (26) but not, as far as we know, in other bacteria. Thus, we wonder whether D. vulgaris Hildenborough has another mechanism for nonspecific termination. Further study of whether rho can be knocked out in D. vulgaris Hildenborough and what effect this would have might clarify this issue.
Inferring transcript starts from 5′ RNA-Seq and tiling data.
We extracted mRNA from cells grown in minimal LS4D media and used 5′ RNA-Seq to map the 5′ ends of the RNAs (7, 49). As shown in Fig. 1, the numbers of 5′ RNA-Seq reads show steep peaks (note the log scale). Sometimes we saw a large number of reads at one position and a much smaller number of reads at locations within 1 to 2 nucleotides; those could reflect variations in the initiation of transcription from the “same” promoter, or they might have arisen from minor errors in mapping the 5′ ends of the transcripts. In any case, each local peak corresponds to a potential transcription start. Some of these peaks may reflect degradation products rather than genuine transcription starts, but peaks in 5′ RNA-Seq that correspond to sharp rises in the tiling data should represent genuine transcription starts. Just 2.2% of 5′ RNA-Seq peaks with 20 to 500 reads lie within 30 nucleotides of a sharp rise, while 32% of peaks with over 500 reads do. (We defined a sharp rise as having a local correlation of 0.8 or above in tiling data from rich media.) Most of the peaks with many reads but no corresponding rise in the tiling data lie within highly expressed regions and probably reflect degradation products. In some cases, there are multiple 5′ RNA-Seq peaks near each other and the tiling data show a more complicated or gradual rise, which might reflect multiple start sites, but we cannot rule out the possibility that they represent degradation products.
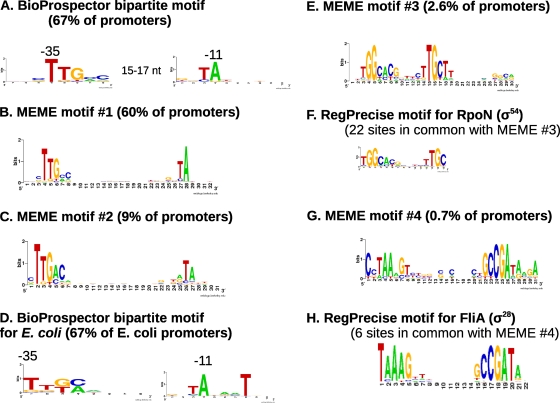
We combined our 5′ RNA-Seq data with the sharp rises in rich media to obtain a preliminary set of 1,618 transcription starts that were likely to be genuine. (For comparison, based on the revised operon predictions below, there should be around 1,900 transcript starts in D. vulgaris Hildenborough.) We searched upstream of these starts for promoter motifs. As shown in Fig. 3, we were able to reconstruct the motifs for σ70, rpoN (also known as σ54) and fliA (also known as σ28). Furthermore, we found a site at two-thirds of these locations, which is the same rate as seen in a compilation of transcript starts in E. coli (22). Thus, the transcript starts that we identified arose primarily from the initiation of transcription and not from RNA degradation. The D. vulgaris Hildenborough genome contains one other sigma factor, rpoH, but we were unable to detect this motif, and we did not detect transcription starts at predicted rpoH-dependent promoters (6, 34), so we suspect that rpoH does not have significant activity under our growth conditions.
Fig. 3.
Promoter motifs. We show motifs from analyses of the −40 to +1 regions of 1,618 moderate-confidence D. vulgaris Hildenborough transcription starts as determined using BioProspector (27) (A) and MEME (4) (B, C, E, and G). For comparison, we also show a motif determined by analysis of 370 known promoters in E. coli K-12 (22) with BioProspector (D) and motifs from RegPrecise (34) (F and H) for alternative Desulfovibrio sigma factors that were inferred by comparative genomics. Each motif is shown as a sequence logo; at each position, the height of a nucleotide is proportional to its information content in bits (11).
To predict which of the 5′ RNA-Seq peaks correspond to genuine transcription starts, we used a machine-learning approach that took into account the number of reads, the correspondence with tiling data, and the presence of a promoter-like sequence. Because we did not have any known promoters with which to train our predictor, we used high- and low-confidence subsets of the data, according to the other features, to train a model for each feature. We then combined the models into a naive Bayesian classifier. The classifier selected 1,124 of the 13,822 peaks as high-confidence transcription starts with a log odds ratio of 4 or higher (e4 ∼ 55). When we randomized the data, the same model predicted just 31 promoters, so we estimate that 3% (31/1,124) of these transcription starts represent false positives.
When we compared the locations of these high-confidence transcription starts in the 5′ RNA-Seq data and the tiling data, the 5′ RNA-Seq peak tended to be at +20 relative to the center of the rise in the tiling data (Fig. 2E). The central tendency confirms that most of the transcription starts are genuine. If 60 nucleotides of hybridization were required for a strong signal, we would expect a overlap of +30, so the location at +20 suggests that hybridization of 50 of 60 nt suffices for a signal.
Revising gene models.
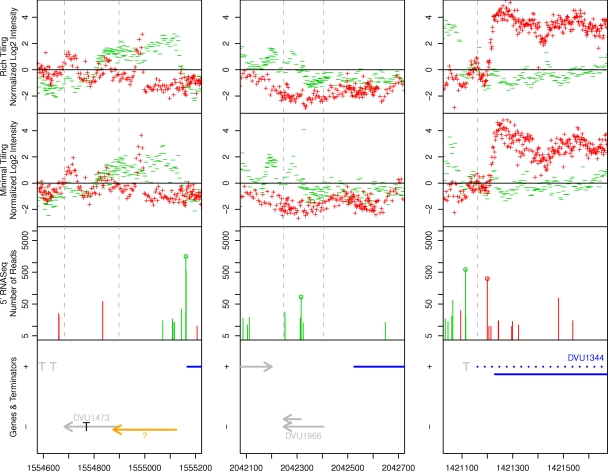
The tiling data suggested that there were numerous errors in the genome annotation, as 246 putative proteins from the original annotation were expressed on the wrong strand and lacked homology to other proteins (see, e.g., DVU1640 and DVU1642 in Fig. 1). Furthermore, we sometimes found open reading frames with homology support on the expressed strand (see, e.g., the DUF497 genes in Fig. 1). We found other suspicious patterns in the tiling data as well, such as strong terminators within putative genes, genes that were expressed only near their 3′ ends, and genes whose transcripts began downstream of their annotated start codons (Fig. 4). Together, these results showed that we needed to reconsider the genome annotation.
Fig. 4.
Examples of modified protein annotations. We show data and modifications to the annotation for three regions of the genome. Dashed vertical lines show the extents of the original gene annotations; the other plotting symbols are as described for Fig. 1. The left panel shows that DVU1473 contains a terminator, while an ORF in another reading frame is expressed from start to stop. That ORF does not belong to a known family but is homologous to other proteins, so it replaced DVU1473 in our annotation. The middle panel shows that only the C-terminal part of DVU1966 is transcribed; the upstream-most start codon that is consistent with the data shown would reduce the ORF to just 22 amino acids, so we removed it from our annotation. The right panel shows that DVU1344, as originally annotated (horizontal dotted line), begins upstream of its promoter; we selected a new start codon downstream of the promoter.
To complement the transcript data, we used peptide spectra for D. vulgaris Hildenborough from shotgun proteomics determinations within the ENIGMA project. As shown in Fig. 2F, most genes from known families were detected in the tiling data, and a majority of the genes from known families were also detected by proteomics. The other genes, which are harder to annotate, were much more likely to be detected by tiling than by proteomics, probably because the proteins encoded by those genes tend to be less highly expressed and shorter, which reduced the number of peptides that could be detected. We also reexamined the annotation of the genome by homology, as additional genomes from the (rather broad) genus of Desulfovibrio have been sequenced since the original annotation and as there have also been improvements to the gene family databases. We used two automated gene finders that consider homology information (CRITICA [3] and RAST [2]) as well as several types of BLAST.
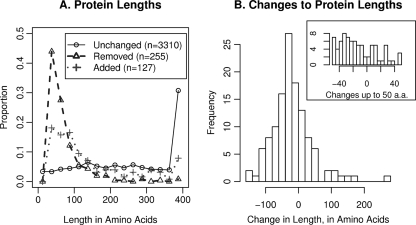
Overall, we made 505 corrections to the genome annotation. We removed 255 putative proteins: 154 were expressed primarily on the wrong strand, 44 were expressed (in the tiling data) only for a small 3′-terminal portion, 31 had internal terminators, and 26 were replaced by overlapping ORFs in another reading frame that had homology or proteomics support. Proteins with suspicious transcript structures were retained if they were detected by proteomics or had homology support, but this was not common. For example, of the putative protein-coding genes that were expressed primarily on the wrong strand, just 22% had homology support (compared to 90% of other proteins) and just 6% were detected by proteomics (compared to 55% of other proteins). As shown in Fig. 5A, most of the removed proteins were relatively short, with a median length of 54 amino acids, but we removed 43 putative proteins of 100 or more amino acids. We added 128 proteins, including 62 that were identified by both CRITICA and RAST. Of the new proteins, 32 (including 9 signaling or regulatory proteins, 3 stress resistance proteins, and 2 enzymes) had informative annotations. A total of 13 of the new proteins were not identified by any gene calling program or were originally annotated as pseudogenes; 4 of the 13 were detected by proteomics and their spectra were validated by inspection (Liu and Redding-Johanson, personal communication), and the other 9 had strong homology support. Finally, we changed 123 start codons. We moved 35 of them upstream, mostly due to proteomics. A few start codons were moved well upstream after examining genes with long gaps between the transcript start and the start codon and checking for conservation of the intervening sequence. We moved 88 start codons downstream, usually because the gene's transcript started downstream of the original start codon. As shown in Fig. 5B, many of the changes to the start codon were quite large, with a median absolute difference of 37 amino acids. Overall, 80% of proteins in our revised annotation were covered from start codon to stop codon by transcripts in the tiling data, and 54% of proteins were detected in the proteomics data.
Fig. 5.
Lengths of proteins. (A) The distribution of lengths of unchanged, removed proteins, and added proteins. Values above 400 are shown in the rightmost bin at 400. (B) The distribution of changes in length for the 123 proteins whose start codons were modified.
We were surprised at extent of these corrections and the lack of agreement between the two automated tools. CRITICA missed 12% of genes in our revised annotation, and RAST missed 7% of genes in our revised annotation. Among the genes predicted by both tools, the start codons differed 32% of the time. Conversely, 0.9% of CRITICA calls and 5.6% of RAST calls were not included in our revised annotation and are likely to represent false positives. As we were rarely able to correct start codons that were too far downstream, we expect that many of the start codons in our revised annotation are still erroneous. An accurate annotation would require proteomics with higher coverage or targeting to N-terminal peptides (1).
Leaders and UTRs.
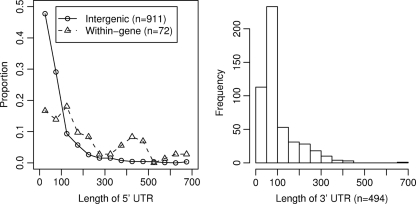
We identified 5′ and 3′ untranslated transcribed regions (UTRs) by checking whether the entire region between a transcript's boundary and the nearest gene was expressed. As discussed above, many transcripts showed nonspecific ends, which made defining the 3′ UTR problematic, so we analyzed only the 3′ UTRs for transcripts with intrinsic terminators. We defined 983 5′ UTRs and 494 3′ UTRs.
One surprise was the presence of “leaderless” promoters, where the transcript began at the first nucleotide of the start codon. Leaderless transcripts were first identified in archaea, but they have been identified in genome-wide studies in various bacteria, including Geobacter sulfurreducens PCA, which, like D. vulgaris Hildenborough, is a deltaproteobacterium (41). However, given the high rate of error in start codon annotations, we wondered whether the leaderless promoters in D. vulgaris Hildenborough were genuine. We checked the start codons for candidate leaderless promoters from a preliminary version of our analysis by asking whether homology extended to the very N-terminal end of the annotation. Of 49 of our preliminary candidates, 43 were confirmed by BLASTp and 21 of those were further confirmed by alignments to known families. The remaining 6 N-terminal regions were not conserved and might be erroneous, but most of the leaderless promoters must be genuine. Our final analysis gave 54 proteins with leaderless promoters out of the 954 proteins corresponding to the beginnings of transcripts with clearly defined starts.
As shown in Fig. 6A, the median length of the 5′ UTR is 55 nucleotides, but some genes have very long 5′ UTRs. Two operons that are central to sulfate reduction have particularly long 5′ UTRs: dsrABD, which encodes three subunits of the dissimilatory sulfite reductase, has a 5′ UTR of 289 nucleotides, and apsB, which encodes a subunit of adenylylsulfate reductase, has a 5′ UTR of 208 nucleotides. However, in general, we could not find a clear pattern for which types of genes had long 5′ UTRs. Among 5′ UTRs of over 100 nucleotides for genes on the main chromosome, about half (106/208) had some conservation in another strain, D. vulgaris Miyazaki B, according to the results of a genome alignment (12). (The Miyazaki B strain is sufficiently divergent from D. vulgaris Hildenborough that there should be no neutral conservation of nonfunctional DNA.) This suggests that many of those 5′ UTRs contain functional elements; however, we cannot be certain that they function as RNA elements rather than as alternative promoters.
Fig. 6.
Lengths of 5′ and 3′ untranslated regions.
For the 494 genes with a confirmed terminator downstream, the median length of the 3′ UTR was 68 nucleotides (Fig. 6B). Of 147 3′ UTRs of over 100 nt, just 16 contained segments that were conserved in D. vulgaris Miyazaki B; thus, we predict that few of those 3′ UTRs contain functional sequences.
Finally, we found little evidence of transcribed regions that are not associated with annotated genes. We found just 26 unannotated transcribed regions with high-confidence promoters, and after removal of the antisense transcripts, this number dropped to just 4. However, both our experimental protocols and our analysis methods are probably biased against RNAs of under 100 nucleotides, so this does not imply that D. vulgaris Hildenborough lacks small RNAs.
Revising operon structures.
Before we began this project, we had predicted operons from the distances between genes on the chromosome, how conserved the proximity of the genes was, whether the genes had similar expression patterns across a large collection of microarray experiments, and whether they were likely to have related functions (13, 39). Here, we used the transcript data to update the operon predictions. We classified each adjacent pair of genes on the same strand as a simple operon pair, as a complex operon pair with an internal operon or internal attenuator, or as a nonoperon pair. (Examples of operons with internal attenuators or internal promoters are shown in Fig. S3 in the supplemental material.) We began with our original predictions (which were intended to determine whether pairs are ever cotranscribed or not) and reclassified pairs with clear signals in our data. Ambiguous examples occurred in cases in which there was a weak drop and then a high-confidence transcription start just downstream of the drop—this could represent a genuine terminator followed by a promoter (but the tiling data lack the resolution to distinguish the drop clearly) or it could represent noise in the tiling data.
Relative to our original predictions, which included 1,558 operon pairs and 838 nonoperon pairs, we reclassified 188 nonoperon pairs as simple operons; we reclassified 14 operon pairs as nonoperons; we identified 169 complex operon pairs with internal promoters, about half of which were originally classified as operons; and we identified 17 complex operon pairs with internal attenuators, 12 of which were originally classified as operons. We were surprised at the number of pairs that were reclassified from nonoperons to simple operons. These tended to be widely spaced (median separation of 108 nucleotides) and moderately coexpressed (median Pearson correlation coefficient of 0.19), which explains why they were classified as nonoperon pairs in our original predictions. The wide spacing and the moderate coexpression also suggested that these might contain internal promoters that were missed by our automated analysis. However, only 30 of these 188 pairs had potential internal transcript starts, according to our classifier (log odds values of 0 to 4). Manual examination of 10 randomly selected cases found potential internal promoters for just 2 of the 10. The weak coexpression could have been due to internal promoters that are not active under our growth conditions or to noise in the expression compendium. Comparisons of tiling data from a wide range of growth conditions (26) would be one way to distinguish these alternatives.
Genes that are cotranscribed but also have an intergenic promoter between them show little coexpression (see Fig. S4 in the supplemental material). We suspect that this is because we can identify internal promoters with high confidence only when they are stronger than the upstream promoter so that the upstream gene is transcribed only from the upstream promoter and the downstream gene is transcribed primarily from the intergenic promoter. When there is an internal promoter that is within the upstream gene, however, we see much stronger coexpression (P < 10−4 [Wilcoxon rank-sum test]). Because the expression data were collected with 1 to 2 probes per gene and thus lack spatial resolution, we suspect that this coexpression is an experimental artifact resulting from the fact that the probe for the upstream gene hybridizes to the internal transcript, which does not include the upstream gene's start codon and cannot lead to its expression. Thus, the gene expression data for the upstream gene are misleading. Knowledge of transcript structures would allow better design of gene expression arrays.
Nonspecific antisense transcription.
As mentioned above, the tiling data suggested weak and potentially nonspecific expression of the antisense strand of most genes. The 5′ RNA-Seq data confirmed the nonspecific transcription: 1.4% of the mapped reads began at 3,983 locations within coding regions on the antisense strand of genes in our updated annotation. (For comparison, 33% of the reads corresponded to high-confidence promoters and 15% of the reads began within coding regions on the sense strand.) We then asked whether these 3,983 antisense transcript starts were located at promoter-like signals, as would be expected if they were genuine transcripts and not experimental artifacts (as seen in reference 15). We considered a weak hit (4 bits or more) to any of the four promoter motifs to represent a promoter-like site. A total of 35% of the antisense starts, but only 12% of random locations, were at promoter-like sites (P < 10−15 [Fisher exact test]). In contrast, putative starts within the sense strand of coding regions exhibited little enrichment in promoter signals, which suggests that most of them represent degradation products.
Given that we can detect nonspecific antisense transcription, we wondered how it differs across genes. Tiling data showed less antisense expression of genes that are more highly expressed on this sense strand (Fig. 2C; r = −0.40 [the rank correlation gave similar results]). If we consider only genes that are expected to be essential, then the correlation is −0.60 (P < 10−15), which shows that the effect is not due to misannotated genes or to genes that are not expressed at all. In contrast, 5′ RNA-Seq analysis showed no effect of sense expression (as quantified by tiling) on the rate of antisense transcript starts in reads per kilobase (r = −0.02; P > 0.2).
To investigate this discrepancy, we looked at the density of promoter-like sequences. In most prokaryotes, promoter-like sequences within genes are selected against and occur a bit less frequently than would expected by chance (18), and we hypothesized that promoter-like sequences would be selected against more strongly for more highly expressed genes. To avoid artifacts due to annotation errors or the edges of genes, we considered only longer genes (300 nucleotides or longer) that belong to known families. We found that highly expressed genes contained fewer internal promoter-like sites per kilobase on the sense strand (Spearman's rank correlation coefficient; r = −0.23 [P < 10−15]). However, expression levels had little effect on the rate of occurrence of promoter-like sequences on the antisense strand (r = −0.04; P = 0.06), which is consistent with the pattern determined by 5′ RNA-Seq analysis. Because the rate of promoter-like sequences on either strand is strongly correlated with GC content, we also tested the relationship using partial correlations; the effect of expression levels on sense-strand promoter motifs remained after controlling for GC content (partial r = −0.09; P < 10−15).
We propose that promoter-like sequences on the sense strand are selected against to prevent expression of truncated proteins, while transcription on the antisense strand is suppressed by transcription on the sense strand. Because we see antisense suppression in the tiling data but not in the 5′ RNA-Seq data, it appears that elongation, rather than initiation, is suppressed. Although a promoter on one strand can suppress transcription intiation from the opposite strand, that seems to rely on a specific site where the RNA polymerase pauses (35) and would not occur in most situations. We do not know what suppresses the elongation of antisense transcripts for highly expressed genes. One possibility is that elongation of antisense transcripts is suppressed because the RNA polymerase backtracks when it collides with RNA polymerase on the sense strand (10). Such collisions would occur more frequently for highly expressed genes, and the RNA polymerase on the sense strand might “win” these collisions because translating ribosomes occur closely behind the RNA polymerase on the sense strand and prevent the RNA polymerase from backtracking (40).
DISCUSSION
Evidence-based annotation of proteins.
We identified 505 changes to the protein annotation (corresponding to 15% of proteins), which were far more than we had expected. We have also collected transcript data for Desulfovibrio alaskensis G20 and found a similar number of errors in the original annotation for that species as well (Arkin laboratory, unpublished data; see also reference 20). For comparison, the annotation of Geobacter sulfurreducens PCA was updated recently using transcript data and shotgun proteomics; the updating resulted in only 144 changes (41). Desulfovibrio genomes are rich in GC, which increases the number of spurious long reading frames, and there are relatively few genome sequences for Desulfovibrios, which makes comparative gene-finding tools such as CRITICA less effective, but both of these challenges apply to G. sulfurreducens as well. Our preliminary analysis suggests that many plausible corrections to the G. sulfurreducens annotation remain: we found 39 protein-coding genes in the updated annotation that lack homology support, were not in the proteomics data, and were expressed only on the “wrong” strand. Nine of these “genes” mask unannotated proteins with homology support on the opposite strand. As the tiling and RNA-Seq experiments described in this paper cost less than sequencing a genome did a few years ago, transcriptomics could be used broadly to improve genome annotation, but new tools are needed to automate this process.
We were also surprised at the number of changes we made based on the tiling data that, in retrospect, could have been made based on homology alone. There were 24 proteins that we removed because they lacked homology support and a conflicting frame had homology support, and there were 10 proteins with homology support that were missed in the original annotation and by both RAST and CRITICA. Neither RAST nor CRITICA uses the full range of approaches to detecting protein homology; RAST relies primarily on pairwise protein comparisons to representatives of known families, and CRITICA relies on nucleotide BLAST hits. We found additional proteins by comparing sequences to those of families with PSI-BLAST (44) or HMMer (http://hmmer.janelia.org/), which can find highly diverged members of known families, and also by comparisons to hypothetical proteins that were annotated in other organisms. Faster tools (e.g., HMMer 3 [http://hmmer.janelia.org/] and FastBLAST [38]) should allow more exhaustive searches and hence more accurate automated annotation.
Experimental design.
We used very-high-resolution microarrays, with probes every 2 to 4 nucleotides. We had hoped that such a high density would let us place promoters and terminators very accurately, but this was not possible because of non-full-length hybridization to 60-mer probes. The high density of the arrays was still beneficial, as nearby probes represent a form of replicates, so that replicate arrays are not necessary. Still, it would be more cost-effective to use arrays with probes every 6 to 10 nucleotides. The savings would allow analysis of samples from more sets of conditions, which should make it much easier to identify alternate promoters (26) and would allow the detection of additional transcripts (perhaps a few percent more transcripts per additional condition) (7).
Another key issue is how best to define precise transcript boundaries. We found that untargeted RNA-Seq had too much bias to be useful (data not shown). In contrast, although our 5′ RNA-Seq protocols returned a mixture of true transcript starts and likely degradation products, we were able to identify 1,124 genuine transcription starts at a false-positive rate of a few percent by combining the reads with tiling data and sequence analysis. It is not clear how to identify the precise 3′ ends of bacterial transcripts experimentally, but RNA-Seq protocols are evolving rapidly.
Implications for analyzing gene regulation.
Our revisions to operon structures, along with the 1,124 transcript starts that we identified at nucleotide resolution, should aid the elucidation of gene regulation in D. vulgaris Hildenborough. First, the transcript structures tell us which promoter(s) controls the expression of most genes. Second, transcript starts tell us exactly where to look for sigma factor binding sites, and our data have been used to expand the regulons of the sigma factors rpoN and fliA (http://regprecise.lbl.gov/RegPrecise/). Third, because repressing sites tend to overlap the region from −35 to +1, while activating sites tend to be upstream of the −35 box (9), promoter locations would help to interpret transcription factor binding sites. For example, a preliminary analysis of computationally identified regulatory sites from RegPrecise suggested that several uncharacterized motifs act as repressors. We have also used the transcript starts to help us interpret data on where transcription factors bind in the genome (L. Rajeev and A. Mukhopadhyay, unpublished data)—these methods identify broad regions around the binding site, and knowing where the promoter is helps to focus the search for the motif.
Conclusions.
We combined tiling microarrays, 5′ RNA-Seq, and proteomics to reannotate the genes and transcripts of D. vulgaris Hildenborough. We corrected hundreds of errors in the genome annotation, but many more errors probably remain, particularly in the identification of start codons. We identified 1,124 transcription starts at nucleotide resolution and found that D. vulgaris Hildenborough prefers a motif different from that preferred by its E. coli counterpart. Many transcripts appear to have nonspecific 3′ ends. Finally, we found nonspecific transcription of the antisense strands of protein-coding genes in both the tiling and the 5′ RNA-Seq data; elongation of these nonspecific antisense transcripts seems to be suppressed by transcription of the sense strand. All of our results, including raw data, processed results, modifications to the annotation, and source code, are available at our website (http://genomics.lbl.gov/supplemental/DvHtranscripts2011/); the data are also available at the Gene Expression Omnibus database (GSE29560).
Supplementary Material
ACKNOWLEDGMENTS
We thank Alyssa M. Redding-Johanson for reanalyzing previously published proteomics data regarding the six-frame translation of the genome. We thank John-Marc Chandonia, Evelyn Dora Szakal, and Allen Simon for providing proteomics data, and we thank the ENIGMA protein complexes group for providing protein fractions. We thank Marcin Joachimiak for designing the tiling array. We thank Judy Wall, Terry Hazen, and their research groups for advice on working with D. vulgaris Hildenborough.
This work conducted by ENIGMA—Ecosystems and Networks Integrated with Genes and Molecular Assemblies—was supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy (contract DE-AC02-05CH11231).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 12 August 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Aivaliotis M., et al. 2007. Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res. 6:2195–2204 [DOI] [PubMed] [Google Scholar]
- 2. Aziz R. K., et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Badger J. H., Olsen G. J. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512–524 [DOI] [PubMed] [Google Scholar]
- 4. Bailey T. L., Elkan C. 1995. Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Mach. Learn. 21:51–80 [Google Scholar]
- 5. Bubunenko M., Baker T., Court D. L. 2007. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 189:2844–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chhabra S. R., et al. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho B. K., et al. 2009. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 27:1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ciampi M. S. 2006. Rho-dependent terminators and transcription termination. Microbiology 152:2515–2528 [DOI] [PubMed] [Google Scholar]
- 9. Collado-Vides J., Magasanik B., Gralla J. D. 1991. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 55:371–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crampton N., Bonass W. A., Kirkham J., Rivetti C., Thomson N. H. 2006. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 34:5416–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darling A. C., Mau B., Blatter F. R., Perna N. T. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dehal P. S., et al. 2009. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38(Database issue):D396–D400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Hoon M. J., Makita Y., Nakai K., Miyano S. 2005. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 1:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dornenburg J. E., DeVita A. M., Palumbo M. J., Wade J. T. 2011. Widespread antisense transcription in Escherichia coli. mBio 1:e00024–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gish W., States D. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266–272 [DOI] [PubMed] [Google Scholar]
- 17. Güell M., et al. 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326:1268–1271 [DOI] [PubMed] [Google Scholar]
- 18. Hahn M. W., Stajich J. E., Wray G. A. 2003. The effects of selection against spurious transcription factor binding sites. Mol. Biol. Evol. 20:901–906 doi:10.1093/molbev/msg096 [DOI] [PubMed] [Google Scholar]
- 19. Halasz G., et al. 2006. Detecting transcriptionally active regions using genomic tiling arrays. Genome Biol. 7:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hauser L. J., et al. 2011. The complete genome sequence and updated annotation of Desulfovibrio alaskensis G20. J. Bacteriol. 193:4268–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heidelberg J. F., et al. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554–559 [DOI] [PubMed] [Google Scholar]
- 22. Hershberg R., Bejerano G., Santos-Zavaleta A., Margalit H. 2001. PromEC: an updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 29:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hertz G. Z., Stormo G. D. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15:563–577 [DOI] [PubMed] [Google Scholar]
- 24. Kent W. J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kingsford C., Ayanbule K., Salzberg S. 2007. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 8:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koide T., et al. 2009. Prevalence of transcription promoters within archaeal operons and coding sequences. Mol. Syst. Biol. 5:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu X., Brutlag D. L., Liu J. S. 2001. Bioprospector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001:127–138 [PubMed] [Google Scholar]
- 28. Lovley D. R., Phillips E. J. 1994. Reduction of chromate by Desulfovibrio vulgaris and its c(3) cytochrome. Appl. Environ. Microbiol. 60:726–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marchler-Bauer A., et al. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mukhopadhyay A., et al. 2006. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188:4068–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mukhopadhyay A., et al. 2007. Cell-wide responses to low-oxygen exposure in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189:5996–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muyzer G., Stams A. J. M. 2008. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6:441–454 [DOI] [PubMed] [Google Scholar]
- 33. Nie L., Wu G., Zhang W. 2006. Correlation between mRNA and protein abundance in Desulfovibrio vulgaris: a multiple regression to identify sources of variations. Biochem. Biophys. Res. Commun. 339:603–610 [DOI] [PubMed] [Google Scholar]
- 34. Novichkov P. S., et al. 2010. Regprecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38(Database issue):D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palmer A. C., Ahlgren-Berg A., Egan J. B., Dodd I. B., Shearwin K. E. 2009. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol. Cell 34:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perocchi F., Xu Z., Clauder-Münster S., Steinmetz L. M. 2007. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 35:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peters J. M., et al. 2009. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 106:15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Price M. N., Dehal P. S., Arkin A. P. 2008. FastBLAST: homology relationships for millions of proteins. PLoS One 3:e3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Price M. N., Huang K. H., Alm E. J., Arkin A. P. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Proshkin S., Rahmouni A. R., Mironov A., Nudler E. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328:504–508 doi:10.1126/science.1184939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qiu Y., et al. 2010. Structural and operational complexity of the Geobacter sulfurreducens genome. Genome Res. 20:1304–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Redding A. M., Mukhopadhyay A., Joyner D. C., Hazen T. C., Keasling J. D. 2006. Study of nitrate stress in Desulfovibrio vulgaris Hildenborough using iTRAQ proteomics. Brief Funct. Genomic Proteomic. 5:133–143 [DOI] [PubMed] [Google Scholar]
- 43. Rutherford K., et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 44. Schäffer A. A., et al. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29:2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Selinger D. W., et al. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18:1262–1268 [DOI] [PubMed] [Google Scholar]
- 46. Shilov I. V., et al. 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6:1638–1655 [DOI] [PubMed] [Google Scholar]
- 47. Sorek R., Cossart P. 2010. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 11:9–16 [DOI] [PubMed] [Google Scholar]
- 48. Wall J. D., Krumholz L. R. 2006. Uranium reduction. Annu. Rev. Microbiol. 60:149–166 [DOI] [PubMed] [Google Scholar]
- 49. Wurtzel O., et al. 2010. A single-base resolution map of an archaeal transcriptome. Genome Res. 20:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamamoto N., et al. 2009. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou A., et al. 2010. Hydrogen peroxide-induced oxidative stress responses in Desulfovibrio vulgaris Hildenborough. Environ. Microbiol. 12:2645–2657 [DOI] [PubMed] [Google Scholar]
- 52. Zhou J., et al. 2011. How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat. Rev. Microbiol. 9:452–466 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.