Abstract
Salmonella enterica serovar Typhimurium can utilize molecular hydrogen for growth and amino acid transport during anaerobic growth. Via microarray we identified H2 gas-affected gene expression changes in Salmonella. The addition of H2 caused altered expression of 597 genes, of which 176 genes were upregulated and 421 were downregulated. The significantly H2-upregulated genes include those that encode proteins involved in the transport of iron, manganese, amino acids, nucleosides, and sugars. Genes encoding isocitrate lyase (aceA) and malate synthase (aceB), both involved in the carbon conserving glyoxylate pathway, and genes encoding the enzymes of the d-glucarate and d-glycerate pathways (gudT, gudD, garR, garL, garK) are significantly upregulated by H2. Cells grown with H2 showed markedly increased AceA enzyme activity compared to cells without H2. Mutant strains with deletion of either aceA or aceB had reduced H2-dependent growth rates. Genes encoding the glutamine-specific transporters (glnH, glnP, glnQ) were upregulated by H2, and cells grown with H2 showed increased [14C]glutamine uptake. Similarly, the mannose uptake system genes (manX, manY) were upregulated by H2, and cells grown with H2 showed about 2.0-fold-increased [14C]d-mannose uptake compared to the cells grown without H2. Hydrogen stimulates the expression of genes involved in nutrient and carbon acquisition and carbon-conserving pathways, linking carbon and energy metabolism to sustain H2-dependent growth.
INTRODUCTION
Salmonella enterica serovar Typhimurium is an important bacterial pathogen, and it is implicated in a majority of the documented food-borne gastroenteritis cases in humans (47). The metabolic flexibility of S. Typhimurium allows it to survive in diverse environmental conditions both outside and within the host (19, 30). An important factor augmenting the survival capability of this bacterium in macrophages and in animal hosts is its ability to utilize molecular hydrogen produced within the host as an energy source (36, 37, 57).
Uptake-type (H2-oxidizing) hydrogenases are considered catalysts enabling auxiliary energy source use for the generation of a proton gradient across the cell membrane (53, 54). The electrons generated in the process are passed along bacterial electron transport chains to terminal electron acceptors such as fumarate, nitrate, sulfate, CO2, or O2 (54), while the “sidedness” of the H2-splitting reaction generates protons in a way to promote a proton potential across the cytoplasmic membrane. The electrochemical potential thus generated (32) can be utilized by the cells for work, such as energizing transport of nutrients against a gradient and various other cellular processes. S. Typhimurium oxidizes molecular hydrogen by the H2-oxidizing activity of three NiFe-containing respiratory hydrogenases—Hya, Hyb, and Hyd (58). The respiratory hydrogenases are important for the virulence of S. Typhimurium (37), as the host colonic flora produces the highly diffusible H2 gas (36).
Hydrogen can be an important energy source for bacteria growing in an environment where high-energy organic substrates are limiting (53). The availability of H2 in a nutrient-limited condition such as the competitive environment within the host intestinal tract could therefore be instrumental to the survival of salmonellae. We recently studied the effects of exogenous H2 on the anaerobic growth of S. Typhimurium in a nutritionally challenging medium (32). Our study showed that addition of H2 significantly augments the growth of S. Typhimurium in a culture medium containing amino acids as the only carbon source. This H2-mediated growth augmentation is mainly due to the enhanced ability of the bacteria to acquire amino acids from the medium and is largely facilitated by the membrane proton motive force (PMF) generated by the Hyb hydrogenase (32). This caused us to examine whether the cells have mechanisms to increase carbon acquisition when oxidizing H2.
Microarray studies of salmonellae have documented their metabolic flexibility when facing altered availability of a nutrient (23) or by mutation of a specific metabolic factor (17, 33). In our previous study, we observed that in addition to energizing the uptake and transport processes for increased carbon (amino acid) acquisition, H2 stimulates the expression of the proteins TonB and ExbD. The TonB-ExbD system is involved in transducing the PMF to the outer membrane and thus energizes the transport of nutrients across the membrane (44). This led us to herein further investigate the transcriptional roles of H2 availability of S. Typhimurium. In this study, we identified potential nutrient acquisition-associated genes that are linked to H2 metabolism, and we then performed focused physiology studies on how H2 stimulates the acquisition and conservation of carbon by S. Typhimurium growing under carbon limitation.
MATERIALS AND METHODS
Strains, growth conditions, and reagents.
Wild-type (WT) Salmonella enterica serovar Typhimurium ATCC 14028s strain JSG210 (57) was used for the microarrays and real-time PCR-based validation of the microarray data. In addition, aceA and aceB single-deletion strains were used for physiological experiments based on the microarray results. Single-deletion mutants were constructed using the lambda Red system as previously described (11, 57). The deletions were confirmed by PCR using primers complementary to the regions flanking the deleted genes and by sequencing across the deletions (Georgia Genomics Facility, University of Georgia). The strains and plasmids used in this study are listed in Table 1, and the primers used are listed in a supplementary table (see Table S1 in the supplemental material).
Table 1.
Strains and plasmids used in this study
| Strain/plasmid | Genotype/descriptiona | Reference |
|---|---|---|
| S. enterica serovar Typhimurium strains | ||
| JSG210 | ATCC 14028s (WT) | 57 |
| RLK3 | JSG210ΔaceA::FRT (ΔaceA) | This study |
| RLK4 | JSG210ΔaceB::FRT (ΔaceB) | This study |
| Plasmids | ||
| pCP20 | Ampr; contains flippase gene for λ Red mutagenesis | 11 |
| pKD46 | Ampr; contains λ Red genes γ, β, and exo | 11 |
| pKD4 | Kanr; contains kan cassette | 11 |
FRT, flippase recombinase recognition target.
Strains were maintained in Luria-Bertani (LB) broth or on LB agar (LBA) plates. Experiments were performed in CR-Hyd medium (2, 6) containing bacteriological peptone (0.5%, wt/vol), Casamino Acids (0.2%, wt/vol), thiamine (0.001%, wt/vol), MgCl2 (1 mM), (NH4)6Mo7O24 (1 μM), and NaSeO3 (1 μM). The medium was supplemented with sodium fumarate (0.5%) as a terminal electron acceptor. No sugar was added, but 5 μM NiCl2 was included in the medium. Cells were grown at 37°C anaerobically with or without H2. Anaerobic conditions with H2 were established by sparging sealed 165-ml bottles with N2 for 15 min and then with anaerobic mix (10% H2, 5% CO2, and 85% N2) for 20 min; more H2 was then injected to bring the volume of added H2 to 20% partial pressure. Cells were grown anaerobically without H2 in 165-ml bottles by sparging with N2 for 15 min and then injecting the sealed bottles containing cells with CO2 to 5% partial pressure (32).
RNA isolation.
Total bacterial RNA for DNA microarrays and real-time quantitative PCR was isolated from the test (20% H2 added to the medium) and control (no added H2) cultures (A600 = 0.4) of WT by following a previously described method (33) with some modifications described herein. The cultures were treated with 0.15 volumes of ethanol-phenol (95%:5%, vol/vol) to protect the mRNA in the samples. The cells were then pelleted (8,000 × g; 10 min; 4°C), suspended in lysis buffer (10 mM Tris, 1 mM EDTA, 0.5 mg/ml lysozyme) and treated with 1 ml of 10% SDS for 2 min at 64°C. The suspensions were treated with 11 ml of 1 M sodium acetate, pH 5.2, and then with an equal volume of phenol and incubated at 64°C for 6 min, followed by rapid chilling on ice. The suspensions were centrifuged (10,000 × g; 10 min; 4°C); the aqueous supernatant was treated with an equal volume of chloroform and again centrifuged (10,000 × g; 5 min; 4°C). Nucleic acid was then precipitated by treating the aqueous supernatant with 0.1 vol of 3 M sodium acetate and 1.0 vol of cold isopropanol, pelleted by centrifugation (10,000 × g; 25 min; 4°C), and washed with 80% ethanol. The pellet was resuspended in 1 ml of nuclease-free water and 500 U of RNasin Plus RNase inhibitor (Promega, Madison, WI), 250 U of RQ1 RNase-free DNase (Promega, Madison, WI), 20 μl of 1 M Tris (pH 8.3), and 10 μl of 1 M MgCl2. The mixture was incubated at 37°C for 30 min. RNA was extracted by treating once with phenol, once with phenol-chloroform (50:50, vol/vol), and twice with chloroform. The RNA was precipitated with 1.0 vol of isopropanol and 0.1 vol of 3 M sodium acetate, washed once with 80% ethanol, dried, and resuspended in nuclease-free water. The RNA samples were checked for quantity and quality by agarose gel electrophoresis and UV spectrometry at A260/280 and A260/230.
DNA microarrays.
The multiserotype microarray contained 5,660 PCR products covering 95% of all genes of Salmonella enterica serovar Typhimurium strain LT2, S. Typhimurium strain SL1344, Salmonella enterica serovar Typhi strain CT18, S. Typhi strain Ty2, Salmonella enterica serovar Paratyphi A strain SARB42, and Salmonella enterica serovar Enteritidis strain PT4. Each gene probe was dissolved in 50% dimethyl sulfoxide (DMSO) and spotted onto Corning Ultra-GAPS glass slides (catalogue no. 40015; Corning). Each glass slide contained triplicate identical arrays. Methods were as previously described by Lawhon et al. (33). Briefly, cDNA was synthesized from a total of 50 μg RNA and labeled with Cy3- or Cy5-conjugated dUTP using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) and random hexamers. The labeled probes were then incubated with 0.1 M NaOH at 65°C for 10 min, followed by the addition of HCl to a final concentration of 0.1 M. The labeled nucleotides were further purified using the PCR purification kit from Qiagen (Qiagen Inc., Valencia, CA), following the manufacturer's instructions. Equal volumes of labeled probes from the test (20% H2 added to the medium) and control (no added H2) samples were mixed with an equal volume of hybridization solution (50% formamide, 10× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.2% SDS). Array slides were incubated in prehybridization buffer (25% formamide, 5× SSC, 0.2% SDS) at 42°C for 45 min. Labeled probes were added to the hybridization buffer simultaneously and incubated at 95°C for 5 min and then hybridized to the arrays at 42°C for 18 h. The hybridized array slides were washed in numerous steps using buffer I (2× SSC, 0.1% SDS at 42°C) and buffer II (0.1× SSC, 0.1% SDS at room temperature [RT]), followed by rapid rinses in distilled H2O and 95% ethanol, respectively. Three independent microarray experiments were performed for the analysis of each sample. Each microarray experiment was performed in duplicate, with the second chip hybridized with dyes swapped in order to minimize dye bias for each sample analyzed. Array slides were scanned using a ScanArray 5000 laser scanner (GSI Lumonics). Signals were recorded with ScanArray 3.0 software, and signal intensities were quantified using the QuantArray 3.0 software package (Packard BioChip Technologies, Billerica, MA). Spots were analyzed by adaptive quantitation and subsequently statistically analyzed using WebArray (56). The following parameters were used: background subtraction was performed using the “half” method, print-tip Loess normalization was employed within arrays, and scale normalization was used between arrays. Genes showing expression ratios more than 2.0 were considered upregulated, and those showing ratios less than 0.5 were considered downregulated.
Real-time quantitative PCR.
The microarray data were validated by real-time quantitative PCR of six upregulated and six downregulated genes selected from the microarray-identified differentially altered genes. RNA obtained from the test (20% H2 added to the medium) and control (no added H2) samples for real-time PCR were the same as those used for DNA microarrays. First-strand cDNA was synthesized from 200 ng purified RNA samples using random hexamers and Moloney murine leukemia virus SuperScript III reverse transcriptase (Invitrogen) at 42°C for 50 min. The iCycler iQ real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) and iQ SYBR green supermix (Bio-Rad) were utilized for real-time PCR of control and test cDNAs. The expression level (threshold cycle value [CT]) of each sample was normalized using DNA gyrase B (gyrB) as an internal control. The relative fold change in gene expression for each sample was determined using the 2−ΔΔCT method described previously (34). The gene-specific primers used for real-time PCR are listed in Table S1 in the supplemental material.
Growth curves and endpoint growth yields.
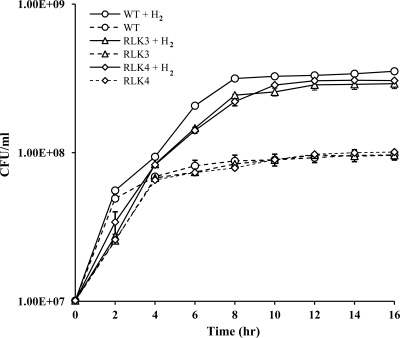
In order to determine the involvement of AceA and AceB on the H2-facilitated growth of Salmonella, growth curves and endpoint growth assays were performed. Sealed 165-ml bottles containing 15 ml CR-Hyd medium with 0.5% sodium fumarate (as described above) were inoculated with 1.0 × 107 wild-type or deletion mutant S. enterica serovar Typhimurium cells. Cells were grown anaerobically with or without 20% H2 for 24 h at 37°C with shaking at 200 rpm. A600 (optical density at 600 nm [OD600]) was measured after growth in order to determine cell number. An A600 of 1.0 corresponds to 6.74 × 108 CFU/ml for the strains used. Standard curves of A600 versus CFU/ml (plate counts) confirmed that the A600 was proportional to the viable cell number within the OD range used herein, including for final yield (i.e., saturation growth) numbers. All growth rate and yield studies were performed three times or more, with results similar to those shown (see Fig. 3).
Fig. 3.
Comparison of H2-facilitated growth of S. enterica serovar Typhimurium strains JSG210 (WT), RLK3 (ΔaceA), and RLK4 (ΔaceB) in CR-Hyd medium.
Enzyme activity assays.
The AceA and AceB activities of the H2-added and H2-absent cultures were compared by the spectrophotometric enzymatic assay methods provided by the Sigma-Aldrich Co. with modifications described herein. Cells were grown in CR-Hyd medium (without glucose, supplemented with 0.5% sodium fumarate and 5 μM NiCl2) with and without added H2, in triplicates for each condition. After the cultures reached an A600 of 0.4, cells were pelleted (8,000 × g; 10 min; 4°C) and suspended in 1 ml of 30 mM imidazole buffer (pH 6.8 for AceA assay; pH 8.0 for AceB assay). The suspensions were sonicated and then centrifuged (10,000 × g; 10 min; 4°C) to obtain cell extracts. Total protein in each sample was quantitated using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific Inc., Rockford, IL) following the manufacturer's instructions. The assays were performed at 25°C, and 4 μg of total protein from each sample was used for individual assays.
Preparation of [14C]glutamine.
[14C]glutamine was synthesized in vitro from 14C-uniformly labeled glutamic acid using purified glutamine synthetase. The reaction mixture contained 50 mM imidazole (pH 7.15), 100 mM NaCl, 40 mM MgCl2, 30 mM NH4Cl, 10 mM ATP, 18 μCi [14C]glutamic acid (specific activity, >50 mCi/mmol; Perkin Elmer, Boston, MA), and 40 μg/ml purified Helicobacter pylori glutamine synthetase. The reaction mixture was incubated for 3 h at 37°C and was quenched by raising the temperature to 85°C for 15 min. A 1-μl portion of the quenched sample was spotted onto a thin-layer chromatographic plate (0.25 mm silica gel; Analtech, Newark, DE) and was developed using tert-butanol-acetic acid-water (25:12:10) for 6 h. Spots corresponding to cold glutamate and glutamine controls were visualized using ninhydrin staining. Regions estimated to contain [14C]glutamic acid and [14C]glutamine were scraped from the plate and quantified using liquid scintillation spectrometry. The conversion of [14C]glutamic acid to [14C]glutamine was determined to be 97% ± 2.5% (n = 4).
[14C]glutamine uptake assay.
Cells were grown in CR-Hyd medium under conditions as previously described in this paper (without glucose, supplemented with 0.5% sodium fumarate and 5 μM NiCl2). Cultures were grown with and without added H2 and in triplicates for each condition. After the cultures reached an A600 of 0.1, all bottles were sparged with N2 for 8 to 10 min at RT; the vials were then injected with CO2 to 5% partial pressure. The OD600 of the cultures was unchanged during the sparging period, and the activities (see Table 3) are expressed based on the cell number determined at the time of the transport assay. [14C]glutamine prepared as described above was injected into the bottles to a final concentration of 0.2 μCi/ml of growth medium. [14C]glutamine uptake by the cultures was measured at 5, 10, 15, and 60 min following a previously described method (42).
Table 3.
Comparison of [14C]glutamine uptake activities of S. enterica serovar Typhimurium JSG210 grown with versus without H2a
| Presence of H2 | [14C]Glutamine uptake [(cpm × 103)/108 cells] atb: |
|||
|---|---|---|---|---|
| 5 min | 10 min | 15 min | 60 min | |
| + | 9.1 ± 0.7* | 13.1 ± 1.3* | 19.6 ± 2.7* | 96.1 ± 2.3* |
| − | 7.4 ± 0.3 | 10.4 ± 0.6 | 13.3 ± 1.6 | 66.6 ± 4.2 |
Cells were grown with and without added H2 to an A600 of 0.1, and the H2 present in the cultures was removed by sparging the culture bottles with N2. 14C-labeled glutamine was added to a final concentration of 0.2 μCi/ml, and uptake was measured in the absence of H2, so the uptake could be compared for the two types of cultures without the H2-PMF influence.
*, significantly increased activity compared to the H2-absent condition (P, <0.01, n = 3).
14C-labeled d-mannose uptake assay.
14C-labeled d-mannose was obtained from Moravek Biochemicals (Brea, CA; specific activity, 53 mCi/mmol). The procedure and the final concentration of 14C-d-mannose used were the same as described for the [14C]glutamine uptake assay. Uptake was measured at 1, 2, 5, and 10 min after the addition of 14C-labeled d-mannose. The uptake assays as well as other physiological experiments were repeated, with results obtained similar to those reported herein.
Microarray data accession number.
The complete microarray data set has been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE29739.
RESULTS AND DISCUSSION
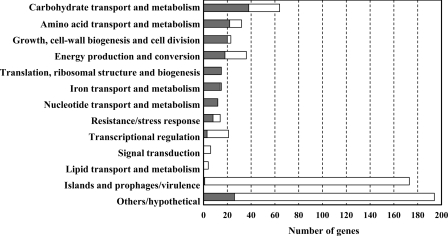
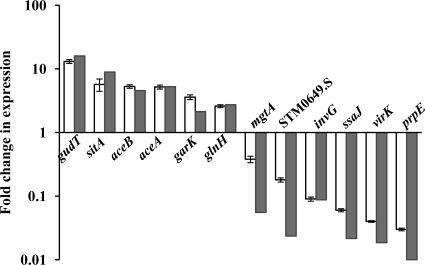
Microarrays revealed a significantly altered H2-dependent expression of 597 S. enterica serovar Typhimurium genes when cells were cultured anaerobically in CR-Hyd medium. Of the significantly altered genes, 176 were upregulated and 421 were downregulated. Of the significantly H2-upregulated genes, a considerable proportion is classified as being involved in transport and metabolism (Fig. 1). Interestingly, a significant percentage of the genes downregulated by the H2-grown salmonellae are associated with virulence. Real-time quantitative PCR of selected microarray-identified genes with expression changes showed a significant correlation between the gene expression detected by the microarrays and separately by real-time quantitative PCR (correlation coefficient [R2] = 0.96), thus validating the microarray data (Fig. 2). DNA gyrase B (gyrB) was used as an internal control to normalize the expression levels of the genes used for real-time PCR validation of the microarray data. Another validation of the data is the strong correlation between previously described H2-upregulated proteins TonB and ExbD (32) and their H2-affected transcript levels reported herein.
Fig. 1.
H2-affected differential expression of genes belonging to functional groups in S. enterica serovar Typhimurium. Bars represent percentage of differentially expressed genes belonging to each group. White bars indicate the proportion of downregulated genes, and gray bars indicate the proportion of upregulated genes.
Fig. 2.
Real-time quantitative PCR of selected differentially expressed genes for validation of microarray data. Bars represent relative fold change in expression of selected microarray-identified genes in S. enterica serovar Typhimurium cells grown with H2 compared to that in cells grown without H2. Gray bars indicate values obtained from real-time quantitative PCR, and white bars represent values obtained from microarrays. For a comparison of the expression ratios obtained by microarrays and real-time PCR, see Table S2 in the supplemental material.
The genes that are significantly upregulated by H2 include those that encode proteins involved in the transport and metabolism of iron, amino acids, purine, pyrimidine and nucleosides, and sugars (Table 2). Additionally, genes encoding isocitrate lyase (aceA) and malate synthase (aceB), both involved in the carbon-conserving glyoxylate pathway, and the d-glycerate pathway-associated genes (garK, garL, garR), also involved in glyoxylate metabolism, are significantly upregulated in cells grown in the presence of H2 (Table 2). Surprisingly, the expression of the hyd hydrogenase was downregulated (3.0-fold) by H2 while the hyb hydrogenase gene was not altered by H2 exposure (see Table S3 in the supplemental material). For the discussion relevant to our study, a representative table of selected microarray-identified upregulated genes is shown in this study. A full set of the 597 genes affected by H2 is provided in Table S3.
Table 2.
Microarray-identified H2-stimulated select genes in S. enterica serovar Typhimurium
| Locus IDa | Genea | Functiona | Expression ratiob (fold change) |
|---|---|---|---|
| Carbon transport and metabolism | |||
| Carbohydrates | |||
| STM2962 | gudT | d-Glucarate permease | 5.7 |
| STM4077/78 | yneA/B | Putative sugar transport protein | 3.6/3.0 |
| STM2960 | gudD | d-Glucarate dehydratase | 3.5 |
| STM3557 | ugpB | Glycerol 3-phosphate transport protein | 2.6 |
| STM2190 | mglB | Galactose transport protein | 2.4 |
| STM3884 | rbsB | d-Ribose transport protein | 2.4 |
| STM1830/31 | manX/Y | PTS, mannose-specific enzyme IIAB-IIC | 2.3/2.4 |
| STM4325 | dcuA | Anaerobic dicarboxylate transport protein | 2.2 |
| STM0685 | nagE | PTS, n-acetylglucosamine-specific enzyme IIABC | 2.2 |
| STM4074 | ego | Putative aldose transport protein | 2.2 |
| STM4075/76 | ydeY/Z | Putative sugar transport protein | 2.1/2.1 |
| Amino acids | |||
| STM0791 | hutH | Histidine ammonia lyase | 4.3 |
| STM3106 | ansB | l-Asparaginase II | 3.8 |
| STM3709 | kbl | Glycine acetyltransferase | 2.8 |
| STM0890 | artI | Arginine transport system | 2.6 |
| STM3708 | tdh | Threonine 3-dehydrogenase | 2.6 |
| STM0828-30 | glnQ/P/H | Glutamine high-affinity transporter | 2.0–2.6 |
| STM2071-78 | hisA-I | Histidine metabolism | 2.0–2.6 |
| STM4007 | glnA | Glutamine synthetase | 2.3 |
| STM2555 | glyA | Serine hydroxymethyltransferase | 2.3 |
| STM4477 | pepA | Aminopeptidase A | 2.3 |
| STM0002 | thrA | Aspartokinase I, bifunctional enzyme | 2.0 |
| Purine/pyrimidine/nucleotides | |||
| STM0756 | nadA | Quinolinate synthetase A | 6.4 |
| STM4452 | nrdD | Anaerobic ribonucleoside-triphosphate reductase | 3.4 |
| STM0012/13 | dnaK/J | DNA biosynthesis | 3.2/2.2 |
| STM0757 | pnuC | NMN/NmR transporter | 3.1 |
| STM2277/78 | nrdA/B | Ribonucleoside diphosphate reductase 1, α and β subunits | 2.9/2.3 |
| STM3511 | yhgI | DNA uptake protein | 2.6 |
| STM0523 | allB | Allantoinase | 2.5 |
| STM2807 | nrdE | Ribonucleoside diphosphate reductase 2, α subunit | 2.4 |
| STM2641 | nadB | Quinolinate synthetase B | 2.3 |
| Iron/manganese | |||
| STM0191/93 | fhuA/D | OMP receptor for iron uptake | 15.2/2.0 |
| STM2861/62/63 | sitA/B/C | Salmonella Fe(II)/Mn(II) transporter | 13.0/8.2/3.6 |
| STM4055 | sodA | Manganese-dependent superoxide dismutase | 8.2 |
| STM1737 | tonB | Energy transducer; uptake of substrates | 2.4 |
| STM3158/59 | exbD/B | Uptake of enterochelin; tonB-dependent uptake | 2.0/5.0 |
| Carbon conservation and assimilation | |||
| STM4183 | aceB | Malate synthase | 5.3 |
| STM3248 | garR | Tartronate semialdehyde reductase | 4.9 |
| STM3249 | garL | 2-Dehydro-3-deoxy-galactarate aldolase | 3.9 |
| STM4184 | aceA | Isocitrate lyase | 3.6 |
| STM3247 | garK | Glycerate kinase | 3.3 |
| STM4187 | iclR | Acetate operon transcriptional repressor | 2.9 |
| STM0772 | gpmA | Phosphoglyceromutase 1 | 2.8 |
| STM0210 | cdaR | Carbohydrate diacid transcriptional activator | 2.2 |
| STM1173-79 | flgA-G | Flagellar biosynthesis | 2.2–3.0 |
| STM2082-90 | rfbJ-V | LPS biosynthesis | 2.2–2.6 |
| STM3326 | mtgA | Biosynthetic peptidoglycan transglycosylase | 2.1 |
| STM0310 | ghmA | Phosphoheptose isomerase/LPS biosynthesis | 2.1 |
| STM3326 | mtgA | Peptidoglycan biosynthesis | 2.1 |
| STM1752 | galU | Glucose-1-phosphate uridylyltransferase | 2.0 |
Based on the published genome of S. enterica serovar Typhimurium strain LT2 using the NCBI GenBank database.
Hyphens indicate the range of expression ratio corresponding to the genes indicated in column 1. Slashes indicate the expression ratios for the two or three genes indicated in colulmn 1.
H2 stimulates expression of iron and/or manganese acquisition and transport-associated genes in Salmonella Typhimurium.
Genes encoding iron/manganese-acquisition and transport proteins (fhuA, sitA, sitB, sitC, fhuF) are the most upregulated in Salmonella Typhimurium grown with H2. The CR-Hyd medium (2, 6) used in this experiment contains low iron and manganese, their sole source being the bacteriological peptone that reportedly contains 0.004% iron and less than 0.001% manganese (12). It is well established that bacteria adapt to iron scarcity by upregulating the expression of iron-scavenging and transport proteins; however, they can also downregulate the synthesis of proteins that require iron for enzymatic activity (for a review, see reference 39). Manganese can function as an important alternative to iron for the activation of some enzymes in bacterial cells; an example of this in enteric bacteria is the expression of a manganese-dependent isozyme of superoxide dismutase when the iron levels are low in the growth environment (7, 39). This could be an important adaptive mechanism pathogenic bacteria use to counter the sequestration of iron by the host upon bacterial invasion. It is noteworthy that H2 causes upregulation of the gene sodA (8.2-fold upregulated) that encodes a manganese-binding superoxide dismutase. In addition, the sitABCD genes that encode an important transporter of Mn(II) and Fe(II) (5, 24) are also upregulated by H2 (Table 2). SitABCD is also required for full virulence of S. Typhimurium (5, 22). It has also been suggested that Mn(II) plays an important role in central carbon metabolism, and several enzymes of the carbon metabolic pathway either require Mn(II) for activity or are highly induced by Mn(II) (for a review, see reference 25). The transcription of the genes gpmM and glpX that encode the Mn(II)-dependent enzymes phosphoglyceromutase (GpmM) and fructose-1,6-bisphosphate phosphatase (GlpX), respectively, in S. Typhimurium (see reference 25) were not affected by H2 in our microarray experiments. However, some Mn(II)-dependent enzymes involved in carbon metabolism in S. Typhimurium have likely not been identified. The H2-dependent upregulation of genes involved in Fe(II)/Mn(II) scavenging and transport may also promote maturation of iron-containing respiratory enzymes in the H2 oxidizing pathway.
Several of the highly upregulated genes detected by the microarrays are iron-acquisition and transport-associated genes that encode TonB-dependent proteins in Escherichia coli and Salmonella (NCBI database; http://www.ncbi.nlm.nih.gov/). The TonB protein forms a complex with two other proteins, ExbD and ExbB, that function as a secondary transport system also known as the TonB-ExbD-ExbB system. This system utilizes the cytoplasmic proton motive force (PMF) for the transport of substrates by the TonB-dependent outer membrane transport proteins (TBDTs) in E. coli and other Gram-negative bacteria (45, 49). In addition to its previously suggested role in the uptake of iron complexes and vitamin B12, the TonB-ExbD-ExbB system is now implicated in the transport of other substrates such as nickel, carbohydrates, cobalt, and copper (48). In our previous study, we showed that the expression of tonB and exbD genes as well as the TonB and ExbD proteins was increased by Salmonella Typhimurium when growing in an atmosphere containing H2 (32). The microarray experiments in the current study substantiated upregulation of the tonB, exbD, and exbB genes in H2-added cultures of S. Typhimurium. As we proposed previously, H2-stimulated expression of the TonB-ExbD-ExbB system proteins of S. Typhimurium is likely a way for the bacteria to enhance the uptake of the much-required nutrients such as iron-siderophores, nickel, carbohydrates, vitamin B12, etc.; in this way the increased energy input from the oxidation of H2 can be coupled to macromolecule synthesis and growth.
Microarrays also detected significant H2-dependent upregulation of the DNA synthesis-associated genes nrdD, nrdA, nrdE, and nrdB (3.4-, 2.9-, 2.4-, and 2.3-fold upregulated, respectively). NrdD is an anaerobic ribonucleotide reductase, and NrdAB is an aerobic isozyme. Another isozyme that shows ribonucleotide reductase activity in Escherichia coli is NrdEF (8). It has been shown that NrdEF is a Mn(II)-dependent protein that supports cell replication during iron limitation in E. coli (39), and therefore the possession of the alternative Mn(II)-dependent NrdE enzyme could aid the pathogen to survive iron-limited conditions. However, in our experiments we found that the transcription of both the anaerobic and aerobic reductase genes and the isozyme subunit gene nrdE is significantly stimulated by H2.
H2 stimulates carbon acquisition and transport in Salmonella Typhimurium.
As stated earlier, H2 addition significantly upregulated the expression of several genes that encode proteins involved in the acquisition and transport of carbon sources. The genes encoding glutamine high-affinity transporters (glnH, glnP, and glnQ) were significantly upregulated by H2 exposure, as were the arginine transport proteins encoded by genes artI and artP (Table 2). It was shown earlier that H2 rapidly augments the uptake of amino acids by Salmonella in this same medium/condition; this augmentation in amino acid uptake was attributed to the energization of the transport processes by the proton motive force generated by the oxidation of H2 by S. Typhimurium (32). The microarray data also showed that the mannose uptake protein-encoding genes manX and manY were upregulated (2.3- and 2.4-fold, respectively) by H2 exposure (Table 2).
Physiological experiments were conducted to assess the significance of the carbon source uptake and conservation-related microarray data. In order to eliminate the direct influence of the H2 oxidation-generated PMF on amino acid transport, cells were grown with and without added H2 to an A600 of 0.1 and then the H2 present in the cultures was removed by sparging the culture bottles with N2. The 14C-labeled glutamine and mannose uptake abilities were then tested in the absence of H2, so the uptake could be compared for the two types of cultures without the influence of the H2-generated PMF. By assaying the uptake in H2-absent conditions for both cultures, we intended to detect any change in the uptake rates in the culture as a result only of the expression of the uptake/transport-associated proteins. We found that S. Typhimurium cells pregrown with added H2 exhibited 1.5-fold-increased uptake of 14C-labeled glutamine compared to cells without added H2 (Table 3). Though markedly lower than the 4.0-fold-increased uptake of amino acids when the uptake assay was conducted with H2 in our previous study (32), the increase in glutamine uptake by cells pregrown under an H2-added condition was statistically significant. This result is likely due to the H2-stimulated expression of the glutamine transporters GlnH, GlnP, and GlnQ. The cells pregrown with H2 also had 1.7-, 1.7-, 2.5-, and 2.0-fold-increased 14C-d-mannose uptake at 1, 2, 5, and 10 min, respectively, compared to the cells grown in the absence of H2 (Table 4). Increased uptake of d-mannose in the H2-added condition could be due to the H2-stimulated expression of the mannose uptake proteins ManX and ManY.
Table 4.
Comparison of [14C]d-mannose uptake activities of S. enterica serovar Typhimurium JSG210 grown with versus without H2
| Presence of H2 | [14C]d-Mannose uptake [(cpm × 103)/108 cells] ata: |
|||
|---|---|---|---|---|
| 1 min | 2 min | 5 min | 10 min | |
| + | 25.7 ± 1.2* | 25 ± 1.6* | 113 ± 6* | 116 ± 5* |
| − | 14.8 ± 2.9 | 15 ± 2.4 | 45 ± 1 | 59 ± 4 |
*, significantly increased activity compared to the H2-absent condition (P, <0.01, n = 3).
H2 stimulates the carbon-conserving glyoxylate pathway and the d-glucarate/d-glycerate pathway genes in Salmonella Typhimurium.
In addition to the genes encoding proteins involved in amino acid transport, several sugar acquisition and transport-protein-encoding genes (gudT, gudD, rbsB, mglB, ugpB, manY, nagE) are also upregulated by H2 in S. Typhimurium (Table 2). The genes encoding the enzymes of the glyoxylate pathway, namely, isocitrate lyase (aceA) and malate synthase (aceB), were H2 upregulated 3.6- and 5.3-fold, respectively, as detected by the microarrays and 5.2- and 4.5-fold, respectively, by real-time quantitative PCR. In E. coli, AceA cleaves isocitrate to glyoxylate and succinate (50), and AceB converts acetyl coenzyme A (CoA) and glyoxylate to malate (1). The glyoxylate bypass plays an important role in conserving carbon by avoiding decarboxylation steps; it is especially important when bacteria are growing in a carbon-limited environment and a carbon pool needs to be maintained via the tricarboxylic acid (TCA) cycle to sustain gluconeogenesis and other biosynthetic pathways (10, 28, 29). Under anaerobic conditions, the TCA cycle is not functional; nonetheless, the biosynthetic precursors are maintained by alternative pathways: the reductive pathway producing succinyl-CoA and the oxidative pathway producing 2-ketoglutarate (10). Additionally, carbon flow can be mediated between these pathways by the action of the glyoxylate cycle enzyme AceA, which converts isocitrate to succinate and glyoxylate (9).
In our microarray experiment, we compared the differential gene expression as a result of addition of H2 to the cultures; the two cultures were otherwise grown under the same (anaerobic respiration) condition. Therefore, any change in gene expression observed indicates the response of the bacteria to the H2 added to the growth medium. Gene expression analysis revealed that both aceA and aceB are significantly H2 upregulated. More interestingly, the regulator of the ace operon, iclR, that encodes a repressor protein known to partly regulate the ace operon (51) is upregulated about 3.0-fold by H2 (Table 2).
We thus investigated the physiological expression of the AceA and AceB proteins by enzyme activity assays. Equal amounts of total proteins obtained in crude cell extracts of S. Typhimurium incubated with versus without H2 were compared for enzyme activities by spectrophotometric analyses. The enzymatic activities of AceA and AceB were higher in cells with H2 than in cells without H2 (Table 5). The activity of AceA was, however, markedly higher than that of AceB. We further confirmed the involvement of AceA and AceB in the H2-facilitated anaerobic growth of S. Typhimurium by gene deletion experiments. The aceA and aceB genes were deleted individually using the lambda Red system to produce ΔaceA (RLK3) and ΔaceB (RLK4) mutants (Table 1). The growth of the mutants and the wild type was then compared in CR-Hyd medium (without glucose) in H2-added versus H2-absent conditions. Both mutants exhibited H2-enhanced growth and achieved final yields similar to those for H2-incubated wild-type cells. However, the growth rates of the mutants in the exponential phase were approximately 2.0- to 2.5-fold less than that of the wild type (Fig. 3 and data not shown). In repeated growth experiments, the wild type consistently reached maximum growth yield earlier than the mutants in a way similar to the results shown in Fig. 3. Without H2, growth rates and yields of the two mutants were similar to those of the wild type. These findings suggest that the glyoxylate bypass enzymes aceA and aceB are important for the full-blown H2-enhanced growth of S. Typhimurium. In a carbon-limited growth environment, H2 likely stimulates carbon flow via glyoxylate in order to conserve carbon. Increased AceA activity indicates an increased carbon flow from isocitrate via glyoxylate, thus minimizing the loss of carbon through decarboxylation of isocitrate to 2-ketoglutarate.
Table 5.
Comparison of isocitrate lyase and malate synthase enzyme activities in S. enterica serovar Typhimurium strain JSG210 grown with versus without H2
| Enzyme | Activity (μmol/min/mg)a |
|---|---|
| AceA (isocitrate lyase) | |
| + H2 | 53.0 ± 7.8* |
| No H2 | 9.7 ± 6.9 |
| AceB (malate synthase) | |
| + H2 | 18.9 ± 0.8 |
| No H2 | 11.1 ± 3.7 |
*, significantly increased activity compared to the H2-absent condition (P, <0.01, n = 3).
The carbon-conserving glyoxylate cycle enzymes play important roles in the pathogenesis of several fungal and bacterial pathogens (for a review, see reference 14). Transcriptional profiling of phagocytosed cells showed an upregulation of all the steps of the glyoxylate cycle in Candida albicans (35). In Mycobacterium tuberculosis, isocitrate lyase mRNA expression is increased in response to human macrophages (13, 20), and malate synthase enhances adherence of the bacteria to lung epithelial cells (26). The involvement of isocitrate lyase and malate synthase in the pathogenesis of Salmonella has also been studied, and it was shown that isocitrate lyase is required for persistence of Salmonella during chronic infection but not for acute lethal infection (18, 52). Our finding shows that in a carbon-poor medium the glyoxylate pathway is maximized by H2 so that carbon is conserved. This may be useful for Salmonella survival under H2-rich but carbon-poor conditions such as that expected to occur within the host intestinal tract.
The addition of H2 also stimulates the expression of genes that encode proteins associated with the d-glucarate and d-glycerate utilization pathways (Table 2). The enzyme glycerate kinase (GarK) is central to both of these pathways; in E. coli, it catalyzes the synthesis of 2-phosphoglycerate by utilizing glyoxylate when the latter is available as the main carbon substrate (21, 27). When d-glucarate, d-glycerate, or d-galactarate is available as the carbon source, GarK utilizes the substrate through a catabolic pathway (3). The gene garK is 3.4-fold upregulated by H2 (Table 2). The genes encoding glucarate permease (gudT), glucarate dehydrogenase (gudD), and tartronate semialdehyde reductase (garR) that are required for glucarate uptake and utilization in E. coli (46) are also significantly upregulated by H2 in S. Typhimurium (Table 2). Furthermore, in E. coli, the operons that encode the enzymes of the d-glucarate, d-galactarate, and d-glycerate pathways are commonly regulated by an autogenous regulator, SdaR (43). S. Typhimurium also possesses protein CdaR (carbohydrate diacid transcriptional activator, encoded by cdaR) with a similar function and 97% homology with SdaR. The cdaR gene is 2.2-fold upregulated by H2 (Table 2). Therefore, it is possible that the H2-stimulated expression of the genes of the d-glucarate and d-glycerate pathways is linked to the increased transcription of cdaR by H2.
These findings connect H2 metabolism to increased carbon use via the glyoxylate and the d-glucarate/d-glycerate pathways. During anaerobic growth in CR-Hyd mediun with added H2, Salmonella is faced with a condition whereby energy is adequate but carbon is limited. The increased expression of the carbon-acquisition genes as well as the glyoxylate and the d-glycerate pathway genes is likely a transcriptional response by the bacteria to acquire and conserve carbon while growing in a carbon-deficient condition.
The H2-stimulated up-expression of the d-glucarate utilization pathway in salmonellae might have important implications for survival of the bacteria under carbon-limited conditions within the animal host. d-Glucarate is normally present in the tissues and body fluids of humans (15, 38, 40) and is a major serum organic acid in humans (4). d-Glucarate is also considered to have antitumor and chemopreventive properties (31, 55) and is taken as a dietary supplement. The possession of the glucarate uptake and utilization mechanism could be particularly beneficial to the pathogenic bacteria when growing in the competitive environment within the host intestine, where most of the readily utilizable sugar sources have been exhausted by the host microbial flora. In such a condition, activation of the glucarate utilization mechanism may be expected to increase the survival of the pathogen by permitting use of an alternative carbon source. The presence of a glucarate catabolic pathway in E. coli is considered to be an evolved mechanism for use of glucarate as an alternative carbon source (46).
H2 downregulates the expression of virulence-associated genes in S. Typhimurium in vitro.
The microarrays revealed a 2- to 100-fold downregulation of virulence-associated genes by H2. A majority of the downregulated genes belong to the functional group of prophages and Salmonella pathogenicity islands (SPI). This indicates a much compromised virulence of the strain under the condition tested by the microarrays. However, several genes involved in cell growth which also have important roles in virulence, such as the flagellar biosynthesis-, cell envelope-, and lipopolysaccharide (LPS) biosynthesis-associated genes, are upregulated (Fig. 1; Table 2). The cells are in an in vitro nutrient-limited condition, and it can be predicted that their primary foci under such condition would be survival and cell growth rather than invasion and proliferation. It is noteworthy that several genes associated with LPS and nucleotide biosynthesis were downregulated during intracellular infection (16, 41).
In this study, we observed that H2 exposure causes an upregulation of expression of genes and respective proteins required for the transport and metabolism of necessary nutrients. One major change caused by H2 is increased carbon use, metabolism, acquisition and conservation, linking carbon and energy metabolism to sustain H2-enhanced growth. We propose the results have relevance to in vivo situations where carbon is limited and H2 is abundant (36).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by award number 1R21AI073322 from NIH.
We thank Jennifer Turpin for technical assistance with microarray analyses and Erica F. Miller for the preparation of 14C-glutamine.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Ajl S. J. 1956. Conversion of acetate and glyoxylate to malate. J. Am. Chem. Soc. 78:3230–3231 [Google Scholar]
- 2. Ballantine S. P., Boxer D. H. 1985. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J. Bacteriol. 163:454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blumenthal H. J., Fish D. C. 1963. Bacterial conversion of D-glucarate to glycerate and pyruvate. Biochem. Biophys. Res. Commun. 11:239–243 [DOI] [PubMed] [Google Scholar]
- 4. Blumenthal H. J., Lucuta V. L., Blumenthal D. C. 1990. Specific enzymatic assay for D-glucarate in human serum. Anal. Biochem. 185:286–293 [DOI] [PubMed] [Google Scholar]
- 5. Boyer E., Bergevin I., Malo D., Gros P., Cellier M. F. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohen G. N., Rickenberg H. V. 1956. Concentration specifique reversible des amino acides chez Escherichia coli. Ann. Inst. Pasteur (Paris) 91:693–720 [PubMed] [Google Scholar]
- 7. Compan I., Touati D. 1993. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J. Bacteriol. 175:1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cotruvo J. A., Jr., Stubbe J. 2008. NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 105:14383–14388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Creaghan I. T., Guest J. R. 1977. Suppression of the succinate requirement of lipoamide dehydrogenase mutants of Escherichia coli by mutations affecting succinate dehydrogenase activity. J. Gen. Microbiol. 103:183–194 [DOI] [PubMed] [Google Scholar]
- 10. Cronan J. E., Jr., LaPorte D. 9 November 2006, posting date Tricarboxylic acid cycle and glyoxylate bypass. In Böck A.(ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology (http://www.ecosal.org). ASM Press, Washington, DC: doi: 10.1128/EcoSal.3.5.2 [Google Scholar]
- 11. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Difco Laboratories, Division of Becton Dickinson and Company 1998. Difco manual, 11th ed., p. 383–385 Difco Laboratories, Sparks, MD [Google Scholar]
- 13. Dubnau E., Fonta N. P., Manganelli R., Soares-Appel S., Smith I. 2002. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect. Immun. 70:2787–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunn M. F., Ramírez-Trujillo J. A., Hernandez-Lucas I. 2009. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155:3166–3175 [DOI] [PubMed] [Google Scholar]
- 15. Dutton G. J. 1980. Glucuronidation of drugs and other compounds, p. 83–89 CRC Press, Boca Raton, FL [Google Scholar]
- 16. Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J. C. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 17. Evans M. R., et al. 2011. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol. 11:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang F. C., Libby S. J., Castor M. F., Fung A. M. 2005. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect. Immun. 73:2547–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Foster J. W., Spector M. P. 1995. How Salmonella survive against the odds. Annu. Rev. Microbiol. 49:145–174 [DOI] [PubMed] [Google Scholar]
- 20. Graham J. E., Clark-Curtiss J. E. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. U. S. A. 96:11554–11559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen R. W., Hayashi J. A. 1962. Glycolate metabolism in Escherichia coli. J. Bacteriol. 83:679–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Janakiraman A., Slauch J. M. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146–1155 [DOI] [PubMed] [Google Scholar]
- 23. Kauko T., et al. 2010. Phenotype MicroArray in the metabolic characterisation of Salmonella serotypes Agona, Enteritidis, Give, Hvittingfoss, Infantis, Newport and Typhimurium. Eur. J. Clin. Microbiol. Infect. Dis. 29:311–317 [DOI] [PubMed] [Google Scholar]
- 24. Kehres D. G., Janakiraman A., Slauch J. M., Maguire M. E. 2002. SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3159–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kehres D. G., Maguire M. E. 2003. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 27:263–290 [DOI] [PubMed] [Google Scholar]
- 26. Kinhikar A. G., et al. 2006. Mycobacterium tuberculosis malate synthase is a laminin-binding adhesin. Mol. Microbiol. 60:999–1013 [DOI] [PubMed] [Google Scholar]
- 27. Kornberg H. L. 1966. Anaplerotic sequences and their role in metabolism. Essays Biochem. 2:1–31 [Google Scholar]
- 28. Kornberg H. L. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 99:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kornberg H. L., Krebs H. A. 1957. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179:988–991 [DOI] [PubMed] [Google Scholar]
- 30. Kwon Y. M., Ricke S. C. 1998. Survival of a Salmonella typhimurium poultry isolate in the presence of propionic acid under aerobic and anaerobic conditions. Anaerobe 4:251–256 [DOI] [PubMed] [Google Scholar]
- 31. Lajolo C., et al. 2010. Calcium glucarate inhibits DMBA-induced oral carcinogenesis in the hamster: histomorphometric evaluation. Anticancer Res. 30:843–849 [PubMed] [Google Scholar]
- 32. Lamichhane-Khadka R., Kwiatkowski A., Maier R. J. 2010. The Hyb hydrogenase permits hydrogen-dependent respiratory growth of Salmonella enterica serovar Typhimurium. mBio 1(5):e00284–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lawhon S. D., et al. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633–1645 [DOI] [PubMed] [Google Scholar]
- 34. Livak K., Schmittgen J. T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25(4):402–408 [DOI] [PubMed] [Google Scholar]
- 35. Lorenz M. C., Bender J. A., Fink G. R. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maier R. J. 2005. Use of molecular hydrogen as an energy substrate by human pathogenic bacteria. Biochem. Soc. Trans. 33:83–85 [DOI] [PubMed] [Google Scholar]
- 37. Maier R. J., Olczak A., Maier S. S., Soni S., Gunn J. 2004. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect. Immun. 72:6294–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marsh C. A. 1963. Metabolism of D-glucuronolactone in mammalian systems. II. Conversion of D-glucuronolactone into D-glucaric acid by tissue preparation. Biochem. J. 87:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin J. E., Imlay J. A. J. A. 2011. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol. Microbiol. 80:319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matsui M., Fukuo A., Watanabe Y., Wanibe T., Okada M. 1977. Studies on the glucaric acid pathway in the metabolism of D-glucuronic acid in mammals. IV. Fluorometric method for the determination of glucaric acid in serum. Chem. Pharm. Bull. 20:845–848 [DOI] [PubMed] [Google Scholar]
- 41. Maurer J. J., Doggett T. A., Burns-Keliher L., Curtis R., 3rd 2000. Expression of the rfa, LPS biosynthesis promoter in Salmonella typhimurium during invasion of intestinal epithelial cells. Curr. Microbiol. 41:172–176 [DOI] [PubMed] [Google Scholar]
- 42. Mehta N., Benoit S. S., Mysore J. V., Sousa R. S., Maier R. J. 2005. Helicobacter hepaticus hydrogenase mutants are deficient in hydrogen-supported amino acid uptake and in causing liver lesions in A/J. mice. Infect. Immun. 73:5311–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Monterrubio R., Baldoma L., Obradors N., Aguilar J., Badia J. 2000. A common regulator for the operons encoding the enzymes involved in d-galactarate, d-glucarate, and d-glycerate utilization in Escherichia coli. J. Bacteriol. 182:2672–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ollis A. A., Manning M., Held K. G., Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 73:466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Postle K. 1993. TonB protein and energy transduction between membranes. J. Bioenerg. Biomembr. 25:591–601 [DOI] [PubMed] [Google Scholar]
- 46. Roberton A. M., Sullivan P. A., Jones-Mortimer M. C., Kornberg H. L. 1980. Two genes affecting glucarate utilization in Escherichia coli K12. J. Gen. Microbiol. 117:311–382 [DOI] [PubMed] [Google Scholar]
- 47. Rodrigue D. C., Tauxe R. V., Rowe B. 1990. International increase in Salmonella enteritidis: a new pandemic? Epidemiol. Infect. 105:21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schauer K., Rodionov D. A., de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33:330–338 [DOI] [PubMed] [Google Scholar]
- 49. Skare J. T., Ahmer M. M., Seachord C. L., Darveau R. P., Postle K. 1993. Energy transduction between membranes: TonB, a cytoplasmic membrane protein, can be chemically crosslinked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302–16308 [PubMed] [Google Scholar]
- 50. Smith R. A., Gunsalus I. C. 1954. Isocitritase: a new tricarboxylic acid cleavage system. J. Am. Chem. Soc. 76:5002–5003 [Google Scholar]
- 51. Sunnarborg A., Klumpp D., Chung T., LaPorte D. C. 1990. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J. Bacteriol. 172:2642–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tchawa Yimga M., et al. 2006. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74:1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vignais P. M. 2007. Hydrogenases and H+-reduction in primary energy conservation. Results Probl. Cell Differ. 45:223–244 [DOI] [PubMed] [Google Scholar]
- 54. Vignais P. M., Colbeau A. 2004. Molecular biology of microbial hydrogenases. Curr. Issues Mol. Biol. 6:159–188 [PubMed] [Google Scholar]
- 55. Webb T. E., Abou-Issa H., Stromberg P. C., Curley R. C., Jr., Nguyen M. H. 1993. Mechanism of growth inhibition of mammary carcinomas by glucarate and the glucarate: retinoid combination. Anticancer Res. 13(6A):2095–2099 [PubMed] [Google Scholar]
- 56. Xia X., McClelland M., Wang Y. 2005. WebArray: an online platform for microarray data analysis. BMC Bioinformatics 6:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zbell A. L., Maier R. J. 2009. Role of Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 75:1456–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zbell A. L., Benoit S. L., Maier R. J. 2007. Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiology 153:3508–3516 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.