Abstract
Carolacton, a secondary metabolite isolated from the myxobacterium Sorangium cellulosum, disturbs Streptococcus mutans biofilm viability at nanomolar concentrations. Here we show that carolacton causes leakage of cytoplasmic content (DNA and proteins) in growing cells at low pH and provide quantitative data on the membrane damage. Furthermore, we demonstrate that the biofilm-specific activity of carolacton is due to the strong acidification occurring during biofilm growth. The chemical conversion of the ketocarbonic function of the molecule to a carolacton methylester did not impact its activity, indicating that carolacton is not functionally activated at low pH by a change of its net charge. A comparative time series microarray analysis identified the VicKRX and ComDE two-component signal transduction systems and genes involved in cell wall metabolism as playing essential roles in the response to carolacton treatment. A sensitivity testing of mutants with deletions of all 13 viable histidine kinases and the serine/threonine protein kinase PknB of S. mutans identified only the ΔpknB deletion mutant as being insensitive to carolacton treatment. A strong overlap between the regulon of PknB in S. mutans and the genes affected by carolacton treatment was found. The data suggest that carolacton acts by interfering with PknB-mediated signaling in growing cells. The resulting altered cell wall morphology causes membrane damage and cell death at low pH.
INTRODUCTION
Streptococcus mutans, a facultative anaerobic, Gram-positive microorganism, is thought to be the main causative agent of dental caries (43, 49, 79). Within the dental plaque, a complex biofilm community consisting of up to 500 different species of microorganisms, S. mutans can compete in its ecological niche due to its extreme aciduricity and acidogenicity, genetic transformability, and a repertoire of bacteriocins (58). Pathogenic bacteria forming biofilms within the human body are a severe health problem due to their low susceptibility to conventional drug treatments such as, e.g., antibiotics. Therefore, new substances that will erase bacteria living in biofilms or reduce their pathogenicity are urgently needed.
Carolacton, a secondary metabolite isolated from the myxobacterium Sorangium cellulosum, was shown to have only minor effects on planktonically growing bacteria but effectively kills S. mutans biofilm cells (33). The three-dimensional structure of carolacton was elucidated by Jansen et al. (28). Carolacton is a macrolide ketocarbonic acid. LIVE/DEAD staining of carolacton-treated biofilms of S. mutans proved that profound membrane damage was caused by carolacton. The final biofilm mass and the growth rate of biofilms were only marginally reduced by carolacton treatment (33). Carolacton-treated biofilm cells had an abnormal cell shape and an increased chain length, which is suggestive of cell wall changes and a defect in cell division. Carolacton has a sigmoidal dose-response curve. At a carolacton concentration of 10 nM, 35% membrane damage already was observed, and between 53 nM and 53 mM the membrane damage was constantly approximately 60% (33). The molecular target in the cell must be present in a very low copy number, since it is saturated at a very low carolacton concentration. This suggests that a signaling pathway rather than a metabolic enzyme is affected.
Signaling in S. mutans occurs, like in other Gram-positive bacteria, mainly via protein phosphorylation (14, 17). Bacteria sense and respond to changes in the environment via two-component signal transduction systems (TCSs) and, as recently discovered, via serine/threonine protein kinases (STPKs). Key virulence traits of S. mutans, e.g., acid tolerance, bacteriocin production, and biofilm formation, are controlled via one or several of its 14 TCSs (39). Environmental signals are sensed by the membrane-bound histidine kinase (HK), which is thereby autophosphorylated at a histidine residue. Transfer of the phosphate group to a conserved asparagine residue of the cognate response regulator (RR) induces or represses the transcription of target genes (73). Cross regulation between certain histidine kinases and noncognate response regulators was postulated (12). Kunze et al. (33) showed that the competence regulon comCDE, which plays an essential role in biofilm formation, virulence, adaptation to acidic conditions, and genetic competence, is not the primary target of carolacton, since a ΔcomD mutant was only slightly less sensitive than the wild type. However, a reduction of the competence-stimulating peptide (CSP)-induced comX promoter activity by carolacton was shown (33).
Among the TCSs of S. mutans, the VicKRX system plays a key role in maintaining the cell membrane and cell wall integrity. VicKRX controls the expression of fructosyltransferases, glucosyltransferases, and glucan binding proteins, indicating an essential role in biofilm formation (36, 70). Since a VicR mutant is not viable, this response regulator is apparently an essential gene (70, 81). A recent study revealed that PknB, a serine/threonine protein kinase which regulates competence development, bacteriocin production, and cell wall metabolism in S. mutans, may also modulate the activity of VicKRX (3). It was generally believed that signal transduction in prokaryotes is mediated mainly by histidine protein kinases. Recent studies showed that STPKs and their associated and genetically linked serine/threonine protein phosphatases (STPPs), which until now have been found mainly in eukaryotes, also display an alternative signal transduction system in prokaryotes (14). The N-terminal kinase domains of bacterial STPKs are predicted to be located in the cytoplasm, followed downstream by a central membrane domain and an extracellular C-terminal sensory part. Signal transduction is mediated through autophosphorylation of the STPK at a serine or threonine residue (hence the terminology) and a subsequent transfer of the phosphate group to target proteins in order to modulate their activity. So far, little information and no general prediction of these target proteins is available (16, 30, 47, 53). STPKs regulate various cellular functions, including stress responses (52), biofilm formation (26), sporulation (32), and metabolic and developmental processes (44, 51). In the genome of S. mutans, a single STPK gene, named pknB, was found (26).
The first aim of this study was to investigate the effect of carolacton on biofilms of S. mutans in more depth. To this end, the release of proteins and DNA into the supernatants of carolacton-treated biofilms was determined during biofilm growth. Since growth of S. mutans in unbuffered media is accompanied by a strong acidification and the maximum carolacton activity correlates with a rapid drop in pH, we also tested the effect of pH on the activity of carolacton. Using a chemically modified carolacton (carolacton methylester), we analyzed the effect of the net charge of the molecule on its biological activity. Second, a time series analysis of the transcriptome of carolacton-treated S. mutans biofilms was performed to elucidate the effect of carolacton on the genetic level and to identify strongly regulated genes and pathways. Finally, the sensitivity of mutants with deletions in potential primary target genes of carolacton was studied in more detail.
MATERIALS AND METHODS
Strains, media, and growth conditions.
S. mutans wild-type strain UA159 (ATCC 700610) and the histidine kinase-deficient mutants (kindly provided by C. Levesque, University of Toronto) (39) were grown routinely in Todd-Hewitt broth (THB) (Becton Dickinson, Heidelberg, Germany). For the mutants, 10 μg/ml erythromycin (Sigma-Aldrich, Taufkirchen, Germany) was added to the medium. The medium for the PknB-complemented strain contained 10 μg/ml erythromycin and 20 μg/ml chloramphenicol (Sigma-Aldrich, Taufkirchen, Germany). For biofilm growth, 0.5% sucrose (Sigma, Taufkirchen, Germany) was added to THB (THBS). All media were degassed by flushing with nitrogen, and experiments were carried out at 37°C in an anaerobic chamber (Don Whitley Scientific, Shipley, England) which provided an atmosphere of 80% N2, 10% H2, and 10% CO2. The phosphate-buffered media were prepared according to standard procedures (66).
Deletion and complementation of pknB.
For the construction of a pknB-deficient mutant, the PCR ligation mutagenesis approach (35) was used to replace the pknB-gene by an erythromycin resistance cassette via double homologous recombination. Primers pknB1 and pknB2 (see Table S1 in the supplemental material) were used to amplify the 5′ flanking region of pknB, introducing an AscI restriction site. To amplify the 3′ flanking region, the primers pknB3 and pknB4 were used, thereby introducing a restriction site for FseI. The erythromycin resistance cassette was amplified from plasmid pALN122 (76) using primers ERMFor and ERMRev, containing the restriction sites for AscI and FseI, respectively. After digestion with the appropriate restriction enzyme(s), the three amplicons were ligated together and transformed in S. mutans UA159 according to the method of Li et al. (40). Mutants were selected on THB agar plates containing erythromycin (10 μg/ml) and verified by PCR using primers (pknB1 and pknB4) flanking pknB. The altered phenotype of the ΔpknB mutant (3, 26) was analyzed under a microscope. For long-term storage of the mutant, a single colony was used to inoculate 10 ml of THB containing 10 μg/ml erythromycin. One milliliter of the exponentially grown culture was added to 200 μl of sterile glycerin in a cryotube and stored at −80°C. For all experiments precultures were inoculated directly from the freezer stock to minimize secondary mutations, as reported previously (3).
For the complementation of the mutant, the entire pknB gene, including the natural ribosomal binding site, was PCR amplified from genomic DNA of S. mutans UA159 using primers PknBFor and PknBRev. Subsequently the PCR product was cloned blunt ended into the SmaI restriction site of replicative vector pIB166 (5) bearing a constitutive, synthetic promoter and transformed into Escherichia coli DH5α. After verification of the vector sequence by DNA sequencing, the construct was transformed via electroporation according to the procedure of Buckley et al. (8) into the pknB-deficient strain. The selection of positive clones was carried out on THB agar plates containing erythromycin (10 μg/ml) and chloramphenicol (20 μg/ml).
Construction of mutants with deletions of atlA, mutacin V, SMU.609, and comX.
The deletion mutants were constructed using the same strategy as described above for the ΔpknB mutant. The primers used are listed in Table S1 in the supplemental material.
Carolacton.
Carolacton, derived from a bioreactor fermentation of Sorangium cellulosum (28), was dissolved in methanol at a concentration of 1 mg/ml. Based on this stock solution, different dilutions in methanol were made. The resulting methanol concentrations in media did not exceed 2.5% (vol/vol).
Biofilm experiments.
For the time series experiment, an overnight culture of UA159 in THB was diluted to an optical density at 600 nm (OD600) of 0.05 in THBS medium. Eight hundred microliters of this dilution was transferred into each well of a 24-well polystyrene plate (Greiner Bio-One, Frickenhausen, Germany) and grown anaerobically for 3.5 h at 37°C. Each time point and condition (treated/untreated) tested was represented in 12 wells in parallel. After 3.5 h, the supernatant was removed and fresh THBS medium supplemented with 0.25 μg/ml carolacton (or an equal volume of methanol as a control) was added. At 20, 40, 60, 120, and 300 min after carolacton addition, samples were collected. For sampling, the supernatant was completely removed, fresh medium (200 μl/well) and RNA Protect (Qiagen, Hilden, Germany) (400 μl/well) were added, and the biofilms were scraped off using a cell scraper (Nunc, Langenselbold, Germany). The cells from 10 to 12 different wells representing the same experimental condition were pooled. Subsequently, biofilm cells were harvested by centrifugation (6000 rpm, 10 min) and washed twice with nuclease-free water (Sigma, Taufkirchen, Germany). After centrifugation, cells were frozen or immediately used for RNA extraction.
For endpoint determination, carolacton dissolved in methanol (final concentration of 0.25 μg/ml) was added under sterile conditions to the wells of a 24-well polystyrene microtiter plate and incubated until all of the methanol was evaporated. For the control wells, an equivalent volume of methanol was used. An overnight culture of S. mutans grown in THB was diluted to an OD600 of 0.01, and 800 μl was transferred into each well. Biofilms were grown anaerobically for 8 h at 37°C, and cells were harvested and prepared as described for the time series experiment. Control wells were prepared in the same way and used for LIVE/DEAD staining to control biofilm inhibition by carolacton.
Quantitative reverse transcription-PCR (RT-PCR).
Primers for quantitative PCR (qPCR) were designed using the Primer 3 software (http://frodo.wi.mit.edu) to generate amplicons in the range of 100 to 150 bp. All primers were purchased from MWG Eurofins Operon (Ebersberg, Germany). One microgram of total RNA was reverse transcribed using the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The cDNA was subsequently diluted 1:20 for use in PCRs. For the amplification, the QuantiTect Sybr Green kit (Qiagen, Hilden, Germany) was used. Fifteen-microliter reaction mixtures with primer concentrations of 0.25 μM were run in the Light Cycler 480 (Roche, Germany). To determine the primer efficiencies, serial dilutions of pooled cDNA were measured in triplicate. Each sample was measured in triplicate, and each experiment was performed at least three times. For data normalization, an RNA spike (specific for the red fluorescent protein DsRed) and the housekeeping gene SMU.1331 was used. SMU.1331 was found in the microarray analysis not to be differentially expressed under the experimental conditions used. Data analysis was performed as described by Pfaffl (61).
RNA isolation.
Harvested cells were resuspended in 200 μl of lysozyme-mutanolysin solution (15 mg/ml lysozyme and 500 U/ml mutanolysin) and incubated in a shaker (1,100 rpm) at 25°C for 45 min. Subsequently the mixture was added to 700 μl of RLT buffer (originating from the Qiagen RNeasy kit) and 50 mg of acid-washed glass beads (125-μm diameter) in a 15 ml Falcon tube and vortexed for 3 min. After centrifugation (6,000 rpm, 1 min), the supernatant was transferred to an Eppendorf tube and centrifuged again (13,000 rpm, 2 min). The resulting supernatant was transferred to a new tube, and 470 μl of ethanol was added and carefully mixed by pipetting. For the following steps, the RNeasy kit (Qiagen, Hilden, Germany) was used. Two DNase I treatments, on a column (10 U) and in solution (5 U), were performed to remove all genomic DNA. The RNA was purified according to the procedure described in the RNeasy kit (Qiagen, Hilden, Germany). The integrity of RNA extractions was analyzed on a denaturing formaldehyde agarose gel. The samples were proven to be free of DNA contamination by PCR with 3 different primer pairs specific for randomly chosen genes of S. mutans.
Labeling procedure and hybridization.
For the microarray experiments, a customized array was used as described previously (82). The array design was performed using the eArray platform (Agilent, Böblingen, Germany). Each array consists of 15,000 spots, and 8 arrays are located on one slide (8 × 15K design). Three different antisense probes (60 bp length) for all open reading frames and intergenic regions of S. mutans UA159 were designed; each probe is present in duplicate randomly distributed over the array. In addition to Agilent positive and negative controls, spike-in controls for RFPexpress and GFPmut2 were implemented on the array.
Two micrograms of total RNA was labeled using the ULS labeling kit (Kreatech, Netherlands). The degree of incorporation was checked to be in the range of 1.0 to 3.6 using a NanoDrop spectrophotometer. Dye swap samples were mixed and fragmented for 30 min at 60°C according to the instructions of the hybridization kit from Agilent (Böblingen, Germany). Hybridization was carried out according to the recommendations of the manufacturer. The samples were hybridized to the arrays for 16 h in a hybridization oven (Agilent) at 65°C under rotation. Subsequently the arrays were washed according to the Agilent protocol. For scanning, the Agilent DNA microarray scanner was used at settings recommended by Agilent.
Data processing.
The feature extraction of the microarray scans was performed using commercially available software from Agilent. For the secondary analysis of data, including the identification of differentially expressed genes, the R package (http://www.r-project.org) LIMMA was applied. For dye-specific within-array normalization of the data, the Lowess algorithm (13) was used. For between-array normalization, the quantile normalization method (7) was applied. From the log-transformed (to base 2) intensities the fold change was calculated by subtracting the overall normalized and background-corrected value for the untreated sample from the value for the treated sample.
Genes with a threshold value of absolute log2 fold change above or equal to 0.8 (corresponding to a numerical value of 1.74) and a P value below 0.01 were considered to be differentially expressed. Principal-component analysis (PCA) was performed on the samples using the Princomp function of the R programming language. K-means clustering of regulated genes was performed using the freeware Cluster 3. For the number of clusters, 9 was chosen as the starting value for the algorithm, and the correlation coefficient was used as the similarity measure. For the visualization of the expression curves of the different clusters, the mean of the log2 fold changes of all genes belonging to one cluster was calculated.
LIVE/DEAD staining.
For the viability assays using the commercially available LIVE/DEAD staining dyes (Invitrogen, Darmstadt, Germany), biofilms were grown for 20 h in black optical microtiter plates (Nunc, Langenselbold, Germany). The supernatants of the biofilms were completely removed, and biofilm cells were washed twice with 0.85% NaCl solution.
One hundred microliters of staining solution (15 μl of SYTO 9 and 15 μl of propidium iodide dissolved in 10 ml 0.85% NaCl solution) was added per well and incubated for 15 min at room temperature in the dark. Green and red fluorescence values were determined using a Victor 3 fluorescence plate reader (Perkin-Elmer, Rodgau, Germany). The green/red ratio was calculated, and the inhibition was determined relative to untreated reference cells as described previously (33). The mean of the green/red ratio of the treated samples was divided by the mean of the green/red ratio of the controls and multiplied by 100, giving the percentage of relative viability. Subtraction of this value from 100% results in the percentage of inhibition.
Construction of a β-galactosidase reporter strain (S. mutans MR15) and β-galactosidase assay of biofilm supernatants.
The entire E. coli lacZ gene, including the natural ribosomal binding site, was PCR amplified using genomic DNA of E. coli strain BL21 as the template with primers lacZFor and lacZRev. After purification, the PCR product was blunt-end cloned via the SmaI restriction site in vector pIB166 (5) and transformed into E. coli DH5α. PIB166 bears the constitutive promoter P23 derived from Lactococcus lactis and is replicative in S. mutans.
Positive clones were selected on chloramphenicol-containing LB agar and verified by sequencing of plasmid DNA. Subsequently the plasmid was transformed into S. mutans UA159 according to the procedure of Li et al. (40), and the strain was functionally tested for hydrolysis of the substrate ortho-nitrophenyl-β-d-galactopyranoside (ONPG). For the β-galactosidase assay, the supernatants of biofilms were collected and centrifuged (13,000 rpm, 2 min). Eight hundred microliters of cell-free supernatant was mixed with 240 μl of 5× Z buffer (0.3 M Na2HPO4, 0.2 M NaH2PO4, 0.05 M KCl, 0.005 M MgSO4, and 0.25 M β-mercaptoethanol) and 160 μl of ONPG (4 mg/ml, dissolved in water). The mixture was incubated at 30°C for 2 h, and the optical density was measured at 420 nm using a spectrophotometer. Medium treated by the same procedure was used for blank measurements.
SDS-PAGE of biofilm supernatants.
Supernatants of treated and untreated biofilms grown in 24-well polystyrene microtiter plates were harvested and centrifuged (13,000 rpm, 2 min) to remove cells. Trichloric acid (Sigma, Taufkirchen, Germany) was added to a final concentration of 10% (vol/vol), and proteins were precipitated at 4°C for at least 4 h. After centrifugation (13,000 rpm, 15 min, 4°C), the supernatants were removed and protein pellets were redissolved in 100 μl of water and neutralized by addition of 5 μl saturated Tris base solution. Twenty microliters of 6× Laemmli buffer (34) was added, followed by an incubation for 5 min at 95°C. Subsequently, 20 μl of protein solution was loaded on a 12.5% polyacrylamide gel and run at 100 V for approximately 2 to 3 h. Protein bands were visualized using the silver staining procedure as described Blum et al. (6).
Quantification of external DNA (eDNA) in biofilm supernatants.
Supernatants were harvested as described above, and 600 μl of cell-free supernatant was mixed with 200 μl of Tris-EDTA (TE) buffer (pH 7.0) at 4°C. Eight hundred microliters of phenol-chloroform-isoamyl alcohol mixture (25:24:1) was added and extensively mixed (by vortexing). Following centrifugation at 4°C (13,000 rpm, 5 min), the aqueous phase was transferred into a new tube and a second extraction step with 800 μl of chloroform was performed. Seventy microliters of 5 M NaCl and 1,400 μl of ethanol were added to the resulting aqueous phase, and the DNA was precipitated overnight at −70°C. After centrifugation (13,000 rpm, 20 min), the pellet was washed with 75% ethanol, centrifuged again, and dissolved in 25 μl of nuclease-free water (Sigma, Taufkirchen, Germany). The resulting DNA was used as the template for quantitative PCR using three different primer pairs specific for sequences randomly distributed over the genome of S. mutans (specific for SMU.1997, SMU.1331, and SMU.22). Each experiment was repeated three times with DNA derived from three independent experiments and extractions. The ratios of fold change between treated samples and untreated controls were calculated.
Chemical synthesis of carolacton methylester.
Carolacton (1 mg, 2.13 μmol) was dissolved in a solution of methanol and diethyl ether (1:1, 1 ml). Diazomethane gas was bubbled through the solution while stirring at 0°C for 1 h until the yellow color persisted. The excess diazomethane and solvent were removed under a stream of nitrogen. Diazomethane was generated by addition of potassium hydroxide solution (1.08 M in methanol-H2O [1:1, 4 ml]) to a stirred Diazald solution (0.52 M diethyl ether-diethyleneglycol monoethyl ether [1:1, 4 ml]) in a screw-cap glass vial with polytetrafluoroethylene (PTFE) tubing attached. The diazomethane evolved, which is yellow, was allowed to pass through a trap to avoid addition of by-products and then further into the glass vial containing the reaction mixture. The product was purified and analyzed by high-pressure liquid chromatography-mass spectrometry (HPLC-MS), showing the complete absence of carolacton.
Microarray data accession numbers.
The microarray data have been deposited at NCBI-GEO (accession no. GSE26274 and GSE26441).
RESULTS
Carolacton disturbs the membrane integrity of S. mutans biofilm cells, as previously determined by staining with propidium iodide (33). To gain new insights into the effects of carolacton, we determined the amounts of released proteins and DNA in the supernatants of carolacton-treated biofilm cells during growth. In addition, a strain of S. mutans constitutively expressing the intracellular enzyme β-galactosidase from E. coli was constructed (MR15 [Table 1]). The specific β-galactosidase activity of enzyme released into the supernatants of carolacton-treated biofilms was used for quantification of membrane damage.
Table 1.
Strains and plasmids
| Strain or plasmid | Genotype or characteristics | Reference or source |
|---|---|---|
| S. mutans strains | ||
| UA159 | Wild-type ATCC 700610 | ATCC |
| ΔpknB mutant | ΔpknB::Ermr | This study |
| Complemented pknB mutant | ΔpknB::Ermr harboring pIB166-pknB | This study |
| Mutacin V deletion mutant | ΔMutacin V::Ermr | This study |
| ΔSMU.609 mutant | ΔSMU.609::Ermr | This study |
| ΔatlA mutant | ΔatlA::Ermr | This study |
| ΔcomX mutant | ΔcomX::Ermr | This study |
| MR15 | UA159 harboring pIB166-LacZ | This study |
| HK11 | ΔSMU.486::Ermr | 39 |
| HK10 | ΔSMU.577::Ermr | 39 |
| HK6 | ΔSMU.660::Ermr | 39 |
| HK4 | ΔSMU.928::Ermr | 39 |
| HK8 | ΔSMU.1009::Ermr | 39 |
| HK7 | ΔSMU.1037c::Ermr | 39 |
| HK3 | ΔSMU.1145::Ermr | 39 |
| HK12 | ΔSMU.1548::Ermr | 39 |
| HK5 | ΔSMU.1814::Ermr | 39 |
| HK9 | ΔSMU.1965::Ermr | 39 |
| ΔvicK mutant | ΔvicK::Ermr | 39 |
| ΔciaH mutant | ΔciaH::Ermr | 39 |
| ΔcomD mutant | ΔcomD::Ermr | 39 |
| E. coli strains | ||
| DH5 | Cloning strain | Stratagene |
| BL21(DE3) | Expression strain | Stratagene |
| Plasmids | ||
| pIB166 | E. coli-Streptococcus shuttle vector; Cmr | 5 |
| pALN122 | Ermr | 76 |
| pMRE14 | pIB166-pknB | This study |
| pMRE15 | pIB166-lacZ | This study |
SDS-PAGE analysis of supernatants of carolacton-treated biofilms.
Figure S1 in the supplemental material shows the results of the SDS-PAGE of trichloroacetic acid (TCA)-precipitated supernatants of carolacton-treated biofilms. Beginning from 8 h of growth, a significant increase in protein content was observed for the treated samples. Especially in the low-molecular-weight range, many protein bands appeared in the treated samples that were not detected in the controls. The highest band intensities were found after 12 h of biofilm growth, followed by a slight decrease, which perhaps was caused by growth arrest and proteolytic activity in the supernatants. SDS-PAGE analysis clearly demonstrated a leakage of cytoplasmic proteins from carolacton-treated biofilm cells.
Quantification of eDNA in the supernatants.
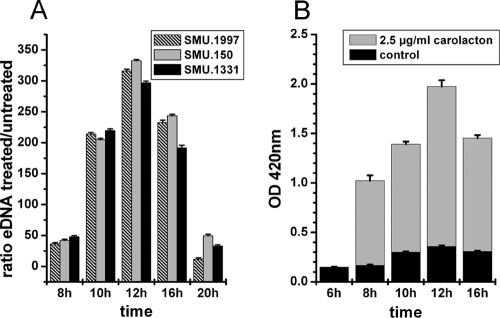
A comparison of external DNA (eDNA) contents in the supernatants of carolacton-treated (2.5 μg/ml) and untreated biofilms was performed using quantitative PCR. Three different primer pairs, specific for regions distributed over the S. mutans genome, were used for quantification. The results are shown in Fig. 1A. After 8 h of growth, the supernatant of carolacton-treated biofilms contained approximately 50 times more DNA than that derived from untreated biofilms. This ratio increased to around 300 after 12 h of growth, followed by a significant decrease to the starting ratio. Nuclease activity in the culture medium may degrade the excess extracellular DNA derived from carolacton-treated cells with increasing incubation time.
Fig. 1.
Carolacton causes leakage of cytoplasmic DNA (A) and proteins (B). (A) Relative quantification of the external DNA (eDNA) of carolacton-treated biofilms. Supernatants of carolacton-treated (2.5 μg/ml) and untreated biofilm cells were analyzed using quantitative PCRs specific for 3 different genes randomly distributed over the S. mutans genome. The means and standard deviations from 3 biological replicates are shown. (B) Carolacton-caused release of intracellular β-galactosidase into the supernatant during biofilm growth of an S. mutans reporter strain (MR15 [Table 1]). The β-galactosidase activity was measured by hydrolysis of the β-galactosidase substrate ortho-nitrophenyl-β-d-galactopyranoside (ONPG) to galactose and o-nitrophenol in the supernatant of carolacton-treated and untreated biofilm cells. The OD420, corresponding to the absorption maximum of o-nitrophenol, is plotted against the time of biofilm growth. Means and standard deviations from 3 biological replicates are shown.
A strong increase of the eDNA amount in the supernatants of carolacton-treated biofilm cells clearly demonstrates the formation of large holes in the bacterial membrane, since DNA is a sterically complex large molecule.
Quantification of membrane damage by a reporter strain (MR15).
For quantification of the amount of carolacton-released protein, an S. mutans reporter strain encoding E. coli β-galactosidase was constructed. The enzyme activity of the released intracellular protein in the culture supernatant was used as a measure of membrane damage and leakage of cytoplasmic content. Determination of β-galactosidase activity is based on the hydrolysis of the enzyme substrate ortho-nitrophenyl-β-d-galactopyranoside to the yellow product ortho-nitrophenol and β-d-galactose. ortho-Nitrophenol is photometrically quantified at 420 nm.
The supernatants of carolacton-treated biofilms and untreated control biofilms were analyzed for their β-galactosidase activities, and the results are shown in Fig. 1B. After 6 h of biofilm growth, no significant difference in the β-galactosidase activities in the supernatants of treated and untreated biofilms was observed. However, beginning from 8 h of growth, the β-galactosidase activity in the supernatant of treated biofilms strongly increased. The highest β-galactosidase activity was observed after 12 h of growth, and then the activity decreased again. For the supernatants of control biofilms, a much smaller increase of β-galactosidase activity was measured over time.
Carolacton is active on growing biofilms at low pH.
The morphological changes caused by carolacton, e.g., increased chain length and larger, bulging cells (33), suggest an influence on cell division and alterations of the cell wall composition. These morphological changes, especially an increase in chain length, were also observed for acid-exposed S. mutans cells (69). The cultivation of S. mutans biofilms in sucrose-containing medium is always accompanied by a strong acidification of the medium, as the cells produce an excess of organic acids during growth. Therefore, the effect of pH on the membrane damage caused by carolacton was evaluated.
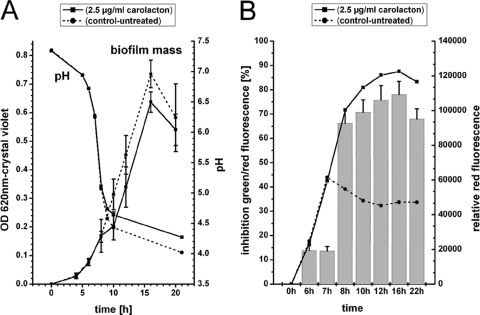
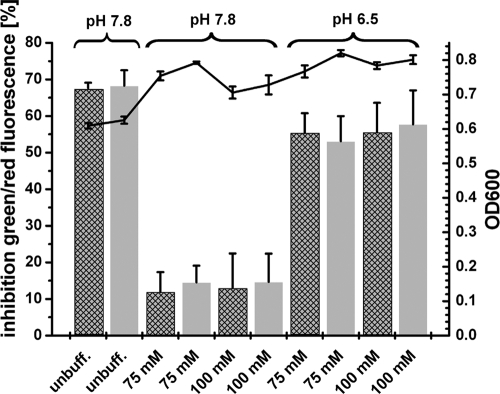
Figure 2A shows the amount of biofilm cells determined by crystal violet staining. There was no difference between treated and untreated cells for the first 8 h. Between 8 h and 10 h, the carolacton-treated biofilms showed a slightly reduced amount of cell mass compared to the controls. This difference could not be compensated during the remaining biofilm cultivation, leading to a slightly reduced amount of cell mass in the carolacton-treated biofilms after 22 h of growth. Figure 2B shows the membrane damage of the biofilms. Again, there is no significant effect of carolacton for the first 7 h. From 8 h onwards, the carolacton-treated biofilm shows 60 to 70% membrane damage (determined by LIVE/DEAD staining) as well as a large increase of red fluorescence, indicative of membrane damage or cell death. Interestingly, the severe drop in pH to pH 5 coincides with the first effect of carolacton; both occur after 8 h. To separate the effects of pH and growth stage on the activity of carolacton, biofilms were grown in buffered THBS medium with either a high (7.8) or a low (6.5) initial pH. As a reference, unbuffered THBS medium (pH 7.8) was used. For medium buffered at pH 7.8, the buffering capacity keeps the pH above pH 5.8 under the chosen experimental conditions during biofilm growth (data not shown). In contrast, in medium buffered at pH 6.5, the final pH values were around 4.3, similar to those found in biofilm cultures in unbuffered media (data not shown). As shown in Fig. 3, carolacton caused approximately 60% inhibition of viability (determined by LIVE/DEAD staining) for the biofilms growing in media that were strongly acidified during growth (unbuffered media and buffered media with an initial pH of 6.5). In contrast, only a very weak inhibition of biofilm viability by carolacton (approximately 10 to 15%) was found in buffered media with an initial pH of 7.8 and a final pH of approximately 5.8. Biofilm growth in the controls was comparable in all media (Fig. 3, line diagram). Thus, the activity of carolacton is tightly coupled to acidification and requires a pH below 5.8. To exclude degradation or inactivation of carolacton at higher pH values, we incubated the pure substance overnight in THBS at pH 7.8 and inoculated the wells with S. mutans the next day. No significant loss of carolacton activity was observed (data not shown). In a further experiment, the growth medium of a 3.5-h-old biofilm was exchanged against different buffers and solutions with pH values in the range of 4.8 to 7.0. Biofilms were not able to grow further in these buffers but remained viable. Addition of carolacton to these buffers caused no inhibition of biofilm viability (data not shown). Thus, we infer that growth is necessary for the inhibitory effect caused by carolacton.
Fig. 2.
Effect of carolacton on biofilm mass (A) and viability (B) and the corresponding pH drop. (A) Biofilm mass and pH during growth of carolacton-treated and untreated samples, as determined by crystal violet staining. The means and the standard deviations from two biological replicates, each consisting of 6 technical replicates, are presented. (B) Inhibition of green/red fluorescence (bar chart), as a measure for viability, and the red fluorescence (line plots), as a measure for membrane damage, for LIVE/DEAD-stained biofilms. For the inhibition of green/red fluorescence, the means and standard deviations from 2 biological replicates, each consisting of 6 technical replicates, are shown.
Fig. 3.
Carolacton activity depends on pH. Carolacton was added to phosphate-buffered (75 and 100 mM) and unbuffered (unbuff.) THBS media before inoculation with S. mutans UA159 to an OD600 of 0.01. Two different carolacton concentrations were tested, 0.25 μg/ml (crosshatched bars) and 2.5 μg/ml (gray bars). The media differed in buffer concentration (75 mM versus 100 mM) and in the initial pH (7.8 versus 6.5). After 20 h, biofilms were washed and stained with LIVE/DEAD viability staining. The inhibition of viability was calculated relative to untreated control wells (see Materials and Methods). Control wells contain the same amount of methanol as used for the carolacton-treated wells but lacked carolacton. Standard deviations were calculated from 3 independent biological replicates, each consisting of 5 technical replicates. The mean of the OD600 is shown as a line graph.
Taken together, the data show that carolacton acts only on growing cells at low pH. There is no significant effect of carolacton on actively growing cells at a pH of above 5.8.
Carolacton damages planktonically growing cells at low pH.
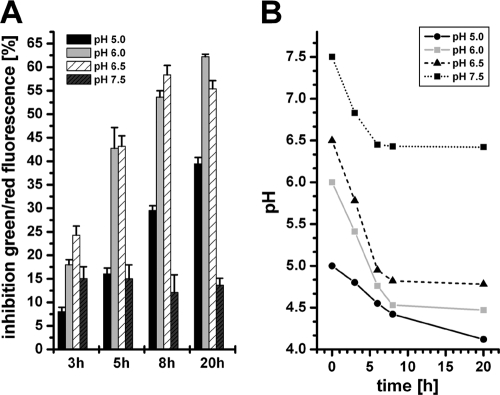
To test if the biofilm-specific activity of carolacton is due to the strong acidification in the biofilm cultures of S. mutans, planktonic cells were cultivated at different pHs in buffered THBY (Todd-Hewitt broth supplemented with 1% yeast extract). The inhibition of viability caused by carolacton was determined after 3, 5, 8, and 20 h of growth (Fig. 4A), and the corresponding pH was determined (Fig. 4B). Inhibition increased with time until 8 h of cultivation; then, no further increase in inhibition was observed. From this time point onwards, the pH also did not drop any further (Fig. 4B). The inhibition of viability for planktonic cells growing at pH 6.0 and 6.5 after 8 and 20 h was comparable to that determined for biofilm cultures (approximately 60%) (33). At a lower initial pH (pH 5.0), the growth rate and final growth yields were severely reduced, and thus the inhibition was weaker. Cultivation of S. mutans in buffered medium that prevent a significant pH drop (pH 7.5) almost completely abolished membrane damage. In conclusion, the growth rate and the pH of the culture are both essential for the strong membrane damage caused by carolacton. Planktonic cells were damaged like their biofilm counterparts as long as they showed similar growth rates and the pH drop in the culture medium was comparable to that of biofilm cultures (50).
Fig. 4.
Sensitivity to carolacton of planktonic cells growing under acidic conditions. Carolacton-treated (2.5 μg/ml) planktonic cells growing in buffered (75 mM phosphate buffer) THBY medium at different initial pH values were cultivated under anaerobic conditions. (A) Inhibition of viability as determined by LIVE/DEAD staining. (B) pH in the cultures during growth.
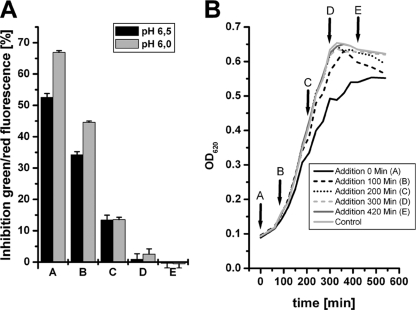
To further demonstrate the influence of the growth rate on the activity of carolacton, the inhibitor was added to the medium at different growth phases of the planktonic cultures growing at an acidic pH. Carolacton was supplemented at the lag phase (condition A), at the early exponential phase (condition B), at mid-exponential phase (condition C), at the onset of stationary phase (condition D), and at the stationary phase (condition E). All cultures were incubated for another 20 h, and inhibition of viability was determined using LIVE/DEAD staining (Fig. 5). Addition of carolacton to proliferating cells (conditions A, B, and C) resulted in an inhibition of viability, while stationary-phase cells (conditions D and E) were not inhibited. The data also show that the damage caused by carolacton was increased if the initial pH was lower, but only for the early stages of the culture (conditions A and B). The data show that there is no significant difference in the activity of carolacton between planktonic and biofilm cells; both are affected while they are growing at low pH.
Fig. 5.
Role of growth phase in the membrane damage caused by carolacton in planktonic culture. Carolacton (2.5 μg/ml) was added at different growth phases (conditions A to E) to planktonic cultures of S. mutans growing in buffered (75 mM phosphate buffer) THBY medium at pH 6.0. and 6.5. (A) Inhibition of viability determined by LIVE/DEAD viability staining. (B) Growth of planktonic cells at pH 6.0.
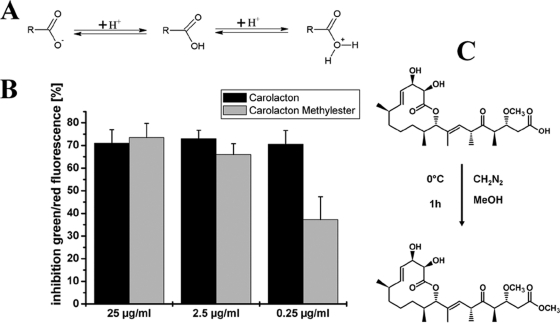
Role of the carboxyl moiety in carolacton activity.
Carolacton is a weak carboxylic acid. To clarify if the pH-dependent activity of carolacton is due to a functional activation of the molecule by a change of its net charge at low pH (Fig. 6A), the ketocarbonic function was chemically converted into a methylester function (Fig. 6C). The resulting methylester was tested in 3 different concentrations for its biological activity against S. mutans biofilms using LIVE/DEAD viability staining. Figure 6B shows that the inhibition of viability caused by the carolacton methylester was the same as that caused by the natural carolacton molecule for the concentrations 2.5 and 25 μg/ml. For the lowest tested concentration of 0.25 μg/ml, the inhibition was slightly reduced to approximately 35%. Thus, it can be assumed that no functional activation of the molecule at low pH occurs and that carolacton is active independent of pH. This is further supported by the detection of significant transcriptional changes already 20 min after carolacton treatment at a pH of well above 6 (see below).
Fig. 6.
Influence of the carboxylic moiety on carolacton activity. (A) Change of the charge of the ketocarbonic function of carolacton upon decreasing pH. (B) Inhibition of biofilm viability by carolacton and the carolacton methylester for 3 different inhibitor concentrations as determined by LIVE/DEAD viability staining. (C) Conversion of carolacton into the corresponding methylester.
Time series transcriptome analysis of carolacton-treated biofilm cells.
To further elucidate the molecular mode of action, we profiled the transcriptional response of S. mutans to carolacton treatment. A time series microarray analysis was performed using carolacton-treated and untreated S. mutans biofilm cells growing under anaerobic conditions. Five different time points (20, 40, 60, 120, and 300 min) following addition of carolacton to a 3.5-h-old biofilm were evaluated for transcriptome changes. In a second approach (endpoint determination), the inhibitor was added to the microtiter plates before the inoculation with bacteria. After 8 h of growth, the transcriptome of biofilm cells was analyzed and compared to that of untreated samples. Below we present the results derived from the time series analysis. The data from the endpoint determination are shown in Tables S2 and S3 in the supplemental material.
Summary of microarray results.
Regulated genes were defined based on their log2 fold change (>±0.8) and on the P value (<0.01). Figure S2 in the supplemental material shows the numbers of carolacton-regulated genes for each time point analyzed. The number of regulated genes increased during carolacton treatment, and at 300 min, 28% of all 1,961 open reading frame (ORFs) of S. mutans were affected, indicating profound changes in cellular metabolism.
The regulated genes were sorted into 9 different clusters according to their transcriptional patterns in the treated versus untreated condition using the k-means algorithm. The correlation coefficient was used as similarity measure to cluster genes which show the same expression profiles. An assignment to the biological functions of the differentially expressed ORFs was made based on the cluster of orthologous groups (COG) for S. mutans of the National Center for Biotechnology Information (NCBI) (77). The means of the gene expression values for the different clusters and an assignment of the cluster genes to their biological functions are presented in Fig. S3 in the supplemental material. Transcriptional changes in prokaryotes occur very fast and are tightly coupled to translation (61). To evaluate the mode of action of carolacton, the fast-responding genes were of particular interest, since they might represent the primary response of the cells to treatment. Accordingly, the different clusters can be divided into two subgroups. Genes of clusters 1, 2, 4, 5, 6, and 9 showed an immediate transcriptional response to carolacton addition, while genes in clusters 3, 7, and 8 were significantly regulated only for the last time point of observation. Thus, regulation of these genes is likely to be a secondary effect of carolacton treatment, showing a general downregulation of cell metabolism-related genes and an upregulation of genes involved in the acid adaptation response.
Taken together, the results indicate that the carolacton-regulated transcriptome reflects changes with an impact on biofilm formation, autolysis, cell shape, cell division, and pyrimidine and histidine metabolism. Regulated TCSs comprised CiaRH, SMU.659/660, SMU.1037c/1038c, VicKRX, RelRS, ComDE and SMU.659/660. VicKRX was the first TCS to respond to carolacton treatment, and genes known to be controlled by VicR were regulated accordingly. The data show on the genetic level that carolacton induces changes in genes that are important for membrane integrity and cell division, two phenotypic traits which have been shown to be impaired by carolacton.
The differential expression of some positively and negatively regulated genes, as identified by microarray analysis, was confirmed using quantitative RT-PCR. The results are shown in Fig. S5 in the supplemental material and indicate a high correlation between the microarray and quantitative PCR data. Genes that were prominently regulated or belong to pathways that were strongly affected by carolacton are discussed in more detail below.
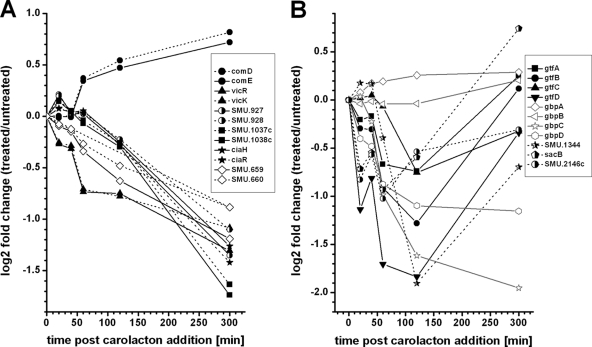
Two-component signal transduction systems.
Due to the important role of TCSs in sensing environmental stimuli, a closer look at carolacton-regulated genes coding for TCSs was taken (Fig. 7A). Six different two-component systems (CiaRH, VicKRX, RelRS, SMU.1037c/1038c, SMU.659/660, and ComDE) were found to be differentially expressed after carolacton addition. Five of them were significantly downregulated, while comDE was the only (slightly) positively regulated TCS. Even though the differential expression of comDE reached values below the cutoff of 0.8 for the log2 fold change, as identified by microarray analysis, the qPCR results (see Fig. S5 in the supplemental material) showed comE to be significantly regulated (log2 fold change of 1.13). Due to these results, the ComDE system was included here. With the exception of VicKRX, the regulation of the TCSs occurred at later time points, beginning 120 min after carolacton treatment. The highest absolute fold change values for all TCS-coding genes were observed at 300 min after carolacton addition. The unique role of VicKRX, as also indicated by the cluster analysis, was further investigated by the examination of genes which are known to be regulated by VicKRX.
Fig. 7.
Effect of carolacton on the expression of TCSs of S. mutans (A) and VicKRX-regulated genes (B) as determined by microarray time series analysis. (A) The differential expression profiles of the response regulators are marked with solid lines, while those of the cognate histidine kinases are marked with dotted lines. (B) Relative expression of genes known to be regulated by the VicKRX system during 300 min of carolacton treatment.
VicKRX-regulated genes.
Senadheera et al. (69) proved that loss of VicK significantly affected genes involved in cell wall metabolism. The strongest regulation was observed for the genes SMU.20, SMU.21, SMU.22, SMU.609, SMU.1344, and SMU.2146c. In addition, it is known (36, 70) that the VicKRX system regulates various glucosyltransferases, fructosyltransferases, and glucan binding proteins. We therefore expected that these genes should also be affected by carolacton treatment. In Fig. 7B their relative expression profiles are shown. With the exception of gbpAB, all genes were found to be downregulated. Thus, not only vicKRX but also the known VicR-regulated genes were strongly downregulated upon carolacton treatment, suggesting that VicKRX plays an important role in the cell response to carolacton. Furthermore VicK-deficient cells show a strongly increased chain length (39), a phenotype which was also observed for carolacton-treated cells (33).
Mutacins, immunity proteins, and bacteriocin-associated genes.
Perry et al. identified 37 CSP-induced genes that were associated with bacteriocin production and controlled via comE (60). In accordance with the observed slight upregulation of the comDE system, these genes were found to be upregulated by carolacton. The relative expression profiles of genes coding for mutacin IV, mutacin V, mutacin VI, and their cognate immunity proteins are shown in Fig. S4 in the supplemental material. Perry et al. (60) identified the product of SMU.925 to be the cognate immunity protein of mutacin V. This gene showed an interesting behavior, as it was regulated opposite to mutacin V for the last time point of observation (log2 fold change of −1.09 at 300 min). To evaluate whether the self-acting autolysin mutacin V could be partly responsible for the membrane damage caused by carolacton, the susceptibility of a mutacin V knockout strain to carolacton was tested (see below).
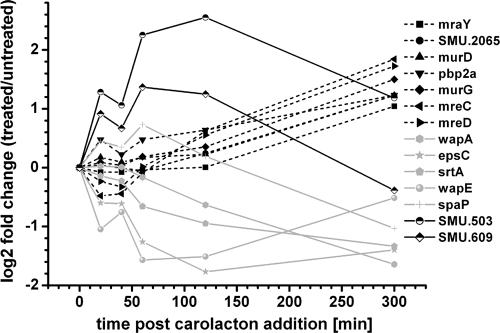
Cell wall and cell membrane metabolism.
Carolacton-regulated genes known to be involved in cell wall metabolism or cell division according to the KEGG categories (http://www.genome.jp/kegg/) are presented in Fig. 8. Genes coding for the cell shape-determining protein MreCD and for enzymes involved in peptidoglycan biosynthesis (murD, murG, pbp2a, mraY, and SMU.2065) were upregulated beginning 60 min after carolacton treatment. The genes coding for the putative murein hydrolase SMU.609 and the membrane protein SMU.503 were instantaneously and strongly upregulated. In contrast, genes coding for surface proteins (wapA, wapE, and spaP) were downregulated. These surface proteins are all substrates of the anchoring enzyme sortase, encoded by the gene srtA, which was also downregulated. Proteins bearing an LPXTG motif at the C terminus are covalently attached to the cell wall by the sortase enzyme. Altered expression of genes coding for surface proteins and sortase must result in changes in the cell surface and might explain the altered morphology of carolacton-treated biofilms (33).
Fig. 8.
Carolacton influences expression of genes involved in cell wall metabolism and cell division. The relative expression profiles for genes coding for membrane proteins (SMU.503 and SMU.609), proteins involved in cell wall metabolism and cell division (MreCD, SMU.2065, MraY, MurDG, and Pbp2a), and surface proteins (SrtA, WapAE, and SpaP), determined by microarray analysis, are shown.
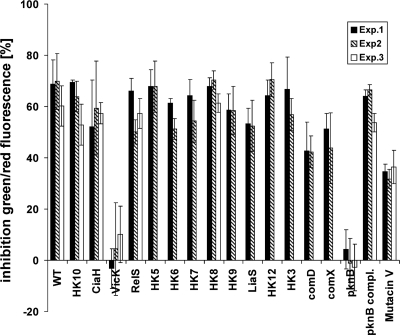
Sensitivity of mutants with deletions of histidine kinases, autolysins, and PknB to carolacton.
Induction of autolysin activity is a key mechanism in the mode of action of many antimicrobials, e.g., β-lactam antibiotics (25). This raises the question of whether autolysin activity could be responsible for the membrane damage of carolacton-treated cells. The genes coding for the autolysins mutacin V and SMU.609 (11) were upregulated upon carolacton treatment. All other autolysin-encoding genes (e.g., SMU.574, SMU.704, SMU.707, and SMU.2147) were not differentially expressed or downregulated. Therefore, SMU.609 and mutacin V knockout mutants were tested for their sensitivity to carolacton. As AtlA was shown to be the major autolysin of S. mutans (1, 2), an atlA-deficient strain was also analyzed (see below).
As shown in Fig. 9, all histidine kinase mutants except the ΔvicK mutant were sensitive to carolacton and showed membrane damage similar to that for the wild type (approximately 60%). However, the insensitivity of the ΔvicK mutant was found to be an artifact of the experimental procedure, likely due to a loosely bound biofilm. In particular, the dead biofilm cells might be easily removed by the washing procedure. Using unwashed biofilms for LIVE/DEAD staining proved the mutant to be sensitive to carolacton treatment. Accordingly, the eDNA content in the supernatant of carolacton-treated ΔvicK biofilm cells was much higher than that in untreated ΔvicK biofilms, indicating that the ΔvicK mutant was sensitive (data not shown). A slightly lower percentage of membrane damage (40%) was found for the ΔcomD mutant, as reported previously (33). Compared to the wild type, the mutacin V deletion strain showed a reduced sensitivity to carolacton (35% versus 60% inhibition). Thus, mutacin V may be partly responsible for the observed membrane damage. The other autolysin-deficient mutants showed a sensitivity comparable to that of the wild type (data not shown). Among the tested deletion mutants, exclusively the ΔpknB mutant was almost insensitive to carolacton treatment, and this mutant was therefore investigated in more depth.
Fig. 9.
Sensitivity to carolacton of mutants with deletions of potential target genes. Mutants of S. mutans deficient for the 13 viable HKs (HK3, HK5 to -10, HK12, LiaS, VicK, CiaH, RelS, and ComD), mutacin V synthesis (Mutacin V), the serine/threonine membrane kinase PknB, and the alternative sigma factor ComX were tested for their sensitivity to carolacton using LIVE/DEAD viability staining of biofilms. The means and standard deviations from at least 5 technical replicates are shown. Each experiment was performed in 2 or 3 biological replicates.
A ΔpknB mutant is insensitive to carolacton.
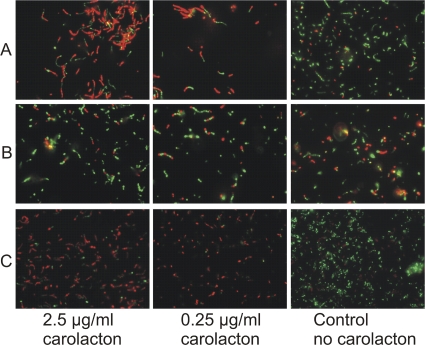
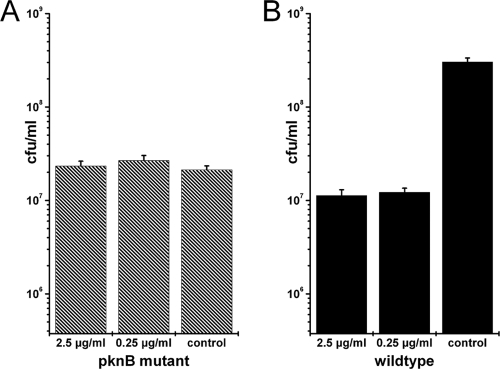
The insensitivity of the ΔpknB mutant to carolacton could be confirmed by independent methods. The differences between carolacton-treated biofilms and controls for the wild type and the ΔpknB mutant were also observed under a fluorescence microscope directly using biofilm cells which were scraped off from the bottom of the microtiter plate (Fig. 10), and this confirmed the results obtained from the fluorescence measurement using a plate reader. The ΔpknB strain complemented in trans showed the same behavior as the wild type using both methods (Fig. 9 and 10). To verify the results from LIVE/DEAD staining, CFU were determined for biofilms that were washed, scraped off, and dispersed by sonication. Data from 9 different technical replicates were used to calculate the standard deviation. Figure 11 shows clearly that for the ΔpknB mutant no significant difference was found between the numbers of colony-forming cells in carolacton-treated and untreated biofilms, confirming the results of LIVE/DEAD staining. In contrast, in the wild type a strong reduction of viable cells was caused by carolacton treatment.
Fig. 10.
Verification of the insensitivity of a pknB-deficient strain to carolacton using LIVE/DEAD staining and fluorescence microscopy. Images of carolacton-treated and untreated biofilm cells of the wild type (A), the ΔpknB mutant (B), and the complemented strain (C) are shown.
Fig. 11.
Verification of the insensitivity of a pknB mutant to carolacton by determination of CFU. CFU of carolacton-treated (2.5 μg/ml or 0.25 μg/ml) and untreated (control) biofilm cells of a pknB-deficient strain (A) and the wild type (B) are shown. Cells derived from 3 different wells of the microtiter plate were diluted independently, and each dilution was plated out on 3 agar plates, resulting in 9 different values for each condition. The standard deviation from these 9 technical replicates is shown. Each experiment was repeated twice, and the figure shows a representative result.
Overlap of carolacton-regulated genes with the PknB regulon.
The pknB deletion mutant was insensitive to carolacton treatment and shared morphological traits with carolacton-treated cells. Even more interestingly, PknB is postulated to modulate the activity of VicR and is an alternative signal transduction system with a strong influence on the acid resistance of S. mutans (3). Therefore, the overlap of carolacton-affected genes with the PknB regulon determined by Banu et al. (3) was investigated (see Tables S4 and S5 in the supplemental material). Thirty-two of the 67 genes identified to be regulated in the pknB deletion strain were similarly regulated after carolacton treatment (48%).
The most strongly regulated genes were found to be regulated in the same direction in both analyses. Genes coding for enzymes involved in cell wall metabolism (mreCD, SMU.609, SMU.984, and SMU.2146c), lipoproteins (SMU.503 and SMU.510), putative bacteriocins (SMU.1895c, SMU.1896c, SMU.277 to SMU.279, SMU.281, SMU.283, and SMU.285) and phosphotransferase systems (SMU.1956 to SMU.1958, SMU.1961, SMU.1962, SMU.2037, and SMU.2038) were all regulated in the same way. Most of the genes that did not overlap between the two data sets code for enzymes involved in carbohydrate or amino acid metabolism. Two significant differences were found between the data sets. Remarkably, the late competence genes SMU.625, SMU.499, SMU.1980 to SMU.1984, and SMU.1987 were downregulated in the ΔpknB deletion strain. These genes were only very slightly regulated in carolacton-treated cells, although comX was upregulated in our analysis. The mutacin V-coding gene SMU.1914c and the genes SMU.1906, SMU.1908 to SMU.1910, and SMU.1912 were downregulated in the pknB knockout strain (consistent with the reduced production of bacteriocins detected in an overlay assay by Banu et al. [3]) but were strongly upregulated after carolacton treatment.
This discrepancy might be a consequence of the different time points analyzed in the two studies. The mutacin genes were upregulated in carolacton-treated cells at later time points (beginning from 120 min), while Banu et al. (3) used cells in the early exponential phase of growth (OD of 0.3) for transcriptome profiling. Moreover, Banu et al. cultivated planktonic cells and analyzed one time point during the exponential growth phase for the transcriptome profiling of the pknB mutant. In contrast, in our approach biofilm cells were used for a time series analysis of carolacton. Therefore, care has to be taken when comparing the two data sets. However, the overlaps between the carolacton-affected genes and the PknB regulon were striking, in particular for the genes encoding enzymes involved in cell wall metabolism.
DISCUSSION
In this study we show that carolacton causes leakage of the cytoplasmic contents (DNA and proteins) of growing cells at low pH. The growth rate and the acidic pH are both crucial factors for the killing of S. mutans caused by carolacton, while the mode of growth (biofilm versus planktonic) plays an indirect role; e.g., the pH drops faster in biofilms than in planktonic culture. Using a time series microarray analysis, a strong influence of carolacton on genes important for cell wall metabolism and pyrimidine metabolism and on the regulons of ComDE and VicKRX was revealed. A mutant lacking pknB is insensitive to carolacton treatment, indicating that PknB is required for cell damage caused by carolacton. We discuss a plausible mode of action of carolacton and further implications derived from these findings below.
Carolacton causes membrane damage.
We demonstrated that carolacton causes leakage of cytoplasmic content by analyzing the extracellular DNA and protein amounts. Furthermore, the release of a large enzyme (β-galactosidase) into the supernatant of carolacton-treated biofilms was quantified. Since β-galactosidase and DNA are sterically huge molecules, the appearance of large holes in the bacterial membrane can be assumed. The bacterial membrane is essential for energy production and pH and nutrient homeostasis and functions as the permeability barrier of the cell. Disturbing the membrane integrity is thus an emerging strategy for the design of new antimicrobial drugs (25). Most conventional antibiotics target the synthesis of proteins, RNA, DNA, folic acids, or peptidoglycan and are therefore active only on growing bacteria (25). Antimicrobials that disrupt the lipid bilayer of the membranes can be promising therapeutic approaches in the case of slow- or nongrowing bacteria (e.g., in persistent infections), as the membrane integrity is also essential for nonproliferating bacteria (25, 80). However, carolacton was proven to act exclusively on proliferating S. mutans cells. This suggests that the disruption of membrane integrity is not caused by direct physical disturbance as observed, e.g., for pore-forming bacteriocins such as nisin (48). Moreover, pore-forming antimicrobials and chemicals that perturb the cell membrane would cause an instantaneous activation of the LiaSFR cell envelope stress response, as shown by Suntharalingam et al. (74). The LiaSFR system is transcriptionally activated by exposure to alkaline shock, detergents, cationic antimicrobial peptides, the bacteriocin nisin, and lipid II cycle inhibitors (46, 63). At no analyzed time point was any significant regulation of liaSFR observed in our study, indicating that carolacton does not directly target the bacterial membrane. One would further expect an increase of membrane damage with increasing amounts of pore-forming agents because they interact in stoichiometric proportions with the membrane components (25, 62). The previous observation that the dose-response curve of carolacton has a sigmoidal shape (33) is not in accordance with such a mode of action.
Most known bacteriocins form pores in the bacterial membrane, leading to an efflux of metabolites, a depolarization of the bacterial membrane, and an partial efflux of cellular ATP (24). The pores formed by most bacteriocins are relatively small, in the range of 0.2 to 2 nm, allowing the passage of hydrophilic solutes with molecular masses of up to 0.5 kDa (24). In the case of carolacton-treated cells, the appearance of large holes was observed, leading to an efflux of huge macromolecules, e.g., DNA and β-galactosidase. A pore-forming mechanism involving a discrete pore size is not consistent with the rather unspecific large holes of carolacton-treated cells.
Carolacton caused leakage of cytoplasmic content but not complete cell lysis. Thus, the remaining cell corpus was stained by crystal violet and contributed to the optical density of the cultures. These classical methods for the determination of biomass do not discriminate between viable and nonviable cells. However, when using viability measurements, e.g., LIVE/DEAD staining or CFU counts, a strong reduction in the amount of viable cells can be observed for carolacton-treated cultures (33). Thus, growth measurements based on total (e.g., live and dead) biomass underestimate the effect of carolacton.
From these observations and studies we infer that the membrane damage caused by carolacton on growing cells of S. mutans at low pH is a secondary effect. Using a chemically modified carolacton molecule (carolacton methylester), we could exclude that the molecule is activated at low pH by a change of net charge, which would be an alternative explanation for the delayed phenotypic response.
Carolacton does not reduce the expression of acid tolerance mechanisms of S. mutans.
In S. mutans the acid tolerance is mediated mainly by the proton-pumping activity of the FoF1 ATPase (64). Moreover the agmatine deiminase system is capable of producing ammonia from agmatine or agmatine-containing peptides, thus neutralizing an excess of protons (22). Furthermore, S. mutans bears a malolactic fermentation system which transforms malate to the weaker acid lactic acid and CO2 to raise the intracellular pH (37). Finally, a branched-chain amino acid (BCAA) system exists in S. mutans, which produces ammonia and ATP for regulating the intracellular pH (38). None of these mechanisms were downregulated at the transcriptional level as a result of carolacton treatment. However, posttranscriptional regulation of the components involved in the acid tolerance might also play a crucial role in the cellular response to acidification. Nevertheless, a reduced acid tolerance is unlikely to be responsible for the killing of carolacton-treated S. mutans cells at low pH, since an upregulation of FoF1 ATPase-coding genes was observed for the last time point of observation. Thus, it is likely that the main acid adaptation system functioned in carolacton-treated S. mutans cells.
Carolacton is unlikely to affect membrane composition.
In addition to these acid tolerance mechanisms, the cell membrane composition play an important role in the ability of S. mutans to withstand acid stress. With decreasing pH, S. mutans alters its membrane composition (18, 56). Fozo and Quivey (18) observed an incremental change from saturated short-chain membrane fatty acids to monounsaturated long-chain fatty acids with decreasing pH. In S. mutans the genes fabDFGHKZ, accACD, SMU.1417c, and bccP code for the essential enzymes of fatty acid biosynthesis (18). Another study by Fozo and Quivey (19) identified the fabM gene product as being responsible for the synthesis of monounsaturated fatty acids and thus essential for survival at low pH. At no analyzed time point did we observe any significant regulation of these genes, indicating that it is quite unlikely that carolacton alters the composition of the cell membranes. Consequently, the pH dependency of membrane damage is likely to be an indirect result of the altered cell wall structure (see below). However, the constraint of potential posttranscriptional regulation of enzymes involved in fatty acid biosynthesis has to be considered.
Mode of action of carolacton.
The data suggest that carolacton affects a signaling mechanism which is central for coordinating cell wall synthesis or remodeling during growth. While this disturbance in cell wall metabolism starts instantaneously after carolacton application, cell death occurs only at low pH several hours later. We hypothesize that it is caused by the inability of the modified cell wall to maintain the intracellular turgor, resulting in bursting of the cell, membrane damage, leakage of cytoplasmic content, and cell death.
The rigidity of the cell wall is provided by a three-dimensional structure of glycan strands cross-linked by peptide bridges (peptidoglycan). The cell wall is the most important shape determinant and prevents the cell from lysis caused by the osmotic pressure (10). This importance for the cell integrity is reflected by the fact that many antibiotics target cell wall metabolism (21, 29). In the cytosol the precursors of peptidoglycan, the disaccharide pentapeptides, are synthesized and then transported to the outside of the cytoplasmic membrane (10). In the following steps of cell wall biosynthesis, penicillin binding proteins covalently attach the precursors to the existing peptidoglycan via a combination of transpeptidation and transglycosylation reactions (15). Inhibitors are known for nearly all the steps of peptidoglycan biosynthesis (9).
A tight control of de novo biosynthesis and remodeling of the cell wall exists in the cell. Perturbations of this dynamic balance often result in lytic phenotypes (23, 27).
Carolacton inhibits biofilms of S. mutans even in nanomolar concentrations (33), which implies that carolacton has a primary target which is present in only a few copies per cell. Compounds that target signaling show a similar behavior. We therefore infer that carolacton disturbs a process which coordinates cell wall metabolism rather than directly inhibiting a certain enzymatic step in the biogenesis of the cell wall. PknB is suggested to play a central role in cell wall remodeling and is known as an alternative signaling system besides the two-component systems (41, 45).
Disturbance in cell wall metabolism may weaken the cell wall. The faster the cell grows, the more crucial are the correct synthesis and remodeling of the cell wall. Lytic and synthetic enzymes function in a delicately balanced interplay during cell wall growth, with the former introducing nicks into the murein structure and the latter inserting new material at these sites (31). Gram-positive bacteria were shown to possess a turgor pressure of up to 50 atm (31). Thus, an enormous tension has to be compensated for by the cell wall. A weakened cell wall will therefore easily break. Thus, at acid pH the weakened cell wall of carolacton-treated cells is likely no longer able to withstand the turgor pressure, and subsequently the membrane integrity is lost and the cytoplasmic content leaks out. Large holes lead to cell death.
The serine/threonine protein kinase PknB is essential for carolacton activity.
One central observation of this study was the insensitivity of the pknB deletion mutant to carolacton treatment, indicating that PknB mediates the deleterious effects of carolacton. Eukaryotic serine/threonine protein kinases were shown to substantially influence cell wall metabolism (41, 67). Maestro et al. (45) and Shah et al. (71) showed that STPKs of S. pneumoniae bind to peptidoglycan and to β-lactam antibiotics which mimic the terminal portions of the peptidoglycan stem peptide. PknB is therefore indicated to play an essential role in the remodeling of the cell wall. PknB was further shown to phosphorylate substrates essentially involved in peptidoglycan biosynthesis (30, 53, 54, 57, 68, 78). Electron microscopic pictures of STPK mutants (4) and the higher susceptibility of STPK mutants to cell wall-acting antibiotics (17) clearly support the role of STPK in cell wall metabolism. Thus, the observed phenotypic changes of carolacton-treated cells and the detected effects on cell wall metabolism-related genes are in accordance with PknB mediating the activity of carolacton.
Furthermore, it was shown that knockout of pknB significantly decreased the expression of genes coding for enzymes involved in pyrimidine metabolism (17, 67). These findings are consistent with our results, as genes coding for enzymes involved in pyrimidine biosynthesis were among the most strongly regulated and fastest-responding genes.
Interestingly, it was shown that the RNA polymerase is among the targets of STPKs in S. pneumoniae (54). This might explain why STPKs act as global regulators of gene expression and influence, e.g., the pyrimidine biosynthesis genes.
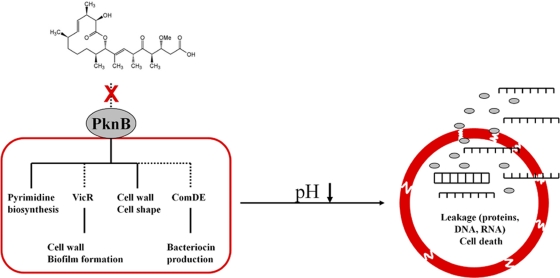
A potential second role of PknB is its cross talk with TCSs. Lin et al. proved that phosphorylation of S. agalactiae CovR (a VicR homologue) by PknB prevented promoter binding of the response regulator (42). Rajagopal et al. (65) showed in vitro a phosphorylation of CovR by STPKs in close vicinity to the aspartate residue phosphorylated by the CovS histidine kinase. Thus, PknB may mediate the effects of carolacton on the cell wall metabolism directly by phosphorylating involved enzymes, in particular those responsible for cell wall remodeling, and/or indirectly via phosphorylation of response regulators, in particular VicR. VicR itself regulates a cascade of downstream genes coding for enzymes involved in cell wall metabolism. The sensitivity of the ΔvicK deletion mutant to carolacton treatment is not a contradiction to the proposed role of VicR in the carolacton mode of action, since the response regulator might be phosphorylated independently from its histidine kinase, e.g., by PknB. A cross-regulation between certain histidine kinases and their noncognate response regulators was postulated by Chong et al. (12), and in particular VicKRX is suggested to cross-communicate with other TCSs because of its global importance in S. mutans (72). Furthermore, potential targets of PknB other than VicR might contribute to the deleterious effects caused by carolacton. The transcriptome of a ΔpknB deletion mutant (3) shows a strong overlap with that of carolacton-treated biofilms. The regulons of VicKRX and ComDE are affected in both. The ΔpknB mutant shows also a phenotype similar to that of carolacton-treated cells, e.g., the sensitivity to low pH. Figure 12 summarizes our current hypothesis regarding the mode of action of carolacton and the likely role of PknB.
Fig. 12.
Working model for the mode of action of carolacton. Carolacton is postulated to interfere with PknB-mediated signaling. Dashed lines represent hypothetical interactions. Disturbance of cell wall metabolism by altered PknB signaling might be mediated either directly by altered phosphorylation of target proteins or indirectly via the VicR cascade. Induction of ComDE is accompanied by an upregulation of bacteriocins and bacteriocin-associated genes. Furthermore, repression of pyrimidine biosynthesis occurs, as STPKs are global regulators of gene expression. Alterations in cell wall composition result in a weakened cell wall, leading to loss of membrane integrity at low pH and leakage of the cytoplasmic contents.
Conclusion and outlook.
We have shown a link between acidification and membrane damage caused by carolacton in the oral microorganism S. mutans. Since acid tolerance is a key virulence trait of S. mutans and is essential for survival and competition in its ecological niche, these findings may have a profound impact on a general strategy aiming to erase biofilm cells in the oral cavity. Acidification plays a key role in the demineralization of the tooth enamel and the appearance of caries. Thus, the aciduricity and acidogenicity of S. mutans are intimately linked to its capacity to cause caries. Furthermore, we showed a strong influence of carolacton on cell wall metabolism-related genes using a microarray approach and provide a plausible mode of action for how carolacton damages S. mutans cells at low pH, based on all the genetic and physiological observations presently available.
PknB mediates the deleterious effects of carolacton, as indicated by the insensitivity of the ΔpknB mutant. To determine if this is caused by direct binding of carolacton to PknB will require experiments with the purified enzyme and phosphoproteome analyses. Eukaryotic-type serine/threonine kinases, which are global regulators of cell metabolism, cell wall biosynthesis, growth, and important virulence traits, may be attractive drug targets. In S. pneumoniae the STPK was selected as one of the lead candidates for the development of a new vaccine (20).
Oxoby et al. (55) identified new bisarylurea derivates as potential S. agalactiae STKP inhibitors, while Szekely et al. (75) investigated inhibitors of Mycobacterium tuberculosis PknB by screening a library of 70.000 molecules. Using in vitro kinase assays, the latter group found hit compounds acting in the nanomolar range. However, using a mycobacterial growth assay, they found no significant inhibition for the tested substances.
STPKs are predicted to be present in two-thirds of all sequenced bacteria and are therefore a universal target (59). Myxobacteria, which have a complex life cycle involving the formation of a fruiting body, have especially large numbers of STPKs. Carolacton was isolated from the myxobacterium Sorangium cellulosum, which hosts 317 STPKs, the highest number reported so far from any prokaryote (47). Thus, the biological role of carolacton may be to regulate one of the STPKs of Sorangium cellulosum during growth or fruiting body formation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rolf Jansen, Herbert Irschik, Heinrich Steinmetz, and other members of the “microbial drugs” group for providing carolacton. We thank Celine Levesque for providing the 13 histidine kinase-deficient mutants of Streptococcus mutans. We also thank Helena Sztajer, Georg Conrads, and Petra Dersch for fruitful discussions. We thank the reviewers, whose comments helped to improve the manuscript.
Michael Reck and Katrin Rutz were supported by the BMBF grant “Medical Systems Biology-MedSys” (FK 0315411A). Jürgen Thomas was funded by the DFG SFB/TRR 51 Roseobacter.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Ahn S. J., Burne R. A. 2006. The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J. Bacteriol. 188:6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahn S. J., Burne R. A. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 189:6293–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banu L. D., et al. 2010. The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infect. Immun. 78:2209–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beltramini A. M., Mukhopadhyay C. D., Pancholi V. 2009. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect. Immun. 77:1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biswas I., Jha J. K., Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blum H., Beier H., Gross H. J. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99 [Google Scholar]
- 7. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 8. Buckley N. D., Vadeboncoeur C., LeBlanc D. J., Lee L. N., Frenette M. 1999. An effective strategy, applicable to Streptococcus salivarius and related bacteria, to enhance or confer electroporation competence. Appl. Environ. Microbiol. 65:3800–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bugg T. D., Walsh C. T. 1992. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat. Prod. Rep. 9:199–215 [DOI] [PubMed] [Google Scholar]
- 10. Cabeen M. T., Jacobs-Wagner C. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3:601–610 [DOI] [PubMed] [Google Scholar]
- 11. Catt D. M., Gregory R. L. 2005. Streptococcus mutans murein hydrolase. J. Bacteriol. 187:7863–7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chong P., Drake L., Biswas I. 2008. LiaS regulates virulence factor expression in Streptococcus mutans. Infect. Immun. 76:3093–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cleveland W. S., Devlin S. J. 1988. Locally-weighted regression: an approach to regression analysis by local fitting. J. Am. Stat. Assoc. 83:596–610 [Google Scholar]
- 14. Cozzone A. J. 2005. Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J. Mol. Microbiol. Biotechnol. 9:198–213 [DOI] [PubMed] [Google Scholar]
- 15. Daniel R. A., Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776 [DOI] [PubMed] [Google Scholar]
- 16. Dasgupta A., Datta P., Kundu M., Basu J. 2006. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology 152:493–504 [DOI] [PubMed] [Google Scholar]
- 17. Donat S., et al. 2009. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J. Bacteriol. 191:4056–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fozo E. M., Quivey R. G., Jr 2004. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70:929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fozo E. M., Quivey R. G., Jr 2004. The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J. Bacteriol. 186:4152–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giefing C., et al. 2008. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205:117–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goo K. S., Sim T. S. 2011. Designing new beta-lactams: implications from their targets, resistance factors and synthesizing enzymes. Curr. Comput. Aided Drug Des. 7:53–80 [DOI] [PubMed] [Google Scholar]
- 22. Griswold A. R., Jameson-Lee M., Burne R. A. 2006. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 188:834–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hadi T., Dahl U., Mayer C., Tanner M. E. 2008. Mechanistic studies on N-acetylmuramic acid 6-phosphate hydrolase (MurQ): an etherase involved in peptidoglycan recycling. Biochemistry 47:11547–11558 [DOI] [PubMed] [Google Scholar]
- 24. Hechard Y., Sahl H. G. 2002. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84:545–557 [DOI] [PubMed] [Google Scholar]
- 25. Hurdle J. G., O'Neill A. J., Chopra I., Lee R. E. 2011. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 9:62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hussain H., Branny P., Allan E. 2006. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 188:1628–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jaeger T., Mayer C. 2008. N-Acetylmuramic acid 6-phosphate lyases (MurNAc etherases): role in cell wall metabolism, distribution, structure, and mechanism. Cell. Mol. Life Sci. 65:928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jansen R., et al. 2010. Carolacton—a macrolide ketocarbonic acid that reduces biofilm formation by the caries- and endocarditis-associated bacterium Streptococcus mutans. Eur. J. Organ. Chem. 2010:1284–1289 [Google Scholar]
- 29. Jovetic S., Zhu Y., Marcone G. L., Marinelli F., Tramper J. 2010. β-Lactam and glycopeptide antibiotics: first and last line of defense? Trends Biotechnol. 28:596–604 [DOI] [PubMed] [Google Scholar]
- 30. Kang C. M., et al. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19:1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. König H., Claus H., Varma A. 2010. Prokaryotic cell wall compounds: strucutre and biochemistry. Springer, Berlin, Germany [Google Scholar]
- 32. Kristich C. J., Wells C. L., Dunny G. M. 2007. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. U. S. A. 104:3508–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kunze B., et al. 2010. Damage of Streptococcus mutans biofilms by carolacton, a secondary metabolite from the myxobacterium Sorangium cellulosum. BMC Microbiol. 10:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 35. Lau P. C., Sung C. K., Lee J. H., Morrison D. A., Cvitkovitch D. G. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193–205 [DOI] [PubMed] [Google Scholar]
- 36. Lee S. F., Delaney G. D., Elkhateeb M. 2004. A two-component covRS regulatory system regulates expression of fructosyltransferase and a novel extracellular carbohydrate in Streptococcus mutans. Infect. Immun. 72:3968–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lemme A., Sztajer H., Wagner-Dobler I. 2010. Characterization of mleR, a positive regulator of malolactic fermentation and part of the acid tolerance response in Streptococcus mutans. BMC Microbiol. 10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Len A. C., Harty D. W., Jacques N. A. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150:1339–1351 [DOI] [PubMed] [Google Scholar]
- 39. Levesque C. M., et al. 2007. Systemic inactivation and phenotypic characterization of two-component systems in expression of Streptococcus mutans virulence properties. Lett. Appl. Microbiol. 45:398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Y. H., Lau P. C., Lee J. H., Ellen R. P., Cvitkovitch D. G. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liebeke M., Meyer H., Donat S., Ohlsen K., Lalk M. 2010. A metabolomic view of Staphylococcus aureus and its ser/thr kinase and phosphatase deletion mutants: involvement in cell wall biosynthesis. Chem. Biol. 17:820–830 [DOI] [PubMed] [Google Scholar]
- 42. Lin W. J., et al. 2009. Threonine phosphorylation prevents promoter DNA binding of the group B Streptococcus response regulator CovR. Mol. Microbiol. 71:1477–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Loesche W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Madec E., Laszkiewicz A., Iwanicki A., Obuchowski M., Seror S. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46:571–586 [DOI] [PubMed] [Google Scholar]
- 45. Maestro B., et al. 2011. Recognition of peptidoglycan and beta-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae. FEBS Lett. 585:357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mascher T., Zimmer S. L., Smith T. A., Helmann J. D. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mengin-Lecreulx D., van Heijenoort J., Park J. T. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moll G. N., Konings W. N., Driessen A. J. 1999. Bacteriocins: mechanism of membrane insertion and pore formation. Antonie Van Leeuwenhoek 76:185–198 [PubMed] [Google Scholar]
- 49. Munson M. A., Banerjee A., Watson T. F., Wade W. G. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mutschler H., Gebhardt M., Shoeman R. L., Meinhart A. 2011. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 9:e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nariya H., Inouye S. 2005. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol. Microbiol. 58:367–379 [DOI] [PubMed] [Google Scholar]
- 52. Neu J. M., MacMillan S. V., Nodwell J. R., Wright G. D. 2002. StoPK-1, a serine/threonine protein kinase from the glycopeptide antibiotic producer Streptomyces toyocaensis NRRL 15009, affects oxidative stress response. Mol. Microbiol. 44:417–430 [DOI] [PubMed] [Google Scholar]
- 53. Novakova L., et al. 2010. Identification of multiple substrates of the StkP Ser/Thr protein kinase in Streptococcus pneumoniae. J. Bacteriol. 192:3629–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Novakova L., et al. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 272:1243–1254 [DOI] [PubMed] [Google Scholar]
- 55. Oxoby M., et al. 2010. Towards Gram-positive antivirulence drugs: new inhibitors of Streptococcus agalactiae Stk1. Bioorg. Med. Chem. Lett. 20:3486–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Papadimitriou K., Pratsinis H., Nebe-von-Caron G., Kletsas D., Tsakalidou E. 2007. Acid tolerance of Streptococcus macedonicus as assessed by flow cytometry and single-cell sorting. Appl. Environ. Microbiol. 73:465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Parikh A., Verma S. K., Khan S., Prakash B., Nandicoori V. K. 2009. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J. Mol. Biol. 386:451–464 [DOI] [PubMed] [Google Scholar]
- 58. Paster B. J., et al. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Perez J., Castaneda-Garcia A., Jenke-Kodama H., Muller R., Munoz-Dorado J. 2008. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc. Natl. Acad. Sci. U. S. A. 105:15950–15955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Perry J. A., Jones M. B., Peterson S. N., Cvitkovitch D. G., Levesque C. M. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 72:905–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pham H. T., Riu K. Z., Jang K. M., Cho S. K., Cho M. 2004. Bactericidal activity of glycinecin A, a bacteriocin derived from Xanthomonas campestris pv. glycines, on phytopathogenic Xanthomonas campestris pv. vesicatoria cells. Appl. Environ. Microbiol. 70:4486–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pietiainen M., et al. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577–1592 [DOI] [PubMed] [Google Scholar]
- 64. Quivey R. G., Kuhnert W. L., Hahn K. 2001. Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:301–314 [DOI] [PubMed] [Google Scholar]
- 65. Rajagopal L., Vo A., Silvestroni A., Rubens C. E. 2006. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol. Microbiol. 62:941–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 67. Saskova L., Novakova L., Basler M., Branny P. 2007. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 189:4168–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schultz C., et al. 2009. Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Mol. Microbiol. 74:724–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Senadheera D., et al. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191:6415–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Senadheera M. D., et al. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187:4064–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shah I. M., Laaberki M. H., Popham D. L., Dworkin J. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smith E. G., Spatafora G. A. Gene regulation in S. mutans: complex control in a complex environment. J. Dent. Res., in press [DOI] [PubMed] [Google Scholar]
- 73. Stock A. M., Robinson V. L., Goudreau P. N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 74. Suntharalingam P., Senadheera M. D., Mair R. W., Levesque C. M., Cvitkovitch D. G. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191:2973–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Szekely R., et al. 2008. A novel drug discovery concept for tuberculosis: inhibition of bacterial and host cell signalling. Immunol. Lett. 116:225–231 [DOI] [PubMed] [Google Scholar]
- 76. Sztajer H., et al. 2008. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190:401–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thakur M., Chakraborti P. K. 2006. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J. Biol. Chem. 281:40107–40113 [DOI] [PubMed] [Google Scholar]
- 79. Thenisch N. L., Bachmann L. M., Imfeld T., Leisebach M. T., Steurer J. 2006. Are mutans streptococci detected in preschool children a reliable predictive factor for dental caries risk? A systematic review. Caries Res. 40:366–374 [DOI] [PubMed] [Google Scholar]
- 80. Van Bambeke F., Mingeot-Leclercq M. P., Struelens M. J., Tulkens P. M. 2008. The bacterial envelope as a target for novel anti-MRSA antibiotics. Trends Pharmacol. Sci. 29:124–134 [DOI] [PubMed] [Google Scholar]
- 81. Wagner C., et al. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 70:6121–6128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xue X., Tomasch J., Sztajer H., Wagner-Dobler I. 2010. The delta subunit of RNA polymerase, RpoE, is a global modulator of Streptococcus mutans environmental adaptation. J. Bacteriol. 192:5081–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.