Abstract
The TonB system of Gram-negative bacteria provides passage across the outer membrane (OM) diffusion barrier that otherwise limits access to large, scarce, or important nutrients. In Escherichia coli, the integral cytoplasmic membrane (CM) proteins TonB, ExbB, and ExbD couple the CM proton motive force (PMF) to active transport of iron-siderophore complexes and vitamin B12 across the OM through high-affinity transporters. ExbB is an integral CM protein with three transmembrane domains. The majority of ExbB occupies the cytoplasm. Here, the importance of the cytoplasmic ExbB carboxy terminus (residues 195 to 244) was evaluated by cysteine scanning mutagenesis. D211C and some of the substitutions nearest the carboxy terminus spontaneously formed disulfide cross-links, even though the cytoplasm is a reducing environment. ExbB N196C and D211C substitutions were converted to Ala substitutions to stabilize them. Only N196A, D211A, A228C, and G244C substitutions significantly decreased ExbB activity. With the exception of ExbB(G244C), all of the substituted forms were dominant. Like wild-type ExbB, they all formed a formaldehyde cross-linked tetramer, as well as a tetramer cross-linked to an unidentified protein(s). In addition, they could be formaldehyde cross-linked to ExbD and TonB. Taken together, the data suggested that they assembled normally. Three of four ExbB mutants were defective in supporting both the PMF-dependent formaldehyde cross-link between the periplasmic domains of TonB and ExbD and the proteinase K-resistant conformation of TonB. Thus, mutations in a cytoplasmic region of ExbB prevented a periplasmic event and constituted evidence for signal transduction from cytoplasm to periplasm in the TonB system.
INTRODUCTION
The TonB system of Gram-negative bacteria mediates the energization of active transport across the outer membrane (OM) by coupling it to the proton motive force (PMF) of the cytoplasmic membrane (CM). For Escherichia coli K-12, it energizes the OM transport of several different iron siderophores and vitamin B12. In more divergent bacterial species, maltodextrins, sucrose, and Ni2+ reflect the tip of the iceberg of nutrients that require TonB system energy transduction for transport across the OM (for reviews, see references 2, 21, 22, 31, 36, and 41). Bacteriophages and many colicins have evolved to make use of the TonB system to enter E. coli (9, 39, 42). Sensitivity to these reagents and ability to transport iron siderophores are the initial phenotypic assays usually applied to TonB system mutants.
TonB, the protein that directly contacts the OM transporters, has a single transmembrane domain (TMD) that anchors it in the CM; ExbD has a similar topology (13, 18, 34, 40). ExbB has three TMDs, with the majority of the protein occupying the cytoplasm, including its carboxy-terminal domain (amino acids 195 to 244) (19, 20). ExbB and ExbD appear to harvest the PMF of the CM and transduce it into TonB conformational change, such that TonB can energize active transport through a TonB-gated transporter (TGT). The carboxy terminus of TonB directly contacts TGTs in the OM through their N-terminal periplasmic region known as the TonB box (8, 32). Other regions of TonB interaction with TGTs exist but have not been identified (10).
Recent studies show that the single TonB TMD, once thought to be part of the proton translocation pathway, is instead serving a structural role, possibly allowing correct positioning of TonB in complex with ExbB and ExbD (44a). Using the PMF and possibly D25 in its TMD, ExbD appears to guide the conformation of the TonB carboxy terminus, although it is not yet clear how the conformational changes of ExbD are manifested (7, 25, 33). ExbB, ExbD, and TonB all make contact with one another in vivo and form a complex in vitro (4, 33). It is not known whether they constitute a stable energy transduction complex in vivo or whether it dynamically assembles and disassembles.
ExbB protein appears to be the scaffold on which TonB and ExbD assemble. In its absence both proteins are proteolytically unstable, while ExbB is stable when expressed by itself (1, 11, 37; K. Baker and K. Postle, unpublished data). A proton pathway through ExbB has been proposed and investigated by site-specific mutagenesis of potential key residues in the second and third TMDs of ExbB (6, 48). No individual residues were identified as essential, while the combination of T148A and T181A inactivated ExbB. Other than that study, and the isolation of second-site suppressor mutations of TonB ΔV17 at ExbB residues V35, V36, and A39, there has not been a mutagenic analysis of this important protein (28).
We scanned the carboxy-terminal cytoplasmically localized tail of ExbB (residues 195 to 244) with cysteinyl residues. None of the mutants affected previously characterized ExbB assembly or ExbB interactions with TonB and ExbD as assayed by in vivo formaldehyde cross-linking (16, 28, 33, 44). Interestingly, D211C and Cys substitutions near the carboxy terminus formed disulfide-linked dimers in the reducing cytoplasmic environment, suggesting that the carboxy-terminal tail was sequestered in a more oxidizing environment. The majority of substitutions also had little effect on ExbB activity. Four substitutions of Cys or Ala at residues N196, D211, A228, and G244 diminished ExbB activity. Three were dominant negative, and all four decreased or prevented the PMF-dependent formaldehyde cross-link between TonB and ExbD (33). Taken together, the results constituted evidence of signal transduction from the ExbB cytoplasmic domain to the periplasmic domains of ExbD and TonB.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in the present study are listed in Table 1. Plasmid pKP878 is a derivative of pKP660, in which ExbB(C25A) and ExbD are expressed from the arabinose promoter (33). ExbB single residue substitutions are derivatives of pKP878 created by PCR mutagenesis as previously described (47). ExbB(C25A, Δ195-243) was made by using 30-cycle extra-long PCR. Both exbB and exbD genes were sequenced for all mutants by the Penn State Genomics Core Facility (University Park, PA) to rule out unintended base changes. Strain KP1517 was created by P1vir transduction of the Δgor-756::kan gene from JW3467-1 into KP1036.
Table 1.
Strain and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| W3110 | F− IN(rrnD-rrnE)1 | 17 |
| RA1045 | W3110 ΔexbD ΔtolQR | 7 |
| KP1036 | W3110 exbB::Tn10 | 1 |
| JW3467-1 | Δ(araD-araB)567 ΔlacZ4787(::rrnB3) λ− Δgor-756::kanrph-1 Δ(rhaD-rhaB)568hsdR514 | EGSCb |
| KP1517 | W3110 exbB::Tn10Δgor-756::kan | This study |
| RA1017 | W3110 ΔexbBD::kan ΔtolQRA | 25 |
| Plasmids | ||
| pKP660 | exbBD expressed from pBAD promoter | 33 |
| pKP878a | ExbB(C25A), ExbD (derived from pKP660) | This study |
The following plasmids, all derived from pKP878, are listed in order of the ExbB mutation they express (indicated in parentheses): pKP816 (Y195C), pKP664 (Y195A), pKP998 (Y195F), pKP817 (N196C), pKP1081 (N196A), pKP818 (V197C), pKP824 (F198C), pKP825 (A199C), pKP826 (R200C), pKP827 (Q210C), pKP828 (I202C), pKP829 (G203C), pKP830 (G204C), pKP831 (F205C), pKP832 (K206C), pKP1084 (A207C), pKP833 (M208C), pKP834 (L209C), pKP835 (210C), pKP1085 (D211C), pKP1378 (D211A), pKP837 (V212C), pKP838 (A213C), pKP839 (A214C), pKP840 (Q215C), pKP841 (V216C), pKP842 (L217C), pKP1086 (L218C), pKP844 (L219C), pKP845 (Q220C), pKP819 (S221C), pKP820 (R222C), pKP821 (D223C), pKP822 (L224C), pKP823 (D225C), pKP846 (L226C), pKP847 (E227C), pKP848 (A228C), pKP849 (S229C), pKP850 (A230C), pKP851 (A231C), pKP852 (A232C), pKP853 (H233C), pKP854 (P234C), pKP855 (V235C), pKP856 (A236C), pKP857 (V237C), pKP858 (A238C), pKP859 (Q239C), pKP860 (K240C), pKP861 (L241C), pKP862 (R242C), pKP1326 (A234C), pKP1091 (G244C), and pKP1346 (Δ195-243).
EGSC, E. coli Genetic Stock Center.
Media and culture conditions.
Luria-Bertani (LB), tryptone (T), and M9 minimal salts media were prepared as described previously (30, 43). Liquid cultures, agar plates, and T-top agar were supplemented with 100 μg of ampicillin/ml and plasmid-specific levels of l-arabinose for the expression of ExbB proteins at chromosomal levels. M9 salts were supplemented with 1.0% glycerol (wt/vol), 0.4 mg of thiamine/ml, 1 mM MgSO4, 0.5 mM CaCl2, 0.2% Casamino Acids (wt/vol), 40 mg of tryptophan/ml, and 1.85 μM FeCl3. Cultures were grown at 37°C with continuous aeration.
Sensitivity to colicins and bacteriophage.
As indirect measures of ExbB activity, strains expressing ExbB variants at chromosomal levels were grown to mid-logarithmic phase and plated in T-top agar on T-agar plates. The arabinose required for chromosomal levels of expression was determined for each mutant by immunoblotting and was present throughout the assay, as was 100 μg of ampicillin/ml. Dilutions of colicin or bacteriophage preparations were spotted onto the bacterial lawns, and zones of clearing were characterized ∼18 h later (24, 35).
[55Fe]ferrichrome transport.
The initial rates of iron transport were determined as described previously (24, 35). RA1017 strains carrying various plasmids were grown in M9 minimal medium supplemented with l-arabinose sufficient for chromosomal levels of ExbB expression (Table 2). Mid-exponential-phase cultures were harvested and suspended in buffer, and the initial rates of [55Fe]ferrichrome transport were determined in triplicate. At the time when transport was initiated, a sample of each culture was precipitated with trichloroacetic acid (TCA) to determine ExbB expression levels. Proteins were visualized on immunoblots of SDS–11% PAGE gels with anti-ExbB antibody (15).
Table 2.
Phenotypic profiles of ExbB cysteine and alanine substitutions
| Strain | ExbBD plasmid (ExbB phenotype) | Ara (%)a | Sensitivity to colicins B, Ia, and M and phage φ80b |
Mean % [55Fe]ferrichrome transport ± SEMc | |||
|---|---|---|---|---|---|---|---|
| ColB | ColIa | ColM | φ80 | ||||
| W3110 | 8, 8, 8 | 7, 7, 7 | 5, 5, 5 | 9, 9, 9 | 106 ± 13 | ||
| RA1017 | T | T | T | T | 0 | ||
| RA1017 | pKP660 (ExbBD) | 0.0 | 5, 5, 5 | 6, 6, 6 | 3, 3, 4 | 8, 8, 8 | 104 ± 16 |
| RA1017 | pKP878 (C25A) | 0.0 | 5, 5, 5 | 6, 6, 6 | 4, 3, 3 | 8, 8, 8 | 100 ± 1 |
| RA1017 | pKP1346 (Δ195-243) | 0.005 | T | T | T | T | 0 |
| RA1017 | pKP816 (Y195C) | 0.00001 | 3*, 3*, 3* | 5, 5, 5 | 3*, 3*, 3* | 8, 8, 8 | 14 ± 4 |
| RA1017 | pKP998 (Y195F) | 0.000002 | 5, 5, 5 | 6, 6, 6 | 4, 3, 4 | 8, 8, 8 | 112 ± 16 |
| RA1017 | pKP817 (N196C) | 0.2 | T | T | T | T | 0 |
| RA1017 | pKP1081 (N196A) | 0.0 | 2, 2, 2 | 3, 3, 4 | T | 7, 7, 7 | 1 ± 1 |
| RA1017 | pKP1085 (D211C) | 0.2 | T | T | T | T | 0 |
| RA1017 | pKP1378 (D211A) | 0.2 | 3, 4, 3 | 6, 6, 6 | 3*, 3*, 3* | 7, 7, 7 | 18 ± 3 |
| RA1017 | pKP848 (A228C) | 0.000002 | 1, 1, U | 1*, 1*, 1* | 2*, 2*, 2* | 7, 7, 7 | 3 ± 1 |
| RA1017 | pKP860 (K240C) | 0.000002 | 5, 5, 5 | 6, 6, 6 | 3, 3, 3 | 7, 8, 8 | 85 ± 1 |
| RA1017 | pKP862 (R242C) | 0.000002 | 6, 6, 6 | 6, 6, 6 | 4, 4, 4 | 8, 8, 8 | 115 ± 4 |
| RA1017 | pKP1091 (G244C) | 0.00005 | 3, 4, 4 | 6, 6, 6 | 3, 4, 4 | 7, 7, 7 | 31 ± 3 |
Various percentages of arabinose were added to the growth medium and assay plate as indicated to maintain a close level of plasmid-encoded wild-type and mutated ExbB with wild-type chromosomal expression.
Maximum 5-fold dilutions of a standard colicin or 10-fold dilution of phage that provided an evident zone of clearing on a cell lawn are indicated. Asterisks indicate partial clearing of the lawn. T, tolerant (insensitive); U, undiluted.
Transport rates are relative to the transport rate of the ExbB(C25A) mutant transporter (100%).
In vivo formaldehyde cross-linking.
Saturated cultures grown overnight in LB medium (30) were subcultured at 1:100 in M9 minimal medium supplemented with arabinose and 100 μg of ampicillin/ml (43). Concentrations of l-arabinose were added as needed to achieve chromosomal levels of plasmid expression. At mid-exponential phase, 0.5A550-ml portions of each culture were pelleted, suspended in 938 μl of 100 mM sodium phosphate buffer (pH 6.8), and treated with 1% (vol/vol) paraformaldehyde for 15 min at room temperature. The samples were suspended in Laemmli sample buffer (LSB) and heated at 60°C or 95°C for 5 min as specified in the each figure legend (16, 23). Cross-linked complexes were resolved on 11% (or 13% in the case of ExbD detection) SDS-polyacrylamide gels and detected by immunoblotting with ExbB-specific and ExbD-specific polyclonal antibodies (15) and TonB-specific monoclonal antibodies (26).
Effect of CCCP on TonB proteinase K accessibility.
W3110 and RA1017 expressing ExbB variants were freshly grown from LB-saturated overnight cultures into M9 medium to mid exponential phase. Cells (0.5A550-ml) were subjected to spheroplast preparation as described previously (27). Spheroplasts were then suspended in 200 mM Tris-acetate (pH 8.2) containing 20 mM MgSO4 and 250 mM sucrose. The samples were then treated with 60 μM CCCP (carbonyl cyanide m-chlorophenylhydrazone) for 1 min. The CCCP-treated spheroplasts were incubated with proteinase K at 25 μg ml−1 for 15 min at 4°C and subsequently treated with 1.0 mM PMSF (phenylmethylsulfonyl fluoride) to inactivate the proteinase K. Inactivated samples were precipitated with an equal volume of 20% TCA and prepared for analysis on 13% SDS-polyacrylamide gels, followed by immunoblot analysis with anti-TonB antibody.
In vivo disulfide cross-linking.
Saturated cultures of strains carrying plasmids encoding ExbB Cys substitutions were grown overnight in LB medium and subcultured 1:100 in tryptone broth (30). Ampicillin at 100 μg/ml and l-arabinose as described in the figure legends was present throughout. Cultures were harvested in mid-exponential phase and immediately suspended in nonreducing LSB containing 50 mM iodoacetamide at 95°C, as described previously (12). Samples were resolved on 13% nonreducing SDS-polyacrylamide gels and immunoblotted with anti-ExbB polyclonal antibody.
RESULTS
Identification of four important residues: ExbB N196, D211, A228, and G244.
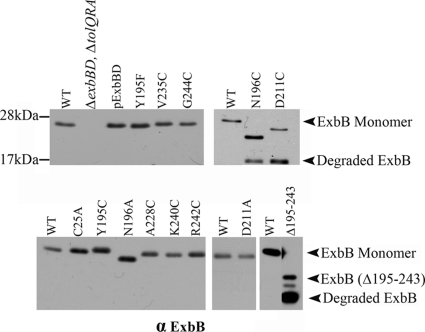
To assess the importance of the carboxy-terminal tail, exbB codons 195 to 243 were deleted by extra-long PCR on a plasmid expressing the exbB-exbD operon under arabinose control. ExbB(Δ195-243) was susceptible to endogenous proteolysis (Fig. 1). Sufficient levels of the full-length ExbB(Δ195-243) could be expressed using 0.2% l-arabinose so that its activity could be determined in RA1017, a ΔexbBD ΔtolQRA strain that eliminated cross talk from the paralogous Tol system (3, 5). ExbB(Δ195-243) failed to support iron transport and was completely tolerant to colicins B, Ia, and M and bacteriophage φ80 (Table 2), suggesting that the carboxy-terminal tail of ExbB was required for activity. Some of the inactivity could also have been due to the presence of proteolytic products that compete with wild-type ExbB, as has been seen for wild-type TonB (29).
Fig. 1.
Steady-state levels of ExbB mutants. Wild-type W3110 (WT), the parent strain for all of the plasmid variants (ΔexbBD ΔtolQRA), the parent plasmid encoding both ExbB and ExbD (pExbBD), and the plasmid-encoded ExbB variants are indicated above each lane of the immunoblot. Bacterial strains expressing plasmid-encoded ExbB variants were grown under conditions identical to those for the iron transport assays in Table 2. Samples with equal numbers of bacteria were precipitated with TCA, electrophoresed on 13% SDS-polyacrylamide gels, and immunoblotted with anti-ExbB antibody. ExbB monomer and proteolytic degradation products are labeled on the right. Positions of mass standards are indicated on the left.
To identify important residues in the carboxy-terminal tail, each was replaced by cysteine on a plasmid template encoding exbB(C25A) exbD, which eliminated the sole native Cys residue. ExbB(C25A) had essentially the same activity as plasmid-encoded wild-type ExbB in terms of iron transport, which is the most discriminative assay (Fig. 1 and Table 2) (24). Most of the Cys substitutions had little effect in the phenotypic assays of colicin sensitivity when expressed at chromosomal levels and were not assayed for iron transport (data not shown). The colicin and bacteriophage assays are the most sensitive and least discriminatory with respect to TonB system activities (24). We focused on the partially and fully inactive Cys substitutions in Y195, N196, D211, A228, and G244 (Table 2). TonB and ExbD levels did not vary significantly in RA1017 expressing the plasmid-encoded ExbB variants, with the exception of decreased ExbD in the presence of N196A. Because the half-life of ExbD was unaffected by ExbB(N196A) (data not shown), it seemed likely that the substitution at exbB(N19A) somehow changed the expression level of the downstream exbD gene. The data for K240C and R242C, which subsequently demonstrated non-wild-type behavior in an in vivo formaldehyde cross-linking assay, were included.
ExbB(Y195C) supported ∼10% of wild-type initial rates of [55Fe]ferrichrome transport; ExbB(Y195A) was completely inactive (Table 2). In contrast ExbB(Y195F) supported ∼100% of the wild-type level, suggesting that an aromatic amino acid at the position 195 was required for the activity.
Although ExbB(N196C) and ExbB(D211C) were completely tolerant to colicins B, D, Ia, and M and phage φ80, these results could not be unambiguously interpreted due to the proteolytic instability of the mutant proteins in an assay that occurs over ∼14 h. With full induction (0.2% arabinose), it was possible to achieve chromosomal levels (Fig. 1), and it was clear that these Cys substitutions did not support [55Fe]ferrichrome transport (Table 2). Substitution of either residue with Ala rendered each protein stable and conferred slight sensitivity to some colicins and φ80, but only 1 and 18% ferrichrome transport, respectively. ExbB(A228C) had reduced sensitivity to phage and colicins and supported only ∼3% [55Fe]ferrichrome transport. ExbB(G244C) had somewhat greater sensitivity to phage and colicins and supported ∼30% [55Fe]ferrichrome transport (Table 1).
The ExbB dimer is a stable subunit of the ExbB tetramer.
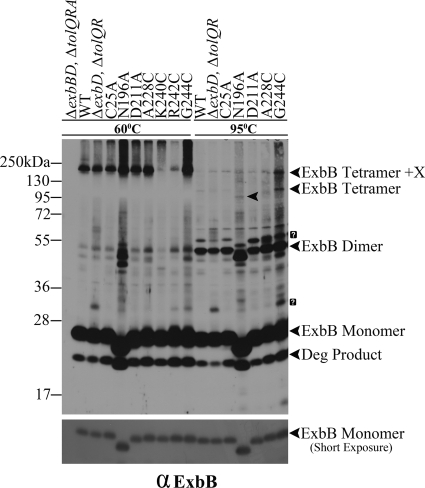
Epitope-tagged ExbB has been shown to form multimers both in vivo and in vitro (16, 37). To determine the effect of the Cys substitutions on ExbB multimerization, it was first necessary to examine the in vivo formaldehyde cross-linking profiles of wild-type chromosomally encoded ExbB. We had previously detected a form of plasmid-encoded, formaldehyde-cross-linked, epitope-tagged T7-ExbB whose apparent molecular mass varied depending on the percentage of the SDS-polyacrylamide gel. Thus, it became known as the ExbB trimer/tetramer (16). It was large enough (∼185 kDa) that it was proposed to include an additional 50 to 100 kDa of unknown protein. We also detected a T7-ExbB dimer in those studies. Because the T7 tag on ExbD in that same study was later shown to artifactually alter ExbD function and cross-linking behavior, the formaldehyde cross-linking profile of untagged ExbB needed to be reevaluated (33).
In Fig. 2, chromosomally encoded ExbB formed the previously observed cross-linked trimer/tetramer-unknown protein(s) complex. In addition, there appeared to also be a dimer of ExbB, although it was difficult to identify the cross-linked dimer among the faint bands in that region. The absence of ExbD did not alter the position or abundance of the trimer/tetramer. A complex of unknown identity at ∼34 kDa appeared in the ΔexbD background. ExbB(C25A) formed the same formaldehyde-linked complexes as chromosomally encoded ExbB.
Fig. 2.
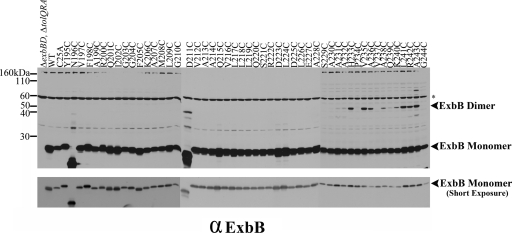
The ExbB dimer is a stable subunit of the ExbB tetramer. The parent strain for all of the plasmid variants (ΔexbBD ΔtolQRA), wild-type W3110 (WT), a strain expressing ExbB from the chromosome (ΔexbD ΔtolQR), and the plasmids expressing the ExbB variants are indicated above each lane of the immunoblot. Strains carrying exbB or exbD plasmids were expressed in the presence of 0.0005% l-arabinose (except for the N196A and G244C substitutions, where 0.001 and 0.002% l-arabinose were used, respectively). Mid-exponential-phase cultures were cross-linked with paraformaldehyde, transferred to LSB (23), and heated for 5 min at either 60 or 95°C. Samples were resolved on a 13% SDS-polyacrylamide gel and immunoblotted with anti-ExbB antibody. The positions of various ExbB complexes are indicated on the right. The arrowhead in the middle of lane N196A at 95°C indicates the position of the ExbB(N196A) tetramer. The positions of mass standards are on the left. A shorter exposure of the same immunoblot to indicate ExbB monomer levels is in the lower panel. Question marks (?) indicate unidentified protein complexes.
Heating of the formaldehyde-cross-linked samples at 95°C rather than 60°C largely abolished the ExbB trimer/tetramer, to be replaced by an ExbB dimer band of equal intensity at ∼52 kDa. Because there was also no significant increase in the level of ExbB monomer, it suggested that the trimer/tetramer had been fully converted into the ExbB dimer by heating at 95°C, which broke some but not all of the formaldehyde cross-links. The only way for this to occur was if the trimer/tetramer was actually a tetramer in vivo (referred to as tetramer+X). A trimer would have been broken down into a monomer plus dimer, decreasing the intensity of the dimer relative to its source trimer and increasing the intensity of the monomer. This result suggested that the ExbB dimers formed by this means were sufficiently strong to survive treatment at high temperature.
ExbB N196A, D211A, A228C, and G244C mutants assemble correctly.
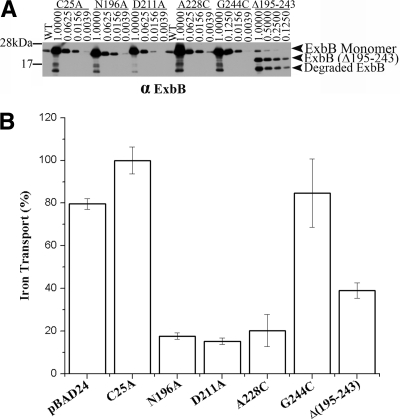
Because ExbB is a multimer, if the ExbB mutants assemble correctly within an otherwise wild-type TonB system, they should be dominant. To test for dominance, expression was fully induced from plasmids encoding ExbB mutants, along with wild-type ExbD, using 0.2% arabinose in strain W3110 (wild type). The single substitutions were overexpressed more than 60-fold compared to wild-type levels. ExbB(Δ195-243) could only be overexpressed 8-fold (Fig. 3 A). Overexpression of ExbB(N196A), ExbB(D211A), and ExbB(A228C) decreased [55Fe]ferrichrome transport to <20% of the wild-type value. ExbB(G244C) did not appear to be dominant, perhaps because it was not highly disabled to begin with. ExbB(Δ195-243) appeared to be the most dominant. Although it reduced iron transport to ∼40% of the wild-type level, it only required about 1/8 as much protein as the other mutants to do so. These results suggested that overexpressed ExbB mutants assembled correctly with the wild-type ExbB and thereby poisoned the tetramers. Because the four missense mutants were relatively stable, proteolytic fragments did not contribute much, if anything, to the dominance effect. Overexpression of wild-type ExbB(C25A) and ExbD from pKP878 did not decrease [55Fe]ferrichrome transport in W3110.
Fig. 3.
Most inactive ExbB mutants are dominant. (A) Expression of plasmid-encoded ExbB mutants and ExbD was fully induced with 0.2% l-arabinose in the wild-type strain W3110 1 h prior to an assay of the iron transport. Immediately prior to the transport assays samples were precipitated with TCA. Dilutions of equal A550-ml samples were electrophoresed on 13% SDS-polyacrylamide gels and immunoblotted with anti-ExbB antibody to determine relative levels of overexpression. The identities of the ExbB variants and the dilutions are indicated above the lanes. The positions of ExbB monomer, ExbB(Δ195-243), and the proteolytic degradation products are noted on the right. Mass standards are indicated on the left. The figure is a composite of two immunoblots. (B) Initial rates of iron transport were determined multiple times in triplicate assays after 1 h of induction with 0.2% l-arabinose. Averages are shown as a percentage of W3110 from which plasmid-encoded ExbB(C25A) and ExbD (pKP878) were overexpressed. The standard deviations are indicated on each bar.
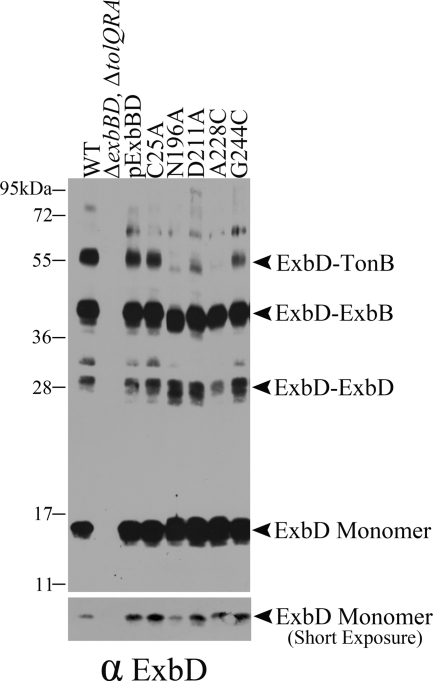
In another test of whether the four ExbB mutants could assemble correctly, strains expressing them were cross-linked in vivo with formaldehyde and immunoblotted with anti-ExbB antibody. Each could form the ExbB tetramer+X and, when heated to 95°C as described above, could also collapse into the same homodimeric form (Fig. 2). This demonstrated that even when expressed in the absence of wild-type ExbB, the four mutants could assemble correctly. The identity of the homodimeric complex was strengthened by the fact that the apparent molecular mass of ExbB(N196A) homodimers was decreased by approximately twice the amount seen for N196A monomers, which migrated aberrantly. The four ExbB mutants could also cross-link normally with ExbD (Fig. 4), and TonB (Fig. 5), suggesting that their inactive phenotypes were not due to a defect in assembly.
Fig. 4.
Inactive ExbB mutants assemble correctly with ExbD. Equivalent numbers of mid-exponential-phase bacteria were cross-linked with paraformaldehyde and evaluated by immunoblotting with anti-ExbD antibody. Wild-type W3110 (WT), the parent strain for all of the plasmid variants RA1017 (ΔexbBD ΔtolQRA), the parent plasmid encoding both ExbB and ExbD (pExbBD), and the plasmids expressing the ExbB variants are indicated above each lane of the immunoblot. The lower panel is a shorter exposure of the upper panel to allow evaluation of monomer ExbD levels. The positions of the ExbD monomer, the ExbD-ExbD dimer, the ExbD-ExbB heterodimer, and the ExbD-TonB heterodimer are indicated on the right. The positions of mass standards are indicated on the left.
Fig. 5.
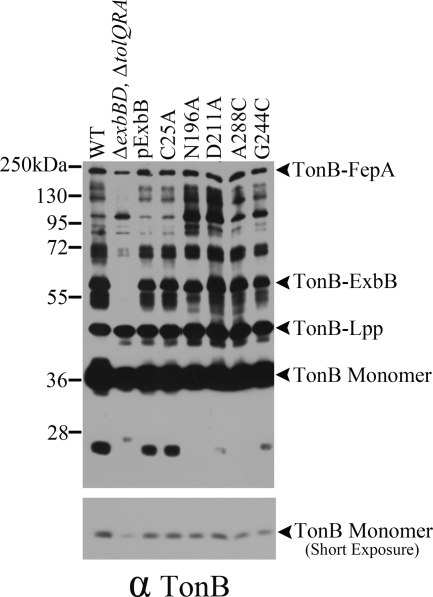
Inactive ExbB mutants assemble correctly with TonB. Equivalent numbers of mid-exponential-phase bacteria were cross-linked with paraformaldehyde and evaluated by immunoblotting with anti-TonB antibody. Wild-type W3110 (WT), the parent strain for all of the plasmid variants RA1017 (ΔexbBD ΔtolQRA), the parent plasmid encoding both ExbB and ExbD (pExbBD), and the plasmids expressing the ExbB variants are indicated above each lane of the immunoblot. The lower panel is a shorter exposure of the upper panel to allow evaluation of the monomer TonB levels. The positions of the TonB monomer, the TonB-Lpp heterodimer, the TonB-ExbB heterodimer, and the TonB-FepA heterodimer are indicated on the right. The positions of mass standards are indicated on the left.
Traces of two additional bands survived the 95°C treatment: the band representing the tetramer+X at ∼180 kDa and a new band at ∼95 kDa (Fig. 2, 95°C). The new band at ∼95 kDa was likely the actual ExbB homotetramer because it was the correct molecular mass for such a species, and the ExbB N196A tetramer exhibited the aberrant migration characteristic of the monomer. This ∼95-kDa band must have arisen due to 95°C disruption of the formaldehyde cross-link between “X” and the ExbB tetramer. ExbB(N196A) tetramer+X did not have a significantly reduced mass compared to the wild-type complex, possibly because the presence of the unknown protein dominated the overall conformation.
The remaining substitutions were cross-linked with formaldehyde and most exhibited no effect on ExbB tetramer formation (data not shown). The two exceptions were ExbB(K240C) and ExbB(R242C). These substitutions, although fully active, formed cross-linked tetramers with much less efficiency (Fig. 2, 60°C) and are discussed in greater detail below. Given the proximity of K240C and R242C in the primary amino acid sequence, these results suggested that formaldehyde-linked tetramer formation occurred through the cytoplasmic tails of ExbB. There was no effect on the formation of formaldehyde cross-linked ExbB complexes with TonB or ExbD for any of the other Cys and Ala substitutions from residues 195 to 244 (data not shown).
The ExbB N196A, D211A, A228C, and G244C substitutions prevent correct interactions between TonB and ExbD periplasmic domains.
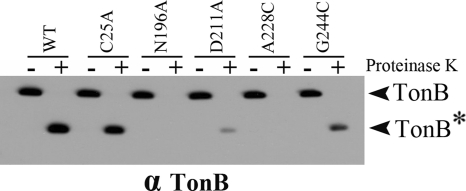
To examine the mechanistic effects of the mutants on TonB energization, the ability of the mutants to support conversion of TonB to the proteinase K-resistant form characteristic of wild-type TonB was examined. When the PMF of spheroplasts is collapsed by the addition of CCCP, wild-type TonB forms a proteinase K-resistant fragment (27), as seen in Fig. 6. The ability of the four ExbB mutants to support formation of this proteinase K-resistant fragment correlated well with the ability of these mutants to support iron transport in Table 2. ExbB(N196A) and ExbB(A228C) supported undetectable amounts of the proteinase K-resistant fragment, ExbB(D211A) supported a very low level, and ExbB(G244C) supported the most. ExbB and ExbD are each required for TonB to form the proteinase K-resistant conformation (14, 27).
Fig. 6.
The levels of proteinase K-resistant TonB are proportional to the activity of the ExbB mutants. RA1017 (ΔexbBD ΔtolQRA) cultures expressing plasmid-encoded ExbB mutant proteins were induced with 0.0005% of l-arabinose except for substitutions N196A and G244C, where 0.001 and 0.002% of l-arabinose were used, respectively, for the expression at chromosomally encoded levels. At mid-exponential phase, the cultures were converted to spheroplasts and treated with the protonophore CCCP, followed by proteinase K for 15 min, or left untreated (−). The samples were resolved on 13% SDS-polyacrylamide gels and immunoblotted with anti-TonB antibody. “TonB*” indicates the position of the TonB proteinase K-resistant fragment. The activity levels for the variants were 100% (ExbB C25A), 1% (ExbB N196A), 18% (ExbB D211A), 3% (ExbB A228C), and 31% (G244C) (see Table 2). In the absence of CCCP, TonB was fully proteolysed by proteinase K, as seen previously (27; data not shown).
The defect in the ExbB mutants not only affected the ability of TonB to undergo a conformational change characteristic of wild-type TonB, it also largely prevented TonB-ExbD formaldehyde cross-linking (Fig. 4). The TonB-ExbD formaldehyde cross-link occurs through the periplasmic domains of these proteins. In the absence of PMF or if either TonB or ExbD carries an inactivating mutation in its TMD, the cross-link does not occur. In Fig. 4, none of the ExbB mutants supports the full degree of TonB-ExbD cross-linking seen with wild-type ExbB. In particular, the least active ExbB N196A and A228C mutants form little if any. Thus, the ExbB mutants prevent the stable energy-dependent interaction between the TonB and ExbD periplasmic domains.
Evidence for tail-tail communication based on disulfide bond formation.
The inability of ExbB(K240C) and ExbB(R242C) to formaldehyde cross-link into tetramers in vivo suggested that dimer and tetramer formation occurred through the carboxy-terminal tail because Lys and Arg are both formaldehyde cross-linkable residues. To further test this idea, the ability of the Cys substitutions to form disulfide-linked dimers was assessed by immunoblot nonreducing SDS-PAGE with anti-ExbB antibodies. Cys substitutions near the extreme carboxy terminus were able to form disulfide-linked dimers to various degrees in strain RA1017 (W3110 ΔexbBD::kan ΔtolQRA), as shown in Fig. 7. The formation of disulfide-linked dimers in the cytoplasmic compartment was unexpected, since it is overall a reducing environment.
Fig. 7.
Cytoplasmic ExbB cysteine substitutions form disulfide-linked homodimers. The parent strain for all of the plasmid variants RA1017 (ΔexbBD ΔtolQRA), wild-type W3110 (WT), the parent plasmid encoding both ExbB and ExbD (pExbBD), and the plasmids expressing the ExbB variants are indicated above each lane of the immunoblot. To achieve chromosomal levels, the plasmid-encoded ExbB Y195C, N196C, D211C, Q215C, D223C, L224C, A228C, and G244C mutants were induced in tryptone broth with 0.00002, 0.005, 0.005, 0.00002, 0.00002, 0.00004, 0.00001, and 0.0001% l-arabinose, respectively. All other mutants were grown in tryptone broth without inducer. Samples were resolved on 13% nonreducing polyacrylamide gels and immunoblotted with anti-ExbB antibody. The figure is a composite of three immunoblots. The positions of the ExbB disulfide-linked dimers and ExbB monomer are indicated on the right. The positions of the mass markers are indicated on the left. The asterisk indicates the nonspecific cross-reactive bands. A shorter exposure of the immunoblot to allow comparison of monomer levels is presented in the lower panel.
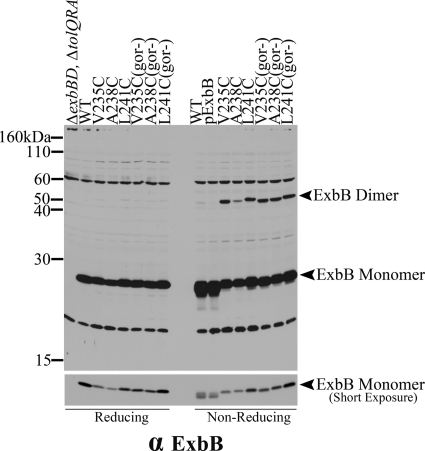
Mutations in the glutathione reductase gene, gor, create a more oxidizing cytoplasm, so that disulfide bonds can be observed in that environment (45). We tested the effect of the gor mutant strain KP1517 on a subset of disulfide bond-forming Cys substitutions. Cultures were harvested in mid-exponential phase, and each sample was divided between gel sample reducing and nonreducing buffers and processed for immunoblot analysis. The level of disulfide-linked dimer did not increase significantly in the gor mutant strain, suggesting that disulfide-linked dimer formation was due to sequestration of the ExbB carboxy terminus away from the reducing cytoplasm (Fig. 8).
Fig. 8.
Dimer formation does not increase significantly in the more oxidizing environment of a strain lacking glutathione reductase. A subset of plasmid-encoded Cys substitutions that form disulfide-linked homodimers was expressed in RA1017 (ΔexbBD ΔtolQRA) and KP1517 (W3110 exbB::Tn10 Δgor-756::kan). l-Arabinose at 0.000005% was added to the growth culture of strains lacking the gor gene (gor−) to induce chromosomal levels of expression. Both reducing and nonreducing sample sets were run on 13% SDS-polyacrylamide gels and immunoblotted with anti-ExbB antibody. RA1017, W3110, the parent plasmid encoding both ExbB and ExbD (pExbBD), and the plasmids expressing the ExbB variants are indicated above each lane of the immunoblot. The lower panel is a shorter exposure of the same image to allow comparison of the ExbB levels.
DISCUSSION
This study was the first mutagenic analysis of any ExbB soluble region, although the cytoplasmic membrane boundary of that region is ambiguous because the boundaries of ExbB TMD 3 are not firmly established. There are three published predictions for the boundaries of ExbB TMD 3: residues 162 to 194 (19), residues 177 to 199 (20), and residues 172 to 194 (48), with the soluble cytoplasmic carboxy terminus concluding at residue 244. Using a 22-residue window, an MPEx (Membrane Protein Explorer [http://blanco.biomol.uci.edu/mpex/]) prediction suggested that the third TMD of ExbB extends from A177 to A199 (data not shown). As more sequences become available, it is clear from comparison of distantly related ExbB sequences (expect values of 4 e−7) that ExbB TMDs 2 and 3 are highly conserved, whereas the sequences of the soluble carboxy termini vary widely. On that basis, the boundaries of TMD 3 would be from residues 173 to 196. Thus, it is not totally clear where the boundary of the cytoplasmic carboxy terminus is. We favor the idea that ExbB TMD3 includes Y195 and N196 because they are highly conserved, even in paralogue TolQ, while the residues from V197 to G244 are not.
Only five of the ExbB residues from 195 to 244 could not be substituted with Cys or Ala and retain activity: Y195, N196, D211, A228, and G244. Although ExbB(Y195C) and ExbB(Y195A) were inactive, ExbB(Y195F) was fully active, suggesting that it was important for packing with another TMD helix. Two substitutions, the ExbB N196C and ExbB D211C substitutions, were proteolytically unstable but were rendered stable by substitution with Ala instead of Cys. D211 is a moderately conserved residue, while A228 and G244 are not conserved. Most interesting was the A228C substitution, which was proteolytically stable and only 3% active in iron transport. It also showed low levels of sensitivity to colicins. This mutation did not affect any known ExbB interactions with itself, ExbD, or TonB. It did, however, prevent the correct interaction between TonB and ExbD periplasmic domains, thus providing the first evidence for a defect in signal transduction from cytoplasm to periplasm in the TonB system. That an Asp and an Ala residue were needed for activity suggested that they played important structural roles required for precise interactions with other cytoplasmic residues. A subset of ExbB Cys substitutions could form disulfide cross-links, suggesting that the extreme carboxy terminus of ExbB was sequestered from the reducing environment of the cytoplasm.
In the present study, formaldehyde cross-linked ExbB tetramers were visualized for the first time by the simple procedure of solubilizing the complexes at 95°C instead of 60°C. Solubilization at 95°C can disrupt formaldehyde cross-links, which suggested that the propensity of ExbB to form dimers must be fairly strong (38, 44). It may be that the ExbB tetramer was actually a dimer of dimers, with one set of the formaldehyde cross-links being refractory to boiling. After long exposures of immunoblots of wild-type ExbB heated at 95°C in the presence of β-mercaptoethanol (our standard sample preparation), we have detected what appear to be ExbB dimers, even though the samples have not been cross-linked (data not shown).
The idea that ExbB may be a dimer of dimers is consistent with the observation that neither ExbB(K240C) nor ExbB(R242C) could efficiently form the tetramer+X complex, likely because neither could form the tetramer. Lys is one of the most active residues for formaldehyde cross-linking in vivo (46). Arg is also formaldehyde cross-linkable, although usually less efficiently than Lys in vivo. Also, the propensity of the Cys substitutions to disulfide cross-link was consistent with the idea that the ExbB carboxy termini were in close contact.
The absence of tetramer+X in R242C suggested that R242 might normally be in a unique environment that somehow supported efficient cross-linking. Prior to the present study, the cross-linking we have seen in the TonB system has been rather inefficient, likely due to the dynamic nature of the interactions. Given that the tetramer+X complex theoretically requires at least four separate formaldehyde cross-links to form, it seems likely that this is actually a stable and abundant form of ExbB.
These studies contradicted a previous study suggesting that T7 epitope-tagged-ExbB might cross-link as a trimer in vivo. It seemed feasible at the time because T7-epitope-tagged ExbD also formed a trimer (16). However, it was recently demonstrated that the T7-epitope tag artifactually changed the behavior of ExbD. Untagged ExbD forms dimers and, unlike T7-epitope-tagged ExbD, does not form trimers but does cross-link to ExbB and TonB (33). In the present study, the true, chromosomally encoded ExbB tetramer without any unknown proteins was observed for the first time in vivo and at its predicted mass of 95 kDa. These data also solidified the idea that there were one or more proteins, up to 85 kDa in mass, that constituted the ExbB tetramer+X.
ACKNOWLEDGMENTS
We thank Kristin Baker for the ExbB half-life data and Fanny Kippelen for engineering many of the mutants. We thank Ray Larsen and members of the Postle lab for critical reading of the manuscript.
This study was supported by grant GM42146 from the National Institute of General Medical Sciences to K.P.
Footnotes
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Ahmer B. M. M., Thomas M. G., Larsen R. A., Postle K. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 177:4742–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Braun V. 2009. FhuA (TonA), the career of a protein. J. Bacteriol. 191:3431–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Braun V. 1989. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J. Bacteriol. 171:6387–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braun V., et al. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun V., Herrmann C. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261–268 [DOI] [PubMed] [Google Scholar]
- 6. Braun V., Herrmann C. 2004. Point mutations in transmembrane helices 2 and 3 of ExbB and TolQ affect their activities in Escherichia coli K-12. J. Bacteriol. 186:4402–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brinkman K. K., Larsen R. A. 2008. Interactions of the energy transducer TonB with noncognate energy-harvesting complexes. J. Bacteriol. 190:421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cadieux N., Kadner R. J. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. U. S. A. 96:10673–10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cascales E., et al. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devanathan S., Postle K. 2007. Studies on colicin B translocation: FepA is gated by TonB. Mol. Microbiol. 65:441–453 [DOI] [PubMed] [Google Scholar]
- 11. Fischer E., Günter K., Braun V. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 171:5127–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghosh J., Postle K. 2005. Disulfide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol. Microbiol. 55:276–288 [DOI] [PubMed] [Google Scholar]
- 13. Hannavy K., et al. 1990. TonB protein of Salmonella typhimurium: a model for signal transduction between membranes. J. Mol. Biol. 216:897–910 [DOI] [PubMed] [Google Scholar]
- 14. Held K. G., Postle K. 2002. ExbB and ExbD do not function independently in TonB-dependent energy transduction. J. Bacteriol. 184:5170–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgs P. I., Larsen R. A., Postle K. 2002. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol. Microbiol. 44:271–281 [DOI] [PubMed] [Google Scholar]
- 16. Higgs P. I., Myers P. S., Postle K. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hill C. W., Harnish B. W. 1981. Inversions between rRNA genes of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 78:7069–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kampfenkel K., Braun V. 1992. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 174:5485–5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kampfenkel K., Braun V. 1993. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 268:6050–6057 [PubMed] [Google Scholar]
- 20. Karlsson M., Hannavy K., Higgins C. F. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8:389–396 [DOI] [PubMed] [Google Scholar]
- 21. Krewulak K. D., Vogel H. J. 2011. TonB or not TonB: is that the question? Biochem. Cell Biol. 89:87–97 [DOI] [PubMed] [Google Scholar]
- 22. Kuehl C. J., Crosa J. H. 2010. The TonB energy transduction systems in Vibrio species. Future Microbiol. 5:1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 24. Larsen R. A., Chen G. J., Postle K. 2003. Performance of standard phenotypic assays for TonB activity, as evaluated by varying the level of functional, wild-type TonB. J. Bacteriol. 185:4699–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larsen R. A., et al. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol. 189:2825–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larsen R. A., et al. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178:1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Larsen R. A., Thomas M. G., Postle K. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809–1824 [DOI] [PubMed] [Google Scholar]
- 28. Larsen R. A., Thomas M. T., Wood G. E., Postle K. 1994. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol. Microbiol. 13:627–640 [DOI] [PubMed] [Google Scholar]
- 29. Mann B. J., Holroyd C. D., Bradbeer C., Kadner R. J. 1986. Reduced activity of TonB-dependent functions in strains of Escherichia coli. FEMS Lett. 33:255–260 [Google Scholar]
- 30. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Noinaj N., Guillier M., Barnard T. J., Buchanan S. K. 2010. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64:43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ogierman M., Braun V. 2003. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J. Bacteriol. 185:1870–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ollis A. A., Manning M., Held K. G., Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 73:466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Plastow G. S., Holland I. B. 1979. Identification of an Escherichia coli inner membrane polypeptide specified by a lambda-tonB transducing phage. Biochem. Biophys. Res. Commun. 90:1007–1014 [DOI] [PubMed] [Google Scholar]
- 35. Postle K. 2007. TonB system, in vivo assays and characterization. Methods Enzymol. 422:245–269 [DOI] [PubMed] [Google Scholar]
- 36. Postle K., Larsen R. 2004. The TonB, ExbB, and ExbD proteins, p. 96–112 In Crosa J. H., Mey A. R., Payne S. M. (ed.), Iron transport in bacteria. ASM Press, Washington, DC [Google Scholar]
- 37. Pramanik A., Zhang F., Schwarz H., Schreiber F., Braun V. 2010. ExbB protein in the cytoplasmic membrane of Escherichia coli forms a stable oligomer. Biochemistry 49:8721–8728 [DOI] [PubMed] [Google Scholar]
- 38. Prossnitz E., Nikaido K., Ulbrich S. J., Ames G. F.-L. 1988. Formaldehyde and photoactivatable cross-linking of the periplasmic binding protein to a membrane component of the histidine transport system of Salmonella typhimurium. J. Biol. Chem. 263:17917–17920 [PubMed] [Google Scholar]
- 39. Rabsch W., et al. 2007. FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J. Bacteriol. 189:5658–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roof S. K., Allard J. D., Bertrand K. P., Postle K. 1991. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J. Bacteriol. 173:5554–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schauer K., Rodionov D. A., de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the “tip of the iceberg”? Trends Biochem. Sci. 33:330–338 [DOI] [PubMed] [Google Scholar]
- 42. Schöffler H., Braun V. 1989. Transport across the outer membrane of Escherichia coli via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol. Gen. Genet. 217:378–383 [DOI] [PubMed] [Google Scholar]
- 43. Shedlovsky A., Brenner S. 1963. A chemical basis for the host-induced modification of T-even bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 50:300–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skare J. T., Ahmer B. M. M., Seachord C. L., Darveau R. P., Postle K. 1993. Energy transduction between membranes: TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302–16308 [PubMed] [Google Scholar]
- 44a. Swayne C. D., Postle K. 2011. Taking the Escherichia Coli TonB transmembrane domain “offline”? Nonprotonatable Asn substitutes fully for TonB His20. J. Bacteriol. 193:3693–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tiebel B., Garke K., Hillen W. 2000. Observing conformational and activity changes of tet repressor in vivo. Nat. Struct. Biol. 7:479–481 [DOI] [PubMed] [Google Scholar]
- 46. Toews J., Rogalski J. C., Clark T. J., Kast J. 2008. Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Anal. Chim Acta 618:168–183 [DOI] [PubMed] [Google Scholar]
- 47. Vakharia-Rao H., Kastead K. A., Savenkova M. I., Bulathsinghala C. M., Postle K. 2007. Deletion and substitution analysis of the Escherichia coli TonB Q160 region. J. Bacteriol. 189:4662–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhai Y. F., Heijne W., Saier M. H., Jr 2003. Molecular modeling of the bacterial outer membrane receptor energizer, ExbBD/TonB, based on homology with the flagellar motor, MotAB. Biochim. Biophys. Acta 1614:201–210 [DOI] [PubMed] [Google Scholar]