Abstract
Purine nucleoside phosphorylase (PNP) is an important enzyme in purine metabolism and cleaves purine nucleosides to their respective bases. Mycobacterial PNP is specific for 6-oxopurines and cannot account for the adenosine (Ado) cleavage activity that has been detected in M. tuberculosis and M. smegmatis cultures. In the current work, two Ado cleavage activities were identified from M. smegmatis cell extracts. The first activity was biochemically determined to be a phosphorylase that could reversibly catalyze adenosine + phosphate ↔ adenine + alpha-d-ribose-1-phosphate. Our purification scheme led to a 30-fold purification of this activity, with the removal of more than 99.9% of total protein. While Ado was the preferred substrate, inosine and guanosine were also cleaved, with 43% and 32% of the Ado activity, respectively. Our data suggest that M. smegmatis expresses two PNPs: a previously described trimeric PNP that can cleave inosine and guanosine only and a second, novel PNP (Ado-PNP) that can cleave Ado, inosine, and guanosine. Ado-PNP had an apparent Km (Km app) of 98 ± 6 μM (with Ado) and a native molecular mass of 125 ± 7 kDa. The second Ado cleavage activity was identified as 5′-methylthioadenosine phosphorylase (MTAP) based on its biochemical properties and mass spectrometry analysis. Our study marks the first report of the existence of MTAP in any bacterium. Since human cells do not readily convert Ado to Ade, an understanding of the substrate preferences of these enzymes could lead to the identification of Ado analogs that could be selectively activated to toxic products in mycobacteria.
INTRODUCTION
Mycobacterium tuberculosis is the etiological agent of tuberculosis (TB), an infectious disease that was diagnosed in more than 9 million individuals and claimed nearly 2 million lives in 2007 (34). An estimated one-third of the world's population is infected with the latent form of the disease, and 10% of these people will develop active TB in their lifetimes. In 2007, TB caused 23% of the estimated HIV deaths (34), and thus, as the global HIV burden and TB-HIV coinfections increase, TB remains a growing health concern. M. tuberculosis strains that are resistant to first- and second-line drugs are also on the rise. According to the March 2010 estimates of the World Health Organization, one in four new cases of TB in northwest Russia was multiple-drug-resistant (MDR) TB, and 58 countries reported at least one incidence of extensively drug-resistant (XDR) TB (35). As drug-resistant TB becomes more prevalent, the array of drugs available to treat this deadly bacterial infection decreases. Thus, there is an urgent need to develop new antituberculosis drugs that have different mechanisms of action than current drugs.
An enhanced basic understanding of the enzymes involved in metabolic processes in mycobacteria could lead to identification of molecular targets for drug discovery. Purine metabolism is an essential process of all living cells, as it generates macromolecules necessary for DNA, RNA, and energy production. Extensive studies of human purine metabolism have led to the development of nucleoside analogs that are currently used to treat cancer. Moreover, differences in purine metabolism between human cells and various infectious agents have been exploited to develop nucleoside analogs used in the treatment of parasitic and viral infections. Since differences in purine metabolism between human and mycobacterial cells exist (20, 24), purine enzymes could be potential targets for the development of nucleoside analogs against TB. Moreover, because purine metabolism is not a target of current TB drugs, nucleoside analogs would likely be active against TB that is resistant to current agents. Furthermore, it is possible that nucleoside analogs could disrupt basic metabolic processes and thus be useful against latent TB.
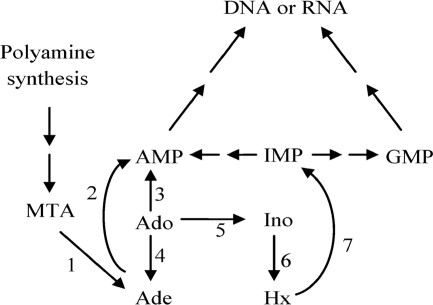
Previous studies have shown the conversion of adenosine (Ado) to adenine (Ade) in M. tuberculosis and Mycobacterium smegmatis cell cultures (9, 23). In human cells, Ado is primarily catalyzed by Ado kinase and Ado deaminase (Fig. 1) and is not readily cleaved to Ade. Therefore, the ability of mycobacteria to cleave Ado indicates a difference in the metabolism of Ado in mycobacteria that could be exploited for drug discovery. There are numerous enzymes known in nature that can cleave Ado. Although most bacterial purine nucleoside phosphorylases (PNPs) (EC 2.4.2.1) can cleave Ado to Ade, it is known that mycobacterial PNP does not accept Ado as a substrate (5, 11, 19). Some bacteria, such as Bacillus subtilis and Bacillus cereus, express an Ado phosphorylase (AdoP) that selectively cleaves Ado (16, 29, 30). A few parasites can also salvage Ado through hydrolases, such as Ado nucleosidase (EC 3.2.2.7) or purine nucleosidase (EC 3.2.2.1). Cleavage of Ado by parasitic 5′-methylthioadenosine phosphorylase (MTAP) (EC 2.4.2.28) has been reported (14, 21). MTAP is primarily found in Eukarya, including trypanosomes, and Archaea (1, 15). Although Pseudomonas isolates were thought to possess MTAP (1), recent work in Pseudomonas aeruginosa showed that the enzyme was a methylthioinosine phosphorylase rather than MTAP (15). The bacterial equivalent of MTAP is 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN or MTA/SAH nucleosidase) (EC 3.2.2.16). According to the UniProt Consortium (32), M. tuberculosis encodes MTAN (Rv0091) and a probable MTAP (Rv0535). Thus, either MTAN or MTAP could be responsible for the Ado cleavage observed in M. tuberculosis.
Fig. 1.
Enzymes involved in adenosine metabolism. (1) 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (not present in human cells) or 5′-methylthioadenosine phosphorylase; (2) adenine phosphoribosyl transferase; (3) adenosine kinase; (4) adenosine phosphorylases (not present in human cells) and most bacterial purine nucleoside phosphorylases; (5) adenosine deaminase; (6) purine nucleoside phosphorylase; (7) hypoxanthine/guanine phosphoribosyl transferase.
Given that the literature indicates that a number of bacterial, parasitic, and mammalian enzymes can cleave Ado to Ade, we investigated Ado cleavage in cell extracts from M. smegmatis, a closely related, fast-growing model of M. tuberculosis. Ado cleavage activity was assayed in all the fractions eluting from our first purification column. This step indicated that two enzymes could cleave Ado. One of the enzymes was identified as MTAP, and the other was an Ado cleavage enzyme that was different from PNP (MSMEG_1701), MTAP (MSMEG_0990), and MTAN (MSMEG_1753). Hence, our work marks the first investigation of two previously uncharacterized Ado cleavage activities in mycobacteria and further illustrates differences in purine metabolism between mycobacterial and human cells. These differences could be exploited for the development of Ado analogs that could be selectively activated to toxic products, thereby exhibiting selective toxicity against M. tuberculosis and forming the basis of a new class of anti-TB drugs.
MATERIALS AND METHODS
Reagents.
The natural nucleosides, nucleobases, and ribose-1-phosphate were purchased from Sigma-Aldrich. 2-Fluoroadenosine, 2-methyladenosine (methyl-Ado), and 9-benzyl-9-deazaguanine were synthesized at Southern Research Institute. Protease inhibitor cocktail was obtained from Sigma and Roche. Sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) minigels, Tris/glycine/SDS running buffer, Coomassie stain, silver stain reagents, low-range molecular weight standards, and Bradford dye reagent were purchased from Bio-Rad Laboratories. EZ-Run prestained protein marker was purchased from Fisher Scientific. Xanthine oxidase (from bovine milk) was purchased from Sigma-Aldrich. Purification columns and molecular weight standards for gel filtration chromatography were purchased from GE Healthcare.
Bacterial strain and growth conditions.
M. smegmatis ATCC 700084 was cultured in Middlebrook 7H9 medium supplemented with oleic acid, albumin, dextrose, and catalase (OADC), and 0.05% Tween 80 at 37°C in a shaking incubator until the optical density at 600 nm (OD600) was 0.4 to 0.6.
Preparation of crude protein extract.
M. smegmatis cells (from a 5-liter culture) were pelleted by centrifugation, and the cell pellet was washed three times in buffer A (50 mM Tris HCl, 1 mM dithiothreitol, and 20% glycerol, adjusted to pH 7.6). The cell pellet was resuspended in buffer A containing protease inhibitor cocktail (Sigma/Roche), and the cells were lysed using a French pressure cell. The resulting suspension was centrifuged at 4°C for 1 h at 100,000 × g. The supernatant was dialyzed overnight against two changes of 1 liter of buffer A plus 150 mM NaCl at 4°C (Spectra/Por 4 Membrane Tubing; 12,000 to 14,000 Da molecular weight cutoff [MWCO]; Fisher Scientific) and filtered using a 0.2-μm-pore-size filter.
Purification of Ado cleavage activity.
A 17-ml aliquot of crude extract was applied to an anion-exchange column (HiTrap Q) and eluted using a linear salt gradient of 150 to 750 mM NaCl in buffer A. Fractions with the most Ado-PNP activity were pooled, and 10 ml of this solution was applied to a hydrophobic interaction column (HiTrap Phenyl). The activity was eluted using a reverse linear gradient from 1 to 0 M ammonium sulfate in 50 mM sodium phosphate buffer (pH 7.5). Fractions with the most Ado-PNP activity were pooled and concentrated to 300 μl, 200 μl of which was loaded onto a size exclusion column (Superdex 200 PG) and eluted with an isocratic run of 150 mM NaCl in buffer A. Similarly, MTAP or PNP activity eluting from the HiTrap Q column was pooled and purified as described above. Protein concentrations of each pooled sample and crude extract were obtained by the Bradford method (7) using bovine serum albumin as a standard.
Activity assays.
Enzyme activity was followed by measuring the formation of product using reverse-phase high-performance liquid chromatography (HPLC). A reaction mixture (50 μl) consisting of 50 mM potassium phosphate buffer (pH 7.5), 50 mM HEPES (pH 7.3), 100 μM substrate, and enzyme was prepared. The reaction was started by the addition of the enzyme. After incubation at 37°C, the reaction was stopped at specific time points by the addition of 50 μl 1 M perchloric acid, and the mixture was neutralized and buffered to pH 7 with a solution containing 3 M KOH plus 0.6 M potassium phosphate buffer. The potassium perchlorate precipitate was removed by centrifugation. For acid-labile substrates, such as 2′-deoxyadenosine and thymidine, the reaction was stopped by boiling for 5 min. For the reverse reaction, the reaction mixture consisted of 200 μM ribose-1-phosphate, 50 mM HEPES (pH 7.3), 200 μM Ade, and enzyme. The substrates were separated from the products using reverse-phase HPLC with a BDS Hypersil C18 column (Keystone Scientific) and a mobile phase consisting of ammonium dihydrogen phosphate and acetonitrile buffer as described previously (23). The substrates and products were detected by their absorbance at 260 nm as they eluted from the column.
The xanthine oxidase-coupled spectrophotometric method of Savarese et al. (27) and Jensen and Nygaard (17) was modified to a 96-well plate format and was used to rapidly detect Ado-PNP, PNP, and MTAP in fractions eluting from purification columns. Ado and MTA cleavage produce Ade, which is then converted to 2,8-dihydroxyadenine by xanthine oxidase, leading to an increase in absorbance at 305 nm. Similarly, mycobacterial PNP converts inosine (Ino) to hypoxanthine (Hx), which is then converted to uric acid by xanthine oxidase, with an increase in absorbance at 293 nm. Each well (200 μl total volume) contained 30 μl of sample, 50 mM potassium phosphate buffer (pH 7.5), 50 mM HEPES (pH 7.3), 0.01 unit of xanthine oxidase, and substrate (2 mM Ado plus 100 μM deoxycoformycin, 100 μM MTA, or 2 mM Ino). The reaction was allowed to proceed at room temperature, and the changes in absorbance at 305 nm and 293 nm were measured over time.
Enzyme kinetics.
The apparent Km (Km app) and Vmax app for nucleoside phosphorolysis and nucleoside synthesis were calculated using the nonlinear regression function of SigmaPlot 2004 (Systat Software, Inc.). The assays were carried out at various concentrations of one substrate and fixed concentrations of the second substrate. The substrate conversions were maintained below 10%. With Ado-PNP, Ado, Ino, Ade, and ribose-1-phosphate concentrations were varied between 40 and 400 μM. Phosphate concentrations were varied between 250 and 5,000 μM. With MTAP, the concentration of MTA was varied between 2 and 10 μM, and the concentration of Ado was varied between 120 and 600 μM. For both MTA and Ado, the phosphate concentration was kept at 50 mM.
Nanoscale liquid chromatography (NanoLC)-tandem mass spectrometry.
Mass spectrometry analysis was conducted at the Targeted Metabolomics and Proteomics Laboratory at the University of Alabama at Birmingham as described previously (25). Briefly, protein bands from SDS-PAGE gels were excised and destained. Following trypsin digestion, peptides were applied to a C18 reverse-phase cartridge, and the eluted peptides were analyzed on an Applied Biosystems-MDS-Sciex (Concorde, Ontario, Canada) 4000 Qtrap mass spectrometer. The tandem mass spectrometry data thus obtained were processed to provide protein identification using an in-house MASCOT search engine (Matrix Science) with the M. smegmatis NCBInr protein database. One missed cleavage site for trypsin was allowed in the analysis.
RESULTS
Identification of adenosine cleavage enzymes.
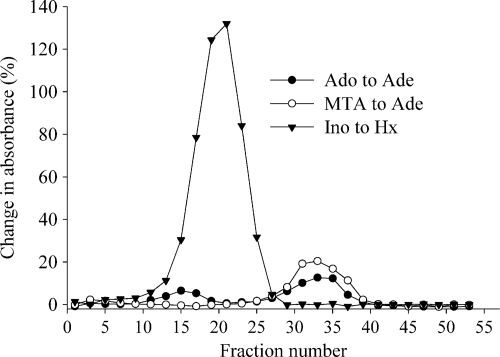
Partial purification of Ado cleavage was achieved by applying a cell extract of M. smegmatis to the HiTrap Q anion-exchange column. Two-milliliter fractions were collected as the proteins eluted with a linear gradient of 150 to 750 mM NaCl, and the fractions were tested for cleavage of Ado, Ino, and MTA by the xanthine oxidase method. Ino cleavage to Hx was seen with maximal activity in fraction 21 (Fig. 2). This fraction had negligible Ado cleavage, which indicated the presence of PNP, since mycobacterial PNP can cleave Ino but not Ado. Two peaks of Ado cleavage were detected, with maximal activity in fractions 15 and 33. MTA cleavage was also seen in fraction 33, suggesting that Ado cleavage in fraction 33 could be due to MTAP and/or MTAN. Fraction 15 had no MTA cleavage, and this indicated the presence of a novel Ado cleavage activity that was different from that of PNP, MTAP, or MTAN.
Fig. 2.
Elution of Ado, MTA, and Ino cleavage activities from M. smegmatis cell extract applied to the HiTrap Q anion-exchange column. M. smegmatis cell extract was applied to the HiTrap Q anion-exchange column and eluted with 150 to 750 mM NaCl gradient. Every other fraction was tested for Ade formation from Ado or MTA and Hx formation from Ino by the xanthine oxidase assay. The formation of Ade and Hx is expressed as the percent change in initial absorbance at 305 nm (for Ado and MTA) and 293 nm (for Ino) after a 2-hour incubation with Ado, MTA, and Ino.
Purification of adenosine cleavage enzymes.
Further purification of fraction 15 (Ado cleavage enzyme) was achieved by using a HiTrap Phenyl hydrophobicity interaction column and a Superdex 200 PG size exclusion column, successively. Superdex fractions were tested for Ado-to-Ino conversion, which would indicate the presence of Ado deaminase. Fractions that contained Ado cleavage activity, but not Ado deaminase activity, were pooled and used for further characterization experiments. Our purification scheme led to the removal of more than 99.9% of total protein and a 30-fold purification of the Ado cleavage activity (Table 1). Although the resulting final fraction had Ado cleavage activity, the protein concentration of that sample could not be determined without using most of the sample, and a single band responsible for Ado cleavage activity could not be identified by Coomassie or silver staining the SDS-PAGE gel. However, since there was no evidence that the sample contained other purine enzymes that could affect substrate or product concentrations, that sample was suitable for further characterization experiments. PNP activity (corresponding to fractions 19 to 23) and MTA cleavage activity (corresponding to fractions 29 to 33) were also further purified by using the HiTrap Phenyl hydrophobicity interaction column, followed by the Superdex 200 PG size exclusion column.
Table 1.
Purification of Ado-PNP from M. smegmatis
| Step | Total protein (mg) | Enzyme activity (nmol/mg·min) | Purification (fold) | Total activity (nmol/min) | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 98 | 0.06 | 1 | 5.9 | 100 |
| HiTrap Q | 3.3 | 0.4 | 7 | 1.4 | 20 |
| HiTrap Phenyl | 0.034 | 1.9 | 32 | 0.056 | 1 |
| Superdex 200 PG | NDa | —b | >32 | 0.021 | 0.4 |
ND, not detected.
—, not determined.
Characteristics of the adenosine cleavage activity.
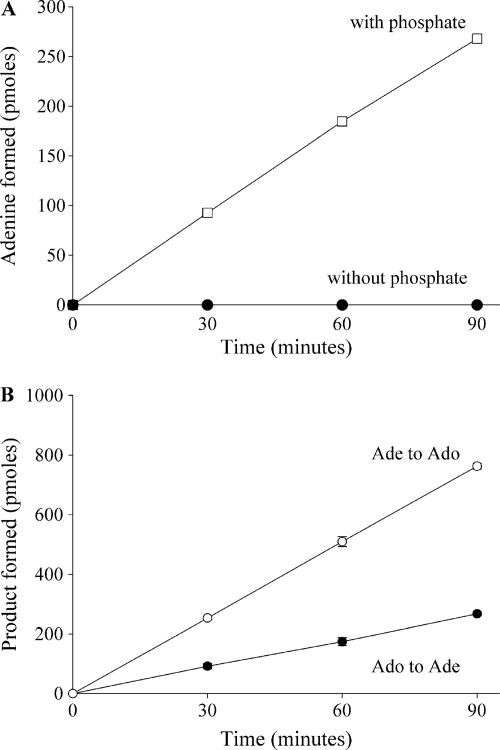
The cleavage of Ado seen in fraction 15 could be due to a hydrolase or a phosphorylase. To determine whether the Ado cleavage enzyme was a member of the hydrolase family, the purified enzyme was incubated in a reaction mixture containing Ado, HEPES (pH 7.3), and water, but no phosphate. Ade formation was not detected under these conditions (Fig. 3A), suggesting that another substrate or cofactor was needed for enzyme activity. When this reaction was repeated in the presence of 50 mM phosphate, Ade formation was seen, indicating that phosphate was required for activity. Additionally, Ado formation was observed when the enzyme was incubated with Ade and ribose-1-phosphate (Fig. 3B). Taken together, these results indicated that the Ado cleavage activity in fraction 15 was due to a phosphorylase that reversibly catalyzed adenosine plus phosphate to adenine plus alpha-d-ribose-1-phosphate.
Fig. 3.
Ado cleavage requires phosphate and is reversible. (A) Fraction 15 was incubated with Ado in the presence or absence of 50 mM phosphate, and Ade formation was monitored by HPLC. (B) The enzyme was incubated with Ado and 50 mM phosphate, and the formation of Ade was monitored by HPLC. The enzyme was also incubated with Ade and ribose-1-phosphate, and the formation of Ado was monitored by HPLC. Each data point represents the mean ± standard deviation for 3 determinations. Some error bars are too small to be seen or are hidden by the symbol.
To establish that the Ado cleavage activity was due to an enzyme other than PNP, MTAP, or MTAN, the activity of the Ado cleavage enzyme with the natural purine or pyrimidine nucleosides was evaluated. The Ado cleavage enzyme accepted Ado, Ino, and guanosine as substrates, with Ado being the best substrate (Table 2). Other substrates included 2′-deoxyadenosine, 2-fluoroadenosine, and methyl-Ado with 58%, 137%, and 31% of the Ado activity, respectively. No activity was detected with xanthosine, pyrimidine nucleosides (cytidine, thymidine, and uridine), MTA, or S-adenosyl-l-homocysteine (SAH). Since MTAP can cleave MTA and MTAN can cleave both MTA and SAH, our data suggested that the Ado cleavage enzyme was neither MTAP nor MTAN. Taken together, our data suggested that the Ado cleavage enzyme was specific for purines and tolerated modifications at the 2′ position on the ribose moiety and the 2 position of the Ade moiety. Thus, our results indicated that the Ado cleavage enzyme is a member of the PNP family. Since a trimeric mycobacterial PNP that can cleave Ino and guanosine but not Ado has been described by others (5, 11, 19), we refer to the Ado cleavage enzyme described in this work as Ado-PNP.
Table 2.
Substrate specificity of M. smegmatis Ado-PNP
| Nucleoside (100 μM) | Enzyme activitya (pmol min−1 ml−1) |
|---|---|
| Purines | |
| Adenosine | 19 ± 2 |
| 2′-Deoxyadenosine | 11.28 ± 0.05 |
| Inosine | 8 ± 4 |
| Guanosine | 6 ± 1 |
| Xanthosine | − |
| 5′-Methylthioadenosine | − |
| S-Adenosyl-l-homocysteine | − |
| Adenosine analogs | |
| 2-Fluoroadenosine | 26.3 ± 0.4 |
| 2-Methyladenosine | 5.8 ± 0.5 |
| Pyrimidines | |
| Cytidine | − |
| Thymidine | − |
| Uridine | − |
Mean ± standard deviation of at least 3 determinations. −, no activity detected.
As shown in Table 2, both Ado and Ino were cleaved by Ado-PNP. Since M. smegmatis expressed high levels of PNP, the Ino cleavage seen in our Ado-PNP sample could be due to contaminating trimeric PNP. If Ado and Ino are both substrates for one enzyme, then Ado cleavage would be inhibited by the presence of Ino. Ado cleavage (100 μM) was inhibited by 45% by 1,000 μM Ino (n = 3), suggesting that both Ino and Ado are substrates of Ado-PNP. Furthermore, 400 μM 9-benzyl-9-deazaguanine, a potent human PNP inhibitor (6, 22), inhibited Ino cleavage by M. smegmatis PNP by more than 75% (data not shown) but had no effect on either Ado or Ino cleavage by Ado-PNP. Taken together, these results indicated that Ado and Ino are both substrates of Ado-PNP.
The native molecular weight of the Ado-PNP was estimated by gel filtration chromatography on the Superdex 200 PG column. A calibration curve was produced based on the UV absorbance spectrum (generated from the Amersham-Pharmacia fast protein liquid chromatography [FPLC] controller LCC-501-Plus) of ferritin, conalbumin, carbonic anhydrase, and RNase A molecular weight standards. Fractions eluting from the column were assayed for Ado cleavage using the HPLC assay. One peak of Ado cleavage activity was seen. Based on the elution volume of the fraction with maximum activity, the molecular mass of Ado-PNP was calculated to be 125 ± 7 kDa (n = 6).
The stability of Ado-PNP at different temperatures was studied by incubating Ado-PNP at 4, 25, 37, or 60°C for 3, 6, or 12 h (data not shown). Ado-PNP activity was assayed at 37°C after each of these incubations. Ado-PNP retained most of its activity at 4, 25, and 37°C over 12 h, but the enzyme was inactive after being incubated at 60°C. To investigate the effect of pH on enzyme activity, Ado-PNP was incubated with Ado at different pH values, which were obtained by adjusting a solution of citric acid and disodium phosphate buffer to pH values of 3 to 8 or by adjusting a solution of sodium diphosphate buffer to pH values of 7 to 11. The formation of Ade at each pH was measured by reverse-phase HPLC, and the optimal pH of the reaction was between 6 and 7 (data not shown). The enzyme was inactive below pH 5. At pH 11, the enzyme had 44% of its activity at pH 7.
Kinetic properties of Ado-PNP.
The apparent steady-state kinetic constants of Ado-PNP were determined for Ado phosphorolysis and synthesis (Table 3). Substrate saturation curves were described by a hyperbolic function (data not shown), indicating that Ado-PNP followed Michaelis-Menten kinetics. Given that the protein concentration of Ado-PNP was not known, discrepancies in enzyme activity could exist between different batches of purified enzyme. Thus, to study the kinetic properties of Ado-PNP, the same batch of purified enzyme was used to calculate the Km app and Vmax app of Ado-PNP. The Km app for Ado (98 ± 6 μM) was higher than that of Ade (43 ± 7 μM). As shown in Table 4, Ino had 43% of the Ado activity at 100 μM substrate. To determine whether the difference in activity was due to substrate binding or catalytic efficiency, the kinetic parameters for Ino cleavage were determined. The Km app for Ino (220 ± 30 μM; n = 3) was more than twice that of Ado. However, there was no difference in Vmax/Km between Ino and Ado. Thus, the difference in Ado and Ino cleavage could be due to a difference in substrate binding.
Table 3.
Apparent steady-state kinetic constants of M. smegmatis Ado-PNP
| Substrate | Cosubstrate | Kma (μM) | Vmaxa (pmol min−1 ml−1) |
|---|---|---|---|
| Nucleoside phosphorolysis | |||
| Adenosine | 50 mM phosphate | 98 ± 6 | 450 ± 10 |
| Phosphate | 200 μM adenosine | 700 ± 100 | 350 ± 20 |
| Nucleoside synthesis | |||
| Adenine | 200 μM ribose-1-phosphate | 43 ± 7 | 1,360 ± 80 |
| Ribose-1-phosphate | 200 μM adenine | 70 ± 10 | 1,470 ± 90 |
Mean ± standard deviation of at least 3 determinations.
Table 4.
Relative activities of M. smegmatis Ado-PNP, MTAP, and PNP
| Nucleoside (100 μM) | Relative activity (%)a |
||
|---|---|---|---|
| Ado-PNP | MTAP | PNP | |
| Purines | |||
| Adenosine | 100 | 16 | − |
| 2′-Deoxyadenosine | 58 | − | − |
| Inosine | 43 | − | 100 |
| Guanosine | 32 | − | 68 |
| Xanthosine | − | − | − |
| 5′-Methylthioadenosine | − | 100 | − |
| S-Adenosyl-l-homocysteine | − | − | − |
| Adenosine analogs | |||
| 2-Fluoroadenosine | 137 | 19 | − |
| 2-Methyladenosine | 31 | − | − |
−, no activity detected.
Characteristics of MTA cleavage enzyme.
As shown in Fig. 2, Ado could be cleaved by a second enzyme, which could also cleave MTA (fraction 33). An SDS-PAGE gel of fractions 29 to 37 had several protein bands, with one band that correlated with MTA cleavage activity (data not shown). This band from fraction 33 was excised from the gel. NanoLC-tandem mass spectrometry analysis suggested the presence of numerous proteins, two of which were MTAP and MTAN. Since sequence homology predicted that M. smegmatis expressed both MTAP and MTAN, it is possible that fraction 33 contained both MTAP and MTAN (the assay used could not differentiate between MTAP and MTAN activities). However, further purification of this fraction led to an enzyme that was active only in the presence of 50 mM phosphate (data not shown), thereby indicating the presence of a phosphorylase and not a nucleosidase.
M. smegmatis MTAP (purified as described above) was also tested for its ability to cleave purines and pyrimidines. MTAP was most active with MTA and also cleaved Ado and 2-fluoroadenosine with 16% and 19% of the MTA activity, respectively (Table 4). No activity was detected with methyl-Ado, suggesting that MTAP was not tolerant of some modifications at the 2 position on the Ade moiety. Similar to human MTAP, cleavage of 2′-deoxyadenosine was not detected with M. smegmatis MTAP. SAH was not a substrate under the conditions tested, indicating that the enzyme was not MTAN. No activity was detected with Ino, guanosine, xanthosine, or the pyrimidine nucleosides. Based on the different substrate specificities for Ado and MTA, Ado-PNP and MTAP are different enzymes.
Human MTAP was shown to cleave Ado with 12.5% of the activity seen with MTA (13). However, human MTAP had a Km of 1.5 ± 0.2 μM with MTA and 760 ± 90 μM with Ado, which corresponds to a 500-fold-higher Km with Ado than with MTA (31). The Vmax with Ado was 62% of the Vmax with MTA and therefore represented an 800-fold-higher catalytic efficiency with MTA than with Ado (31). Hence, Ado is considered to be an inefficient substrate for human MTAP. Since Ado cleavage was observed with M. smegmatis MTAP, we investigated the kinetic properties of the mycobacterial enzyme with MTA or Ado as a substrate in the presence of 50 mM phosphate. M. smegmatis MTAP had a Km app of 1.6 ± 0.2 μM with MTA and 310 ± 40 μM with Ado, with similar Vmax for both substrates, indicating that MTA was a more efficient substrate than Ado for M. smegmatis MTAP.
Using the same size exclusion column and calibration curve discussed above, the native molecular mass of MTAP was calculated to be 64 ± 5 kDa. Since the MSMEG_0990 amino acid sequence predicted a protein of 27 kDa, our data suggested that MTAP expressed in M. smegmatis was a dimer. All previously described MTAPs, including human MTAP, are known to be active as trimers (2, 26) with the exception of Sulfolobus solfataricus MTAP, which is a homohexamer with a molecular mass of 160 kDa (3).
Substrate specificity of PNP.
To compare our Ado-PNP and MTAP results to M. smegmatis PNP, the substrate specificity of M. smegmatis PNP (purified as described above) was studied. Ino was the best substrate for PNP, which also accepted guanosine with 68% of the Ino activity (Table 4). No activity was detected with Ado, 2′-deoxyadenosine, methyl-Ado, 2-fluoroadenosine, MTA, xanthosine, SAH, or the pyrimidine nucleosides. This is consistent with previous reports that mycobacterial PNP is specific for 6-oxopurines (5, 11, 19). Further, the molecular mass of PNP was estimated to be 76 kDa, which is consistent with the description of trimeric M. tuberculosis PNP (5, 8). Taken together, our findings indicated that Ado-PNP, MTAP, and PNP are different enzymes.
DISCUSSION
Ado cleavage to Ade is common in bacteria and parasites. Most bacterial PNPs can cleave both 6-aminopurine and 6-oxopurine nucleosides, but M. tuberculosis PNP is known to accept only 6-oxopurines (5, 11, 19). In this work, we identified two enzyme activities in M. smegmatis that could cleave Ado, and these activities were biochemically determined to be Ado-PNP and MTAP. Either enzyme could account for previous reports of Ado cleavage in M. tuberculosis or M. smegmatis cultures (9, 23). However, because MTAP did not cleave methyl-Ado, only Ado-PNP could be responsible for the methyl-Ado cleavage observed in cell cultures. The Km app values for Ade and ribose-1-phosphate with M. smegmatis Ado-PNP were determined to be 43 and 70 μM, respectively, which are similar to the Km values for Ade and ribose-1-phosphate with Escherichia coli PNP (17) and B. cereus AdoP (30). Our kinetic studies showed that Ado-PNP has a lower Km app for Ade than Ado, suggesting that in vitro, the formation of Ado from Ade is favored. However, as with the other phosphorylases, the equilibrium in vivo is predicted to be in favor of Ado cleavage due to the rapid utilization of adenine and ribose-1-phosphate by other enzymes.
According to Bzowska et al., PNPs can be classified into three groups based on their substrate specificities and quaternary structures: low-molecular-mass homotrimers, high-molecular-mass homohexamers, and others (8). Similar to Bzowska's classification, Pugmire and Ealick have grouped PNPs based on their quaternary structures, and attribute trimeric PNPs to mammals and hexameric PNPs to bacteria, with a few exceptions (26). The authors also mention that some bacteria, such as E. coli, express both a trimeric PNP and a hexameric PNP. In E. coli, hexameric PNP (encoded by deoD) cleaves Ado, Ino, and guanosine (8, 17, 26), whereas the trimeric PNP could cleave all purine nucleosides except Ado and 2′-deoxyadenosine (10, 28). This trimeric PNP is often referred to as xanthosine phosphorylase. Similar to E. coli, B. subtilis and B. cereus also express two nucleoside phosphorylases, namely, inosine-guanosine phosphorylase and AdoP (16, 29, 30). Sgarrella et al. described an Ado-specific phosphorylase (AdoP) in B. cereus and have shown that AdoP could cleave Ado and 2′-deoxyadenosine (30). Ino could also be cleaved by this enzyme with 1.5% of the relative activity of Ado, and no activity was observed with guanosine, xanthosine, or the pyrimidine nucleosides. Utagawa et al., have shown that Enterobacter aerogenes has a PNPase activity that is responsible for the cleavage of Ino at 34% of the relative cleavage of Ado (33). Even though the Ado cleavage enzyme described in our study had a preference for Ado, the enzyme also cleaved Ino and guanosine, and therefore, the enzyme is not AdoP but rather a PNP (purine nucleoside: orthophosphate ribosyltransferase, EC 2.4.2.1). Accordingly, our work suggests that M. smegmatis possesses two PNPs: the trimeric PNP (MSMEG_1701), which is an inosine-guanosine phosphorylase, and Ado-PNP, which is similar in substrate specificity to hexameric E. coli PNP.
Analysis of the amino acid sequence of Ado-PNP could determine the similarity of Ado-PNP to the trimeric PNP, as well as provide the basis for a phylogenetic relationship between the two PNPs. The low expression of Ado-PNP in mycobacteria was a significant hurdle in this work, and unfortunately, we have not yet been able to identify the gene that encodes this enzyme. The purification scheme used in this work yielded an Ado-PNP sample that could be biochemically analyzed because it was free of other purine enzymes. However, the protein concentration of our purified preparation was very low, and we were not able to identify a single band in an SDS-PAGE gel stained with either Coomassie or silver stain reagent. Therefore, more work is needed to identify the gene responsible for this activity. A number of genes that could cleave purine and pyrimidine nucleosides have been identified in the M. tuberculosis genome by sequence homology. Our Ado-PNP substrate specificity results indicated that Ado-PNP was not encoded by PNP, MTAP, or MTAN. Although the enzymes expressed by Rv0535(MTAP) and Rv0091(MTAN) are only tentatively identified, we have cloned and expressed these two genes and have shown that they do express MTAP and MTAN, respectively (unpublished results). Rv3393 has been identified as a probable nucleoside hydrolase (IunH) by sequence homology. We have also cloned and expressed this gene and have shown that it is able to hydrolytically cleave uridine, but not Ado (unpublished results). Finally, Rv2293c has been annotated as encoding a conserved hypothetical protein that participates in the nucleoside metabolic process and could be membrane bound. The PHYRE server was used to generate the predicted secondary structure of Rv2293c, which was then compared to a library of known protein structures (18). While the sequence identity was low, the PHYRE server nonetheless predicted Rv2293c to be a member of either the purine and uridine phosphorylase or the hydrolase superfamily. Although we have attempted to clone and express the enzyme, we have not yet been successful.
The importance of expressing two PNPs in mycobacteria is not known. Mycobacteria could express Ado-PNP, even in small amounts, to supplement PNP, MTAP, and MTAN activities to maximize purine salvage and complement de novo purine synthesis. In the current experiments, PNP activity was much greater than Ado-PNP activity. In our previous studies, we have noted variable expression of Ado cleavage in mycobacterial cell cultures (9, 23), which suggests that Ado-PNP expression may be under the control of an unknown variable. Ado-PNP could be important in sensing and regulating Ado and Ade concentrations, which in turn could affect the expression of the other enzymes shown in Fig. 1. Ado-PNP could be expressed in mycobacteria in response to exogenous Ado from macrophages or to salvage the ribose moiety as a carbon source.
Our work suggests that MTAP could play a role in the metabolism of Ado in M. smegmatis. Although MSMEG_0990 and Rv0535 have been annotated as probable MTAPs in M. smegmatis and M. tuberculosis, respectively, the current work describes the first characterization of a bacterial MTAP. Pseudomonas isolates were thought to possess MTAP (1); however, recent work in P. aeruginosa showed that the enzyme was actually a methylthioinosine phosphorylase instead of MTAP (15). Our preliminary characterization of M. smegmatis MTAP indicated that mycobacterial MTAP may be different from human MTAP. Unlike human MTAP, which is a trimer, M. smegmatis MTAP may be a dimer. Similar to the human enzyme, our kinetic data indicated that MTA was a much better substrate than Ado. However, M. smegmatis MTAP had more than a 2-fold-lower Km with Ado than human MTAP, suggesting that Ado was a better substrate for M. smegmatis MTAP than for human MTAP. Therefore, it is possible that mycobacterial MTAP could selectively activate Ado analogs.
A sequence alignment showed more than 92% sequence identity (196 identical amino acid positions) between MTAPs from M. smegmatis and M. tuberculosis, which suggests that the two mycobacterial MTAPs likely share similar properties. In contrast, there was less than 36% sequence identity (93 identical amino acid positions) between M. tuberculosis and human MTAPs, which predicts that mycobacterial and human MTAPs may differ in substrate specificity and other properties. We have cloned and expressed the Rv0535 gene, which is predicted to express a probable MTAP in M. tuberculosis, and have shown that it is indeed MTAP (unpublished results).
Purine metabolism is an attractive target for TB drug discovery, since a purine-based drug would provide a novel mechanism of action, and therefore, MDR and XDR M. tuberculosis strains would likely be sensitive to Ado analogs. Further, Barrow et al., have shown that methyl-Ado is toxic to latent mycobacteria (4), indicating that an Ado analog could work against both active and latent TB infections. Therefore, our discovery of two adenosine-cleaving activities in mycobacteria could contribute to an anti-TB drug development effort. Since human cells do not express Ado-PNP, these enzyme activities in mycobacteria could be exploited to selectively activate a nontoxic Ado or MTA analog to a toxic product in mycobacterial cells. The differences in substrate specificity between human and parasitic MTAPs have been investigated for antiparasitic drug development (12), and these studies have shown 2′-deoxyadenosine, 3′-deoxyadenosine, and 2′,3′-dideoxyadenosine are poor substrates for human MTAP while Trypanosoma brucei brucei MTAP can cleave these substrates (14). Similarly, a complete structure-activity relationship analysis with these two enzymes could reveal differences in substrate preferences that could be exploited for antituberculosis drug development.
ACKNOWLEDGMENTS
This work was supported by a grant (AI43241) from the National Institutes of Health. Operation of the Targeted Metabolomics and Proteomics Laboratory is supported by grants from the National Center for Complementary and Alternative Medicine and the NIH Office of Dietary Supplements to the Purdue-UAB Botanicals Center for Age-Related Disease (P50 AT00477; C. Weaver, principal investigator [PI]); from the National Cancer Institute to the UAB Center for Nutrient-Gene Interaction (U54 CA100949; S. Barnes, PI); from the National Institute of Digestive, Diabetes and Kidney Disease to the UAB O'Brien Acute Kidney Injury Center (P30 DK079337; A. Agarwal, PI) and the UAB Diabetes Research and Training Center (P60 DK079626; T. Garvey, PI); from the National Institute of Arthritis and Musculoskeletal Disease to the UAB Skin Disease Research Center (P30 AR050948; C. Elmets, PI); and from the UAB Center for Free Radical Biology and the UAB Lung Health Center. Support for the mass spectrometers used in the study came from NCRR Shared Instrumentation grants S10 RR19231 and RR027822.
We thank Stephen Barnes and Mahmoud el Kouni, Department of Pharmacology and Toxicology, University of Alabama at Birmingham, for valuable advice, and Paula Allan, Mary Beth Minyard, and John Anderson, Southern Research Institute, for technical assistance.
Footnotes
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Albers E. 2009. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life 61:1132–1142 [DOI] [PubMed] [Google Scholar]
- 2. Appleby T. C., Erion M. D., Ealick S. E. 1999. The structure of human 5′-deoxy-5′-methylthioadenosine phosphorylase at 1.7 A resolution provides insights into substrate binding and catalysis. Structure 7:629–641 [DOI] [PubMed] [Google Scholar]
- 3. Appleby T. C., Mathews I. I., Porcelli M., Cacciapuoti G., Ealick S. E. 2001. Three-dimensional structure of a hyperthermophilic 5′-deoxy-5′-methylthioadenosine phosphorylase from Sulfolobus solfataricus. J. Biol. Chem. 276:39232–39242 [DOI] [PubMed] [Google Scholar]
- 4. Barrow E. W., et al. 2003. Antimycobacterial activity of 2-methyl-adenosine. J. Antimicrob. Chemother. 52:801–808 [DOI] [PubMed] [Google Scholar]
- 5. Basso L. A., et al. 2001. Purine nucleoside phosphorylase from Mycobacterium tuberculosis. Analysis of inhibition by a transition-state analogue and dissection by parts. Biochemistry 40:8196–8203 [DOI] [PubMed] [Google Scholar]
- 6. Bennett L. L., et al. 1993. Purine nucleoside phosphorylase inhibitors: biochemical and pharmacological studies with 9-benzyl-9-deazaguanine and related compounds. J. Pharmacol. Exp. Ther. 266:707–714 [PubMed] [Google Scholar]
- 7. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 8. Bzowska A., Kulikowska E., Shugar D. 2000. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Ther. 88:349–425 [DOI] [PubMed] [Google Scholar]
- 9. Chen C.-K., et al. 2002. The metabolism of 2-methyladenosine in Mycobacterium smegmatis. Microbiology 148:289–295 [DOI] [PubMed] [Google Scholar]
- 10. Dandanell G., Szczepanowski R., Kierdaszuk B., Shugar D., Bochtler M. 2005. Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene J. Mol. Biology 348:113–125 [DOI] [PubMed] [Google Scholar]
- 11. Ducati R. G., Santos D. S., Basso L. A. 2009. Substrate specificity and kinetic mechanism of purine nucleoside phosphorylase from Mycobacterium tuberculosis. Arch. Biochem. Biophys. 486:155–164 [DOI] [PubMed] [Google Scholar]
- 12. el Kouni M. H. 2003. Potential chemotherapeutic targets in the purine metabolism of parasites. Pharmacol. Therapeutics 99:283–309 [DOI] [PubMed] [Google Scholar]
- 13. Fabianowska-Majewska K., Duley J., Fairbanks L., Simmonds A., Wasiak T. 1994. Substrate specificity of methylthioadenosine phosphorylase from human liver. Acta Biochim. Pol. 41:391–395 [PubMed] [Google Scholar]
- 14. Ghoda L. Y., et al. 1988. Substrate specificities of 5′-deoxy-5′-methylthioadenosine phosphorylase from Trypanosoma brucei brucei and mammalian cells. Mol. Biochem. Parasitol. 27:109–118 [DOI] [PubMed] [Google Scholar]
- 15. Guan R., Ho M.-C., Almo S. C., Schramm V. L. 2011. Methylthioinosine phosphorylase from Pseudomonas aeruginosa. Structure and annotation of a novel enzyme in quorum sensing. Biochemistry 50:1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen K. F. 1978. Two purine nucleoside phosphorylases in Bacillus subtilis. Purification and some properties of the adenosine-specific phosphorylase. Biochim. Biophys. Acta 525:346–356 [DOI] [PubMed] [Google Scholar]
- 17. Jensen K. F., Nygaard P. 1975. Purine nucleoside phosphorylase from Escherichia coli and Salmonella typhimurium. Eur. J. Biochem. 51:253–265 [DOI] [PubMed] [Google Scholar]
- 18. Kelley L. A., Sternberg M. J. E. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 19. Lewandowicz A., et al. 2003. Over-the-barrier transition state analogues and crystal structure with Mycobacterium tuberculosis purine nucleoside phosphorylase. Biochemistry 42:6057–6066 [DOI] [PubMed] [Google Scholar]
- 20. Long M. C., Escuyer V., Parker W. B. 2003. Identification and characterization of a unique adenosine kinase from Mycobacterium tuberculosis. J. Bacteriol. 185:6548–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller R. L., Sabourin C. L., Krenitsky T. A. 1987. Trypanosoma cruzi adenine nucleoside phosphorylase. Purification and substrate specificity. Biochem. Pharmacol. 36:553–560 [DOI] [PubMed] [Google Scholar]
- 22. Parker W. B., Allan P. W., Niwas S., Montgomery J. A., Bennett L. L. 1994. Effect of 9-benzyl-9-deazaguanine, a potent inhibitor of purine nucleoside phosphorylase, on the cytotoxicity and metabolism of 6-thio-2′-deoxyguanosine. Cancer Res. 54:1742–1745 [PubMed] [Google Scholar]
- 23. Parker W. B., et al. 2004. Metabolism of 2-methyladenosine in Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 84:327–336 [DOI] [PubMed] [Google Scholar]
- 24. Parker W. B., Long M. C. 2007. Purine metabolism in Mycobacterium tuberculosis as a target for drug development. Curr. Pharm. Des. 13:599–608 [DOI] [PubMed] [Google Scholar]
- 25. Pressey J. G., et al. 2011. 2D-difference gel electrophoretic proteomic analysis of a cell culture model of alveolar rhabdomyosarcoma. J. Proteome Res. 10:624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pugmire M. J., Ealick S. E. 2002. Structural analyses reveal two distinct families of nucleoside phosphorylases. Biochem. J. 361:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Savarese T. M., Crabtree G. W., Parks R. E., Jr 1981. 5′-Methylthioadenosine phosphorylase. I. Substrate activity of 5′-deoxyadenosine with the enzyme from Sarcoma 180 cells. Biochem. Pharmacol. 30:189–199 [DOI] [PubMed] [Google Scholar]
- 28. Seeger C., Poulsen C., Dandanell G. 1995. Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J. Bacteriol. 177:5506–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Senesi S. 1976. A specific adenosine phosphorylase, distinct from purine nucleoside phosphorylase. FEBS Lett. 64:353–357 [DOI] [PubMed] [Google Scholar]
- 30. Sgarrella F., et al. 2007. Characterization of the adenine nucleoside specific phosphorylase of Bacillus cereus. Biochim. Biophys. Acta 1770:1498–1505 [DOI] [PubMed] [Google Scholar]
- 31. Toorchen D., Miller R. L. 1991. Purification and characterization of 5′-deoxy-5′-methylthioadenosine (MTA) phosphorylase from human liver. Biochem. Pharmacol. 41:2023–2030 [DOI] [PubMed] [Google Scholar]
- 32. UniProt Consortium 2011. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39:D214–D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Utagawa T., et al. 1985. Properties of purine nucleoside phosphorylase from Enterobacter aerogenes. Agric. Biochem. Chem. 49:3239–3246 [Google Scholar]
- 34. World Health Organization 2009. Global tuberculosis control: epidemiology, strategy, financing. WHO report 2009. World Health Organization, Geneva, Switzerland [Google Scholar]
- 35. World Health Organization 2010. Tuberculosis MDR-TB and XDR-TB 2010 report. World Health Organization, Geneva, Switzerland [Google Scholar]