Abstract
Although toxins A and B are known to be important contributors to the acute phase of Clostridium difficile infection, the role of colonization and adherence to host tissues in the overall pathogenesis of these organisms remains unclear. Consequently, we used the recently introduced intron-based ClosTron gene interruption system to eliminate the expression of two reported C. difficile colonization factors, the major flagellar structural subunit (FliC) and the flagellar cap protein (FliD), to gain greater insight into how flagella and motility contribute to C. difficile's pathogenic strategy. The results demonstrate that interrupting either the fliC or the fliD gene results in a complete loss of flagella, as well as motility, in C. difficile. However, both the fliC and fliD mutant strains adhered better than the wild-type 630Δerm strain to human intestine-derived Caco-2 cells, suggesting that flagella and motility do not contribute to, or may even interfere with, C. difficile adherence to epithelial cell surfaces in vitro. Moreover, we found that the mutant strains were more virulent in hamsters, indicating either that flagella are unnecessary for virulence or that repression of motility may be a pathogenic strategy employed by C. difficile in hamsters.
INTRODUCTION
Clostridium difficile is an anaerobic, spore-forming bacterium and the etiologic agent of C. difficile infection (CDI). C. difficile has become the leading cause of infectious diarrhea in hospitalized patients and represents a significant burden to health care systems worldwide (36). Although patients receiving broad-spectrum antibiotics in hospitals are at the highest risk of developing CDI, community-acquired disease and infection in traditionally low-risk populations are increasing (2, 26). This may be attributed to the appearance, in the past decade, of a hypervirulent C. difficile strain known as NAP1/B1/027. This strain is associated with outbreaks worldwide, increased disease severity, and increasing numbers of treatment failures (2, 28, 39).
It is widely accepted that CDI is predominantly a toxin-mediated disease. Two large exotoxins, toxin A (TcdA) and toxin B (TcdB), are the main virulence factors produced by the organism. Both toxins disrupt the actin cytoskeleton of host intestinal epithelial cells, causing a decrease in intestinal barrier function which subsequently leads to fluid accumulation, inflammation, and severe intestinal damage (37). Although the relative importance of each toxin in the pathogenic process has been the subject of recent conflicting reports, it appears that TcdB plays the predominant role (6, 16, 19).
In addition to the toxins, bacterial adherence has been proposed to play a supporting role in the C. difficile pathogenic strategy. Although the contribution of adherence to host tissues is poorly understood, several lines of evidence suggest that colonization may be an important contributor to CDI. For example, patients who are asymptomatically colonized with C. difficile display a decreased risk of acquiring the disease (30). In addition, Sambol et al. showed that precolonizing hamsters with a nontoxigenic strain of C. difficile protected the animals from CDI when they were subsequently challenged with a toxigenic strain (29).
Colonization of host tissues by C. difficile may require a number of cell wall-associated proteins. Of the various putative C. difficile colonization factors described to date, the high- and low-molecular-weight surface layer proteins (SLPs) remain the best characterized. Several studies suggest that these proteins mediate adherence to host cells during infection (5, 15). In addition to the SLPs, 28 other cell surface proteins show homology to that encoded by slpA. These proteins, termed cell wall proteins (CWPs), all contain three copies of the PF01422 cell_wall_binding_2 domain, a motif characteristic of the SLPs that is involved in anchoring the protein to the outer surface of the bacterium (9). Moreover, both Cwp66 and Cwp84 have also been well studied and implicated in host cell adherence and degradation of extracellular matrices (14, 38).
In addition to the CWPs, two C. difficile flagellar proteins, FliC, the flagellin structural monomer, and FliD, the flagellar cap protein, have been reported to be involved in the attachment of the organism to host cells and the intestinal mucus layer (33, 34). Studies of recuperating CDI patients have shown that serum antibody responses against both FliC and FliD are generated, implicating them as important virulence factors during the course of CDI (24, 25). Since flagella have been implicated in the colonization strategy of a number of enterovirulent organisms, we sought to determine the importance of FliC and FliD in C. difficile pathogenesis. To achieve this, we employed the intron II-based ClosTron system (11) to generate fliC and fliD gene interruption mutants, as well as fliC- and fliD-complemented strains using the p84151 modular plasmid (12). We then determined the host cell adherence phenotypes of these strains in vitro in addition to their virulence potential in hamsters.
MATERIALS AND METHODS
C. difficile growth conditions and spore preparation.
C. difficile strains were routinely grown on brain heart infusion (BHI) agar containing 5% sheep blood, 5 mg hemin, and 1 mg vitamin K per liter (Y agar) for 48 h in a Bactron III Anaerobic Chamber (Shel Laboratories) at 37°C. Liquid cultures were produced by inoculating single colonies of C. difficile into BHI medium and incubating them overnight anaerobically at 37°C. C. difficile mutants were grown in standard growth medium, while the complemented C. difficile strains were grown in standard growth medium containing 15 μg/ml thiamphenicol.
C. difficile spores were prepared by mixing 100% ethanol 1:1 with 200 ml 5-day-old anaerobic liquid cultures of C. difficile. After 1 h of incubation aerobically at room temperature, spores were harvested by centrifugation at 6,000 × g for 6 min. Following two washes of the spore-containing pellet with 10 ml sterile sodium/potassium phosphate-buffered (pH 7.2) physiological saline (PBS), the final pellet was suspended in 10 ml Dulbecco's modified Eagle medium. Spores were quantified by performing serial dilutions and spread plating in triplicate on cycloserine-cefoxitin fructose agar (CCFA). Spores were stored for no more than 1 week at −80°C prior to use.
Creation of C. difficile fliC and fliD gene interruption mutants.
C. difficile mutants were produced using the ClosTron system (11). Retargeted pMTL007C-E2 vectors for the fliC mutant Cdi-fliC-260a and the fliD mutant Cdi-fliD-322a were purchased from DNA2.0 Inc. The retargeted vectors were transferred from Escherichia coli CA434 to wild-type 630Δerm C. difficile by conjugation using CCFA plates containing 15 μg/ml thiamphenicol (27). Once colonies appeared on the plates (48 to 72 h), they were restreaked onto CCFA containing 2.5 μg/ml erythromycin to select for bacteria in which the intron had been inserted. Insertions were confirmed by PCR using the EBS universal primer (5′-CGAAATTAGAAACTTGCGTTCAGTAAAC) and a reverse primer (fliC, 5′-CCCTGCTCGAGCTATCCTAATAATTG; fliD, 5′-CCCTGCTCGAGTTAATTACCTTGTGCTTGTG) for each gene.
Creation of C. difficile fliC- and fliD-complemented strains.
The intact fliC and fliD genes, including their respective promoters, were cloned into the p80000 shuttle vector p84151 (12). Briefly, the fliC and fliD genes were amplified using C. difficile 630Δerm genomic DNA as the template for the reaction. The primers used were FliC forward (5′-CCCTGGCGGCCGCAACTTTATGATAGTATGGAGC), FliC reverse (5′-CCCTGCTCGAGCTATCCTAATAATTG), FliD forward (5′-CCCTGGCGGCCGCTAATATATCTAAAGTTGCAC), and FliD reverse (5′-CCCTGCTCGAGTTAATTACCTTGTGCTTGTG). For the fliC complement, the NotI- and XhoI-digested PCR product was ligated into NotI/XhoI-digested p84151. For the fliD complement, the fliD gene was cloned into pCR2.1-TOPO (Invitrogen) according to the manufacturer's protocol. The TOPO-fliD clone was subsequently digested with NotI and XhoI, and the digested fliD gene was extracted from a 1% agarose gel using the Qiagen gel purification kit. This was then ligated into the NotI/XhoI-digested p84151 vector. The fliC and fliD clones were subsequently transformed into CA434 and conjugated into the appropriate mutant as described above. Positive transconjugants were selected for on CCFA plates containing 15 μg/ml thiamphenicol. Positive complements were screened using PCR and phenotypic assays.
Swim agar assays.
Overnight cultures of each of the C. difficile strains were stabbed into 0.175% BHI agar tubes. The swim agar tubes were incubated overnight anaerobically at 37°C, and then swimming ability was visually assessed.
Western immunoblotting.
Overnight cultures of the wild-type, mutant, and complemented strains were normalized by optical density and sedimented by centrifugation at 5,000 × g for 3 min. The cells were subsequently suspended in sample buffer and analyzed by 12.5% SDS-PAGE. The separated proteins were transferred to polyvinylidene difluoride membranes in accordance with standard techniques. The membranes were blocked overnight in 5% skim milk powder dissolved in Tris-buffered saline containing 0.05% Tween 20 (TBSTM) at 4°C. The membranes were then probed with FliC- or FliD-specific IgY antibodies (ZymeFast Inc.) diluted 1:100,000 or 1:10,000 in 5% TBSTM, respectively. Subsequently, a 1:10,000 dilution of anti-chicken IgY conjugated to horseradish peroxidase (Sigma-Aldrich) in 5% TBSTM was added to the membranes. The proteins were visualized using the SuperSignal West Dura extended-duration substrate (Thermo Scientific) according to the manufacturer's instructions and imaged using a Kodak Image Station 2000MM.
Transmission electron microscopy.
Single C. difficile colonies from Y agar plates were suspended in 50 μl of monoethanolamine buffer (pH 10.0). Ten microliters of this suspension was then applied to carbon-coated copper grids (Electron Microscopy Sciences) and allowed to incubate for 2 min at room temperature. Excess liquid was subsequently wicked away using filter paper, and the bacteria were stained by adding 10 μl of 1% phosphotungstic acid (pH 7.0) to the grids for 10 s at room temperature. Excess stain was gently wicked away using filter paper, and the dried grids were examined using a Hitachi H-7650 transmission electron microscope.
Caco-2 cell adherence assay.
The adherence of the C. difficile strains to differentiated human intestine-derived Caco-2 cells was assessed using our previously described protocol (8). Briefly, Caco-2 cells were differentiated in Transwell inserts using the BD Biocoat Intestinal Epithelial Differentiation Environment (BD Biosciences). The cells were subsequently transferred to the anaerobic chamber, and the medium was replaced with overnight prereduced differentiation medium. Bacteria (1 × 106) were added to the apical compartment of the Transwell inserts, and the plates were incubated for 3 h anaerobically at 37°C. The Caco-2 cells were then removed from the chamber and washed 5 times with sterile PBS prior to the extraction of whole genomic DNA from the inserts using the Qiagen DNeasy blood and tissue kit following the manufacturer's protocol for Gram-positive bacteria. Real-time quantitative PCR targeting the C. difficile housekeeping gene for triose phosphate isomerase was used to determine the number of adherent organisms using a standard curve of known concentrations of C. difficile genomic DNA (8). Experiments were performed three times in triplicate for each strain.
Hamster CDI model.
The hamster model of infection used for these experiments was based on the protocol previously described by Lyras et al. (19). The hamster CDI protocol used was reviewed and approved by the University of Calgary Animal Care Committee (protocol M08122). Briefly, 80- to 100-g male Syrian hamsters received 30 mg/kg clindamycin phosphate by gavage on day 0. On day 5, the hamsters were infected by gavage with 103 C. difficile spores. Hamsters were monitored every 4 h, day and night, postinfection and immediately euthanized by CO2 asphyxia as soon as signs of CDI (perianal staining and unresponsiveness) appeared.
To assess the colonization potential of each of the C. difficile mutant strains, cecal contents were collected from euthanized hamsters and weighed. PBS was added to the samples up to 1 ml, and the amount used was recorded. Suspensions were serially diluted in PBS and spread plated on CCFA to determine the number of bacteria per gram of cecal contents. Ceca were subsequently removed from the hamsters and thoroughly washed with PBS. The washed ceca were weighed, suspended to 100 mg/ml in PBS, and homogenized using a Polytron benchtop homogenizer. Suspensions were serially diluted in PBS and spread plated on CCFA to assess the number adherent bacteria per ml of homogenized sample.
Growth curves and Vero cytotoxicity assays.
Overnight cultures of C. difficile were diluted 1:500 into overnight prereduced BHI. Samples were taken from the cultures at various intervals, and their optical density at 600 nm was recorded. At the 24-h time point, bacterial cells were harvested by centrifugation and the resulting supernatant solution was filter sterilized and serially diluted 2-fold in minimal essential medium (MEM). Twenty microliters of these dilutions was subsequently applied to confluent Vero cells (between passages 130 and 150) seeded into 96-well tissue culture plates in MEM containing 10% fetal bovine serum. After 48 h of incubation at 37°C with 5% CO2, the cells were fixed with methanol and Giemsa stained in accordance with standard protocols. After the cells were solubilized with 1% SDS, the results were recorded using a SpectraMax model 340 microtiter plate reader (Molecular Devices) set to a wavelength of 630 nm.
Statistical analyses.
The statistical significance of differences between the numbers of adherent or colonizing bacteria was determined by the Student t test. Statistical analysis of the hamster survival data was performed using the log rank test.
RESULTS
C. difficile fliC and fliD gene interruption mutants lack flagella and are nonmotile.
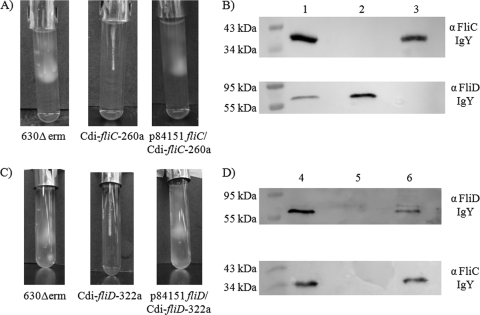
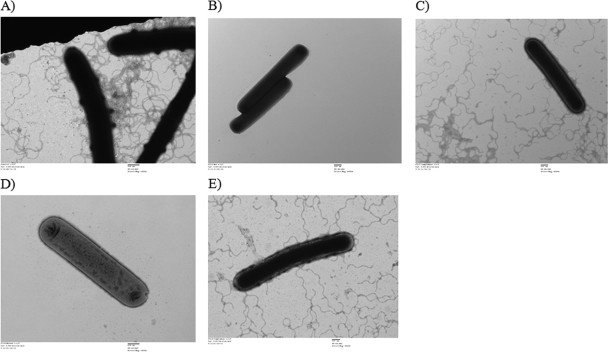
Whereas the wild-type 630Δerm strain and the fliC- and fliD-complemented C. difficile strains (p84151 fliC/Cdi-fliC-260a and p84151 fliD/Cdi-fliD-322a) displayed normal motility, the swim agar assay revealed that the fliC and fliD group II intron insertion mutants (Cdi-fliC-260a and Cdi-fliD-322a) were nonmotile (Fig. 1). As expected, the Western immunoblotting technique revealed that the fliC mutant did not produce detectable amounts of FliC whereas the wild-type 630Δerm and fliC-complemented strains did (Fig. 1B, upper panel). Transmission electron microscopy (Fig. 2) also revealed that the fliC and fliD mutants did not produce any visible flagella, whereas morphologically normal-looking flagella were expressed by the wild-type 630Δerm strain and the fliC- and fliD-complemented strains.
Fig. 1.
Characterization of the wild-type C. difficile 630Δerm strain, fliC and fliD ClosTron interruption mutants, and their respective complemented strains. (A and C) Swim agar assays revealing the motility phenotypes of the wild-type 630Δerm, Cdi-fliC-260a, p84151 fliC/Cdi-fliC-260a, Cdi-fliD-322a, and p84151 fliD/Cdi-fliD-322a C. difficile strains. (B and D) Western immunoblot analyses of the wild-type 630Δerm (lanes 1 and 4), Cdi-fliC-260a (lane 2), p84151 fliC/Cdi-fliC-260a (lane 3), Cdi-fliD-322a (lane 5), and p84151 fliD/Cdi-fliD-322a (lane 6) strains probed with FliC- and FliD-specific IgY.
Fig. 2.
Transmission electron micrographs revealing the lack of flagellum production in C. difficile Cdi-fliC-260a (B) and Cdi-fliD-322a (D) but not in wild-type 630Δerm (A), p84151fliC/Cdi-fliC-260a (C), or p84151 fliD/Cdi-fliD-322a (E).
Although the wild-type 630Δerm strain and the fliC mutant strain produced FliD, the fliC-complemented strain did not (Fig. 1B, lower panel), suggesting that there was a reduction in fliD gene expression when FliC is expressed in trans off a plasmid rather than in cis from the chromosomal gene. Moreover, this observation, in addition to the results of the swim agar experiments (Fig. 1) and the transmission electron microscopy observations presented in Fig. 2, indicates that functional flagella can be produced by C. difficile in the absence of detectable FliD expression. Moreover, Western immunoblot analysis of the fliD mutant revealed that it was incapable of producing either the FliD or the FliC protein, while the wild-type 630Δerm and fliD-complemented strains produced both proteins (Fig. 1D). This suggests that FliD expression is required for the fliC gene to be expressed off the chromosomal gene. Together, these cumulative observations indicate that regulation of expression of the fliC and fliD genes is linked and that eliminating protein expression by either of these genes leads to a loss of both flagellum production and motility.
Absence of FliC and FliD expression results in increased C. difficile adherence to intestinal Caco-2 tissue culture cells.
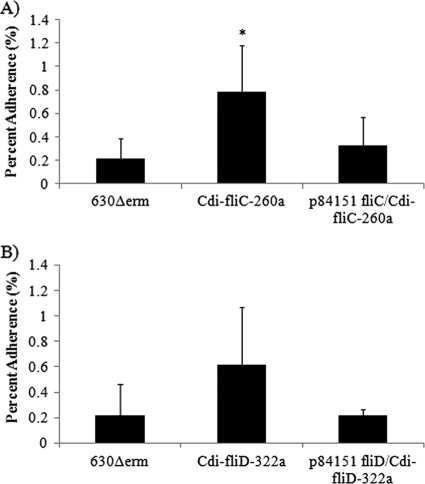
The data presented in Fig. 3 indicate that the fliC mutant strain adhered significantly better (P < 0.05, Student t test) to differentiated Caco-2 cells than did either the wild-type 630Δerm or the fliC-complemented C. difficile strain. The fliD mutant also appeared to adhere better to Caco-2 cells than did the wild-type or the corresponding complemented strain, although in this case, the difference did not reach statistical significance (P = 0.051, Student t test). These results indicate that expression of FliC and FliD and motility are counterproductive to C. difficile adherence to human intestine-derived Caco-2 cells.
Fig. 3.
Adherence of wild-type 630Δerm, Cdi-fliC-260a, and p84151 fliC/Cdi-fliC-260a C. difficile (A) and wild-type 630Δerm, Cdi-fliD-322a, and p84151 fliD/Cdi-fliD-322a C. difficile (B) to differentiated human intestine-derived Caco-2 cells. The error bars represent standard deviations of the means of triplicate determinations. The asterisk indicates a statistically significant difference (P < 0.05) from the adherence of the wild-type 630Δerm C. difficile strain as determined by the Student t test.
FliC and FliD are not important for virulence or intestinal colonization in hamsters.
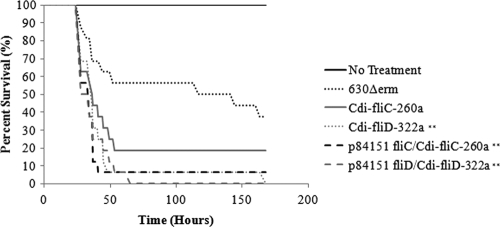
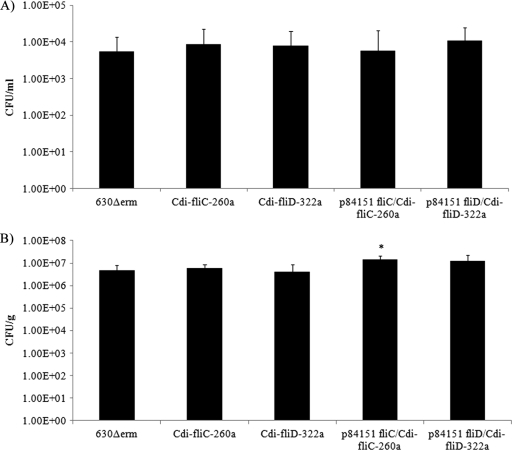
The cumulative results (Fig. 4) of 2 independent hamster CDI experiments revealed that although more animals succumbed to infection with the fliC mutant, there was no significant difference in its virulence in hamsters from that of the wild-type 630Δerm C. difficile strain. In contrast, the data presented in Fig. 4 reveal that the fliC-complemented, fliD mutant, and fliD-complemented strains displayed significantly (P < 0.01, log rank test) greater virulence in hamsters than did the wild-type 630Δerm strain. We therefore performed serial dilution plating of the cecal contents or the washed homogenized cecums from euthanized hamsters to determine if the intron insertions in the fliC and fliD genes affected the number of bacteria at this primary site of infection. The results (Fig. 5) revealed no significant differences in the number of fliC or fliD mutant bacteria or their respective complemented strains either associated with the cecal contents or attached to the cecal epithelium relative to the cecal contents or epithelia recovered from hamsters infected with the wild-type 630Δerm strain. These results suggest that neither FliC nor FliD expression is required for cecal colonization of hamsters.
Fig. 4.
Virulence of the fliC and fliD interruption mutants in the hamster CDI model. Cumulative results from 2 independent experiments plotting the survival of clindamycin-treated hamsters infected with 103 C. difficile spores. Statistically significant differences (**) from hamsters infected with the wild-type 630Δerm strain were determined using the log rank test (P < 0.01).
Fig. 5.
Numbers of C. difficile CFU recovered from the homogenized ceca (A) and the cecal contents (B) of infected hamsters. The error bars represent the standard deviations of the means of at least 3 independent determinations. Statistically significant differences (P < 0.05) from the results obtained with ceca from hamsters infected with wild-type 630Δerm were determined using the Student t test.
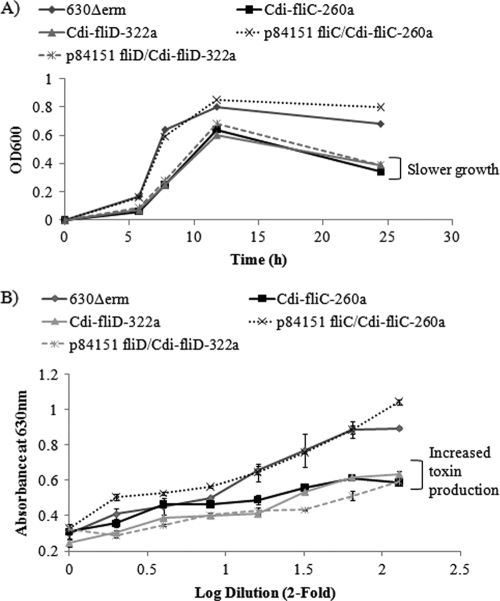
Flagellar mutant strains produced greater amounts of toxin in vitro.
The bacterial growth curves presented in Fig. 6 reveal that the fliC and fliD mutant strains and the fliD-complemented strain grew more slowly than both the wild-type 630Δerm and fliC-complemented strains. Toxin production by C. difficile is known to occur in the late log or early stationary phase of growth (13). When samples from cultures in early stationary phase (24 h) were assessed for cytotoxicity on Vero cells, we observed a correlation between slower bacterial growth rates and increased toxin titers (Fig. 6B). These results suggest that the increased virulence of the mutant C. difficile strains in hamsters may have resulted from their ability to express more toxins earlier in the infection process than the wild-type 630Δerm strain.
Fig. 6.
(A) Growth kinetics of the C. difficile wild-type 630Δerm strain, the fliC and fliD interruption mutants, and the complemented strains. OD600, optical density at 600 nm. (B) Vero cytotoxicity in filter-sterilized supernatants prepared from 24 cultures of each of these strains.
DISCUSSION
Flagella are important for the pathogenic strategy of many bacteria. They promote invasiveness in organisms such as Proteus mirabilis and Burkholderia pseudomallei (7, 22) and are involved in regulating virulence gene expression in Vibrio cholerae (10, 32). Flagellum-mediated motility has been shown to promote biofilm formation in several bacterial species, including Pseudomonas aeruginosa and Listeria monocytogenes (4, 17). Further, in organisms such as Helicobacter pylori and Campylobacter jejuni, flagella mediate adherence of the organisms and colonization of host tissues (23, 40). Flagellated strains of C. difficile are known to adhere significantly better to the mouse cecum in vivo, and this is thought to be mediated by mucosal association of two of the C. difficile flagellar proteins, FliC and FliD (33). Nevertheless, the role of these two proteins in pathogenesis has yet to be confirmed.
With the recent introduction of the ClosTron gene interruption system, it is now possible to use a mutational approach to determining the roles of known or putative virulence factors in C. difficile pathogenesis. Previously, Twine et al. described the production of a fliC mutation in C. difficile; however, this mutant was never evaluated in any biological systems (35). In the present study, we characterized and assayed fliC and fliD gene interruption mutants and their respective fliC- and fliD-complemented strains in both in vitro and in vivo systems. Interruption of either gene led to a loss of both flagellum production and motility.
Further, Western immunoblot analysis of both the mutant and complemented strains not only revealed that only FliC expression was required for the production of functional flagella but also revealed intriguing insights into the regulation of flagellar protein biosynthesis and assembly in C. difficile. First, the FliD protein was expressed in the fliC mutant strain, indicating that cap protein is produced in the presence or absence of FliC (Fig. 1B, lane 2). Second, for FliC to be expressed from the chromosomal fliC gene, fliD gene expression is required (Fig. 1D, lane 5), suggesting that FliD exerts a positive effect on fliC gene expression. Finally, FliD expression from the chromosomal fliD gene is attenuated when FliC is expressed in trans from a plasmid (Fig. 1B, lane 3), revealing that FliC may repress fliD gene expression. In Gram-negative bacteria, flagellum production and regulation of motility involve more than 40 different genes (1, 20, 31). More work is clearly needed to further understand the regulation of flagellar expression and function in C. difficile.
When tested in an in vitro adherence model, both the fliC and fliD mutants demonstrated an increase in adherence to differentiated Caco-2 tissue culture cells. We postulate that the increase in the adherence of the mutant strains might be due to more rapid settling of the bacteria onto Caco-2 cells due to their lack of motility. This suggests that, in vitro, FliC and FliD are not required for and may actually interfere with the adherence of the organisms to host cells. However, FliC and FliD have previously been reported to bind to murine mucus (33), which is absent from the differentiated Caco-2 cell surface. Therefore, the Caco-2 adherence experiments presented here do not exclude the possibility that FliC and FliD have a role in the C. difficile colonization of epithelial cell surfaces which include a mucus layer.
When hamsters were infected with the fliC and fliD gene interruption mutants and their respective complemented strains, more animals succumbed to infection than those infected with the wild-type 630Δerm strain. Of interest, the complemented strains were more virulent than the wild-type 630Δerm strain. However, due to the lack of antibiotic selection, it is possible that the plasmid harboring the complemented genes was lost during the infection of hamsters, thereby allowing these strains to revert to the mutant phenotype in vivo.
Hamsters are exquisitely sensitive to TcdA and TcdB, and injection of the purified forms of these toxins into their ceca is 100% lethal (18). Vero cell cytotoxicity assays conducted using culture supernatants harvested from the fliC and fliD mutants and the complemented strains showed that they produce toxin amounts similar to or greater than those of the wild-type strain in vitro. Assessment of the toxin titers in the hamster cecal contents, however, yielded no observable differences in toxin production (data not shown). Although these data suggest that toxin production may be dependent on flagellar expression, as in other organisms such as Vibrio cholera (10), the observations cannot fully explain the increased virulence of these strains. Moreover, when the metrics of colonization, that is, the presence of bacteria in the cecal contents or associated with the cecal epithelium, were assessed, we detected no differences in the numbers of bacteria upon infection with the wild-type 630Δerm, mutant, or complemented strains, suggesting that these proteins are not essential for the cecal colonization of hamsters.
Together, these results imply that nonmotile C. difficile can reach the site of infection and that FliC and FliD do not appear to be important for establishing infection in hamsters. This is not unprecedented, since nonmotile organisms such as Klebsiella and Shigella are capable of causing infections in other species (3). However, it is possible that C. difficile represses the expression of the fliC and fliD genes, and therefore motility, upon the infection of its host and that this is part of its pathogenic strategy. For example, Yersinia enterocolitica represses flagellum expression in its mammalian host, a mechanism that is thought to allow the organism to evade the host's innate immune system (21). Conversely, artificial expression of flagellin in Y. enterocolitica results in complete attenuation of virulence in a mouse model of infection (21). A similar strategy may be employed by C. difficile once it is inside its mammalian host, and further experiments with hamsters are required to test this hypothesis. Future experiments using less-sensitive hosts, such as mice, to phenotype C. difficile gene interruption mutants may also be important to evaluate the role of C. difficile virulence factors other than its toxins.
ACKNOWLEDGMENTS
This study was funded by the Alberta Ingenuity Centre for Carbohydrate Science. T.C.D. is the recipient of an NSERC Canada Graduate Scholarship.
Strain 630Δerm and the p80000 modular plasmids were kindly provided by Nigel Minton and John Heap.
Footnotes
Published ahead of print on 25 July 2011.
REFERENCES
- 1. Aldridge P., Hughes K. T. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160–165 [DOI] [PubMed] [Google Scholar]
- 2. Ananthakrishnan A. N. 2011. Clostridium difficile infection: epidemiology, risk factors and management. Nat. Rev. Gastroenterol. Hepatol. 8:17–26 [DOI] [PubMed] [Google Scholar]
- 3. Andrade A., Giron J. A., Amhaz J. M. K., Trabulsi L. R., Martinez M. B. 2002. Expression and characterization of flagella in nonmotile enteroinvasive Escherichia coli isolated from diarrheal cases. Infect. Immun. 70:5882–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barken K. B., et al. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331–2343 [DOI] [PubMed] [Google Scholar]
- 5. Calabi E., Calabi F., Phillips A. D., Fairweather N. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70:5770–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carter G. P., Awad M. M., Kelly M. L., Rood J. I., Lyras D. 2011. TcdB or not TcdB: a tale of two Clostridium difficile toxins. Future Microbiol. 6:121–123 [DOI] [PubMed] [Google Scholar]
- 7. Chuaygud T., Tungpradabkul S., Sirisinha S., Chua K. L., Utaisincharoen P. 2008. A role of Burkholderia pseudomallei flagella as a virulent factor. Trans. R. Soc. Trop. Med. Hyg. 102:S140–S144 [DOI] [PubMed] [Google Scholar]
- 8. Dingle T., Mulvey G. L., Armstrong G. D. 2010. A real-time quantitative PCR assay for evaluating Clostridium difficile adherence to differentiated intestinal Caco-2 cells. J. Med. Microbiol. 59:920–924 [DOI] [PubMed] [Google Scholar]
- 9. Emerson J., Fairweather N. 2009. Surface structure of C. difficile and other clostridia: implications for pathogenesis and immunity, p. 157–167 In Bruggemann H., Gottschalk G. (ed.), Clostridia: molecular biology in the post-genomic era. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 10. Gardel C. L., Mekalanos J. J. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heap J. T., Pennington O. J., Cartman S. T., Carter G. P., Minton N. P. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452–464 [DOI] [PubMed] [Google Scholar]
- 12. Heap J. T., Pennington O. J., Cartman S. T., Minton N. P. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79–85 [DOI] [PubMed] [Google Scholar]
- 13. Hundsberger T., et al. 1997. Transcriptional analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur. J. Biochem. 244:735–742 [DOI] [PubMed] [Google Scholar]
- 14. Janoir C., Péchiné S., Grosdidier C., Collignon A. 2007. Cwp84, a surface-associated protein of Clostridium difficile is a cysteine protease with degrading activity on extracellular matrix proteins. J. Bacteriol. 189:7174–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karjalainen T., et al. 2001. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile. Infect. Immun. 69:3442–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuehne S. A., et al. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713 [DOI] [PubMed] [Google Scholar]
- 17. Lemon K. P., Higgins D. E., Kolter R. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189:4418–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Libby J. M., Jortner B. S., Wilkins T. D. 1982. Effects of the two toxins of Clostridium difficile in antibiotic-associated cecitis in hamsters. Infect. Immun. 36:822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lyras D., et al. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCarter L. L. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9:180–186 [DOI] [PubMed] [Google Scholar]
- 21. Minnich S. A., Rohde H. N. 2007. A rationale for repression and/or loss of motility by pathogenic Yersinia in the mammalian host. Adv. Exp. Med. Biol. 603:298–310 [DOI] [PubMed] [Google Scholar]
- 22. Mobley H. L., et al. 1996. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 64:5332–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ottemann K. M., Lowenthal A. C. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70:1984–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Péchiné S., et al. 2005. Immunological properties of surface proteins of Clostridium difficile. J. Med. Microbiol. 54:193–196 [DOI] [PubMed] [Google Scholar]
- 25. Péchiné S., Janoir C., Collignon A. 2005. Variability of C. difficile surface proteins and specific serum antibody response in patients with Clostridium difficile-associated disease. J. Clin. Microbiol. 43:5018–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pituch H. 2009. Clostridium difficile is no longer just a nosocomial infection or an infection in adults. Int. J. Antimicrob. Agents 33:S42–S45 [DOI] [PubMed] [Google Scholar]
- 27. Purdy D., et al. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46:439–452 [DOI] [PubMed] [Google Scholar]
- 28. Rupnik M., Wilcox M. H., Gerding D. N. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 29. Sambol S. P., Merrigan M. M., Tang J. K., Johnson S., Gerding D. N. 2002. Colonization for prevention of Clostridium difficile disease in hamsters. J. Infect. Dis. 186:1781–1789 [DOI] [PubMed] [Google Scholar]
- 30. Shim J., Johnson S., Samore M., Bliss D., Gerding D. 1998. Primary symptomless colonisation of Clostridium difficile and decreased risk of subsequent diarrhea. Lancet 351:633–636 [DOI] [PubMed] [Google Scholar]
- 31. Smith T. G., Hoover T. R. 2009. Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv. Appl. Microbiol. 67:257–295 [DOI] [PubMed] [Google Scholar]
- 32. Syed K. A., et al. 2009. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191:6555–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tasteyre A., Barc M.-C., Collignon A., Boureau H., Karjalainen T. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tasteyre A., et al. 2001. Molecular characterization of fliD gene encoding flagellar cap and its expression among Clostridium difficile isolates from different serogroups. J. Clin. Microbiol. 39:1178–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Twine S. M., Reid C. W., Aubry A., McMullin D. R., Fulton K. M. 2009. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191:7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Viswanathan V. K., Mallozzi M. J., Vedantam G. 2010. Clostridium difficile infection. Gut Microbes 1:234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Voth D. E., Ballard J. D. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Waligora A.-J., et al. 2001. Characterization of a cell surface protein of Clostridium difficile and adhesive properties. Infect. Immun. 69:2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Warny M., et al. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]
- 40. Yao R., et al. 1994. Isolation of motile and nonmotile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883–893 [DOI] [PubMed] [Google Scholar]