Abstract
Mice with the type I interferon (IFN) receptor gene knocked out (IFNAR KO mice) or deficient for alpha/beta IFN (IFN-α/β) signaling clear chlamydial infection earlier than control mice and develop less oviduct pathology. Initiation of host IFN-β transcription during an in vitro chlamydial infection requires interferon regulatory transcription factor 3 (IRF3). The goal of the present study was to characterize the influence of IRF3 on chlamydial genital infection and its relationship to IFN-β expression in the mouse model. IRF3 KO mice were able to resolve infection as well as control mice, overcoming increased chlamydial colonization and tissue burden early during infection. As previously observed for IFNAR KO mice, IRF3 KO mice generated a potent antigen-specific T cell response. However, in contrast to IFNAR KO mice, IRF3 KO mice exhibited unusually severe dilatation and pathology in the uterine horns but normal oviduct pathology after infection. Although IFN-β expression in vivo was dependent on the presence of IRF3 early in infection (before day 4), the IFN-independent function of IRF3 was likely driving this phenotype. Specifically, early during infection, the number of apoptotic cells and the number of inflammatory cells were significantly less in uterine horns from IRF3 KO mice than in those from control mice, despite an increased chlamydial burden. To delineate the effects of IFN-β versus IRF3, neutralizing IFN-β antibody was administered to wild-type (WT) mice during chlamydial infection. IFN-β depletion in WT mice mimicked that in IFNΑR KO mice but not that in IRF3 KO mice with respect to both chlamydial clearance and reduced oviduct pathology. These data suggest that IRF3 has a role in protection from uterine horn pathology that is independent of its function in IFN-β expression.
INTRODUCTION
The obligate intracellular bacterium Chlamydia trachomatis is the most common cause of sexually transmitted bacterial infection in the world and can cause pelvic inflammatory disease and Fallopian tube pathology in women. The cellular paradigm for immune-mediated pathology posits that cytokines produced by persistently infected epithelial cells are necessary and sufficient for the development of pathology during chlamydial infection (49). This theory has generated interest in characterizing the types and classes of genes upregulated during infection (24, 39, 40, 54). In addition to expressing proinflammatory genes, such as those for interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α), infected cells upregulate the type I IFNs (IFN-α and IFN-β), which can signal via the common type I IFN receptor (IFN-α/βR) (29), leading to subsequent induction of a broad assortment of interferon-stimulatory genes (ISGs) (30). Using Chlamydia muridarum and the mouse female genital tract infection models, our laboratory demonstrated that signaling via IFN-α/βR was detrimental to the host during infection, as mice with the gene for IFN-α/β signaling knocked out (IFNAR KO mice) cleared infection sooner than wild-type (WT) mice and developed less oviduct pathology (31). Similar observations were also reported for a lung infection model using C. muridarum (38). These results provide a strong rationale for defining how type I IFNs are produced in the female genital tract and for determining the role of upstream mediators of type I IFN during genital chlamydial infection.
A key difference in the expression of type I IFN genes from that of proinflammatory cytokines like TNF-α and IL-1β is their regulation by interferon-regulatory factors (IRFs). The IRF family of transcription factors consists of nine members (reviewed in reference 46). Three of these transcription factors, namely, IRF3, IRF7, and ISGF3, have been characterized as crucial contributors in the regulation of type I IFNs. IRF3 is constitutively expressed in most cell types and, following activation, is phosphorylated and translocated to the nucleus. Nuclear IRF3 plays an important role in the initial transcriptional upregulation of IFN-β in conjunction with AP-1 and NF-κB (12, 25, 52). IRF7 is expressed at low levels inside most cell types but can be induced severalfold following initial autocrine or paracrine IFN-β signaling (48). IRF7 and ISGF3 (a trimeric complex composed of STAT1, STAT2, and IRF9) (14) bind to promoters with interferon-sensitive response elements (21), leading to induction of interferon response genes (IRGs) and enhanced expression of IFN-β (44). Unlike IRF3, which does not upregulate IFN-α genes, IRF7 has been termed a master regulator of type I IFNs because of its ability to regulate both the IFN-β and IFN-α genes (20). It is important to note that IRF transcription factors also contribute to the transcription of genes other than those for type I IFNs. Many genes that are classified as IRGs can also be induced directly by IRF3 or IRF7 in the absence of type I IFN (1, 11, 16, 17, 22, 50), highlighting the flexible nature of these loosely defined categories. In addition to having a role as a transcription factor, IRF3 plays an important role in cell death. Apoptosis has been shown to require IRF3 during infection with multiple viruses (15, 19, 22, 23, 35, 55). While it is possible that some virally induced cell death is secondary to autocrine IFN-α/βR signaling, in the cases of certain viral and nonviral stimuli, apoptosis can occur via IRF3 directly, independently of the activities of type I IFNs (13, 18, 41, 53). A number of proapoptotic genes, such as those for Zac1, Daax, Erg2, and Scotin, are IRF3 dependent and IFN independent (2), suggesting potential direct involvement of IRF3 in apoptosis.
The critical role of IRF3 in the induction of IFN-β during chlamydial infection has been demonstrated in vitro using multiple Chlamydia species in different cell types (4, 30, 36, 51). However, the role of IRF3 during in vivo infection and its IFN-independent functions during chlamydial infection have not been explored. Using mice deficient for IFNAR, we have shown that type I IFN signaling has a detrimental role during infection with C. muridarum in the mouse genital tract (31). However, it is difficult to pinpoint the relative contributions of IFN-α and IFN-β to this phenotype. Identifying these contributions is especially a problem with the multiple IFN-α subtypes, which preclude the use of single-knockout mice or antibody-mediated neutralization. Since IRF3 is required for IFN-β expression in macrophages infected in vitro with C. muridarum (36), it is expected that IFN-β expression during an in vivo infection would also be IRF3 dependent. Therefore, IRF3 KO mice may provide us with an opportunity to examine the contribution of IFN-β to the phenotype of IFNAR KO mice. With this prospect in mind, our goal in the present study was to characterize the outcome of chlamydial infection in IRF3 KO mice. In contrast to the previously stated hypothesis, our results indicate that the IFN-independent functions of IRF3 during in vivo genital infection outweigh its effect on IFN-β expression, indicating a surprising role for IRF3 in uterine horn protection during chlamydial infection.
MATERIALS AND METHODS
Bacterial stocks and mouse strains.
Chlamydia muridarum, “Nigg” strain, was propagated in Mycoplasma-free McCoy cells grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 μM nonessential amino acids (Invitrogen), 2 mM l-glutamine (Invitrogen), 10% fetal bovine serum (FBS) (Atlas Biologicals), 50 μg/ml gentamicin sulfate (Gemini Bioproducts), and 0.5 μg/ml cycloheximide (Sigma) as previously described (37). The infectious titer of the stock was determined by infecting a fresh McCoy cell monolayer and calculating the number of inclusion forming units (IFU). Numbers of IFU from experimental samples were calculated by infecting a fresh McCoy cell monolayer in 96-well black plates using a previously published protocol (9). IRF3 KO (45) (Stock number RBRC00858) mice and IRF7 KO (20) (stock number RBRC01420) mice in the C57BL/6 background were purchased from Riken BioResource Center, Japan, and bred in-house. All C57BL/6 WT control mice were purchased from Jackson Laboratory (stock number 000664).
In vivo mouse infections.
Seven days prior to infection with C. muridarum, each mouse received 2.5 mg of medroxyprogesterone (Depo-Provera) subcutaneously in 100 μl of phosphate-buffered saline (PBS). Mice were anesthetized with pentobarbital (Nembutal; 200 to 230 μl of a stock diluted 1:10 with PBS) and infected by inoculating 1 × 106 IFU (unless otherwise noted in the figure legends) of C. muridarum in 10 μl SPG buffer (250 mM sucrose, 10 mM sodium phosphate, 5 mM glutamic acid, pH 7.2) directly into the vaginal vault. For experiments featuring antibody-mediated cytokine neutralization, mice were administered 500 U (50 μl) of neutralizing rabbit polyclonal IFN-β antiserum (Fitzgerald, Inc.) at day 0, after the urethra was pulled up with forceps and the antibody injected into the vaginal tissue slowly. Another 100 μl (1,000 U) of antibody was given intraperitoneally to each mouse. Mice were infected 5 h later with an infectious dose of 3 × 105 IFU of C. muridarum. Control mice were given an identical volume of control rabbit serum. Antibody administrations were again performed at day 4 postinfection, when each mouse received 1,000 U intraperitoneally. Mice from both groups were treated similarly with genital swabs for secretions. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences.
Sample collection for IFU determination and cytokine analysis.
Chlamydial colonization was determined by swabbing the cervix and vaginal vault of infected mice with a calcium alginate swab (Spectrum Medical Instruments) at various times postinfection, and numbers of IFU were calculated as described earlier (9). Additionally, genital secretions were collected throughout the duration of infection and subsequently processed for cytokine analysis (9). Briefly, mice were anesthetized with pentobarbital, and absorbent sponges were placed into the vagina for 30 min. Sponges were then removed and stored at −70°C until further processing. Subsequently, cytokines were then eluted from the sponges with a solution containing PBS, 0.5% bovine serum albumin (BSA), and 0.05% Tween 20. This procedure was performed with Spin-X centrifuge tubes (Fisher Scientific number 07-200-385). Sponges were placed in the upper chamber of these tubes in 350 μl of the elution solution and vortexed vigorously for 1 min. Elution was allowed to occur on ice for a further 1 h. Finally, after a second vortexing, samples were centrifuged at 10,000 × g for 5 min at 4°C.
Cytokine analysis.
Enzyme-linked immunosorbent assay (ELISA) kits were used to quantify the protein levels of IFN-β (PBL Biomedical Laboratories kit number 42400-1), Cxcl10 (R&D Systems kit number SMCX100), and IFN-γ (R&D Systems kit number MIF00) in genital tract eluates. Optical densities were measured at 450 nm using a BioTek plate reader. A Luminex-based bead array (Milliplex MPX MCYTO-70K-04; Millipore) was used according to the manufacturer's instructions to measure the levels of TNF-α, MCP-1, and IL-1β.
Tissue processing for flow cytometry and IFU number determination.
For analysis of cell recruitment to the infected site or determination of chlamydial burden in the tissue, infected WT and IRF3 KO mice were euthanized and their genital tracts were excised on day 4 or 7 postinfection depending on the particular experiment. Lower genital tracts were defined as the area from the cervix to the beginning of the uterine horn. Upper genital tracts were defined as the uterine horns and oviducts. Isolated tissues were initially minced thoroughly with surgical scissors and incubated individually in 0.5 ml of dispase (BD Biosciences) for 30 min at 37°C while being rocked. These tissue and cell suspensions were subsequently forced through a 70-μm nylon mesh filter by maceration with a rubber stopper from a syringe. Cell populations were kept separate throughout the remainder of the procedure and not pooled at any time. Red cells were lysed by resuspending the cell pellet in a red blood cell (RBC) lysis solution (140 mM NH4Cl, 17 mM Tris, pH 7.4) for 5 min at 37°C, followed by neutralization with RPMI medium. After centrifugation (350 × g, 5 min, 4°C), cells were resuspended in RPMI medium and passed through a second 70-μm nylon filter. These samples were then used for immediate analysis by flow cytometry as described below. Alternatively, for determination of IFU numbers in the tissue, cell suspensions from the cervix were sonicated, centrifuged to remove debris, and used to infect a fresh McCoy cell monolayer.
Cell analysis by flow cytometry.
Single-cell suspensions (∼106 cells) in PBS supplemented with 0.5% BSA and 2 mM EDTA were incubated with Fc Block (BD Pharmingen; clone 2.4G2) (5 μg/ml) for 10 min at 4°C. This was followed by staining for 20 min on ice with the antibodies Ly-6G-fluorescein isothiocyanate (FITC) (BD Pharmingen; clone RB6-8C5), F4/80-allophycocyanin (APC) (Biolegend, clone BM8), and CD45-peridinin chlorophyll protein (PerCP)Cy5.5 (BD Pharmingen; clone 30-F11) at a titer of 0.25 μg/106 cells. As the Ly-6G-FITC (clone RB6-8C5) antibody stains Gr-1 antigen (Ag) expressed on both neutrophils and macrophages, macrophages were identified as CD45+ F4/80+ Gr-1+ cells and neutrophils as CD45+ F4/80− GR-1+ cells. Cells were also stained with the combinations of corresponding fluorochrome-conjugated isotype control antibodies. Cells were analyzed with a BD FACSAria flow cytometer. Analysis of flow cytometry data was carried out using FlowJo software (Tree Star, Inc.).
Histopathology.
WT control and IRF3 KO mice were sacrificed at day 4, 14, or 45 postinfection depending on the experiment, and their genital tracts were excised and fixed in 10% buffered formalin for a minimum of 24 h. Subsequently, the tissues were embedded in paraffin and longitudinal sections (4 μm) were prepared and stained with hematoxylin and eosin. The sections were evaluated using a previously elaborated semiquantitative scoring system (10) by a veterinary pathologist who was blind to the experimental design. Briefly, acute (with the presence of polymorphonuclear leukocytes [PMNs]) and chronic (with the presence of mononuclear cells, namely, lymphocytes and monocytes) inflammation, epithelial erosion, and dilatation were graded from 0 (absence of the parameter) to 4 (severe lesion). Images were acquired using an Olympus BX40 microscope equipped with a Spot Insight 2 camera.
TUNEL staining.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining was performed as described previously (3), with some modifications for the genital tract tissues. Genital tracts were excised at day 4 postinfection and fixed in 10% buffered formalin for 24 h on a piece of cardboard. The fixed tissues were washed twice in PBS and kept at 4°C. Samples were dehydrated and embedded in paraffin using methyl benzoate (Sigma). Sections were oriented longitudinally, and serial 4-μm-thick sections were cut until the uterine horn lumens were visible bilaterally. For staining, sections were dewaxed, rehydrated in PBS, and processed for TUNEL assay using an in situ cell death detection kit (Roche Diagnostics). Each section was probed with terminal deoxynucleotidyltransferase (TdT) and FITC-labeled precursor in cacodylate-based buffer for 1 h at 37°C, rinsed with 0.05% Tween 20 in PBS 3 times, and then mounted under a ProLong Antifade medium containing DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). Control sections in each slide were treated the same way but did not include TdT for TUNEL specificity. Images were taken using an Olympus IX-81 microscope, equipped with a Hamamatsu ORCA-ER digital camera. Slide book 4.2 software was used for image capturing and analysis. Images were acquired following setting up of the baseline threshold for control sections. For quantification, five images/horn (10 per mouse) were taken, quantified, and averaged (n = 1). Five mice were used per group. Cells that detached into the tube lumen were excluded from quantification. The areas of total nuclear DNA (DAPI positive) and fragmented nuclear DNA (TUNEL positive) in tube walls were masked and measured within each image. The results are presented as percentages of the total nuclear DNA area that was TUNEL-positive nuclear DNA, calculated for individual cells.
Chlamydial staining in paraffin-embedded sections.
The sections were stained with antichlamydial mouse serum at a 1:100 dilution and then with anti-mouse Texas Red antibody (1:1,000). The staining protocol was similar to that of TUNEL staining, and sections were mounted under a ProLong Antifade medium containing DAPI (Invitrogen).
In vitro T cell proliferation.
On day 7, 14, or 21 postinfection, WT and IRF3 KO mice were sacrificed and their iliac lymph nodes were harvested. These nodes were individually macerated through a 70-μm mesh filter and brought into single-cell suspensions at 2.5 × 106/ml in complete medium (RPMI media supplemented with 10% FBS, 100 mM HEPES, 1 mM sodium pyruvate, 2 mM l-glutamine, 100 μM nonessential amino acids, 50 μg/ml gentamicin sulfate, and 50 mM β-mercaptoethanol). In triplicate, 2.5 × 105 cells (100 μl) were added to a 96-well plate with either 100 μl medium, 100 μl medium containing 5 μg/ml concanavalin A (ConA), or 100 μl medium containing 5 μg/ml chlamydial antigen. Chlamydial antigen was prepared by isolating C. muridarum elementary bodies (EBs) from infected HeLa cells by Renografin centrifugation and UV inactivation, as previously described (5). After 96 h, 20 μl of supernatants per well was pulled and replaced with 20 μl alamarBlue. Fluorescence was read 24 h later on a 96-well Synergy II plate reader (BioTek). Triplicate values were averaged to determine the proliferation for each individual mouse. These values were then used to calculate the means and standard deviations for the WT and IRF3 KO mouse groupings. T cell proliferation supernatants were diluted 1:10 for IFN-γ analysis and analyzed undiluted for IL-17 (R&D Systems kit number M1700).
Statistical analysis.
Statistical comparison between the two mouse strains for IFU quantification over the course of infection were made by a 2-way analysis of variance (ANOVA) with the post hoc Tukey test as a multiple-comparison procedure. The Fisher exact test was used for determination of significant differences in the frequencies of hydrosalpinx observed in the two groups. For analysis of pathology scores, the Mann-Whitney rank sum test was performed. A two-tailed t test was performed to analyze the significance of the differences in levels of T cell proliferation, numbers of IFU taken on a single day, TUNEL data, or percentages of cells recruited to the tissues of WT and IRF3 KO mice infected with C. muridarum.
RESULTS
IRF3 KO mice display increased bacterial loads early during infection, although the overall infection course is similar to that of WT mice.
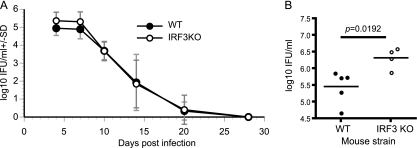
In order to establish the role of IRF3 during chlamydial infection, female WT control and IRF3 KO mice were infected in their genital tracts with C. muridarum. IRF3 KO mice exhibited higher chlamydial colonization in genital swabs from day 4 to day 7 than WT mice (Fig. 1 A). To further confirm that there is increased chlamydial colonization early in infection, we examined numbers of IFU in cervical tissues as described previously (32). IRF3 KO mice had nearly a log increase in the number of chlamydial IFU (P = 0.0192) in lysates prepared from lower genital tract tissues isolated at day 4 postinfection (Fig. 1B). However, increased early colonization in IRF3 KO mice was not maintained and they cleared infection at the same rate as the WT mice (two-way ANOVA, P = 0.067) (Fig. 1A). These results suggest a necessary role for IRF3 in chlamydial control early but not throughout the duration of infection. In contrast to IRF3 KO mice, IRF7 KO mice exhibited no difference in intensity or duration of infection from WT mice during the entire course of infection (data not shown).
Fig. 1.
IRF3 KO mice resolve a primary infection equivalent to that in WT mice despite elevated chlamydial burdens at early time points. (A) Infection courses of WT control (n = 10) and IRF3 KO (n = 12) mice infected with 1 × 106 IFU of C. muridarum (P = 0.067, two-way ANOVA). SD, standard deviation. (B) In an independent experiment, IFU measurements from day 4 postinfection in cervical lysates from individual WT (n = 5) and IRF3 KO (n = 4) mice are represented. The line represents the mean, while each dot represents the value from an individual mouse. An asterisk denotes a statistically significant difference (P < 0.05) as measured by a two-tailed t test.
IRF3 KO mice exhibit increased pathology in the uterine horns following chlamydial infection.
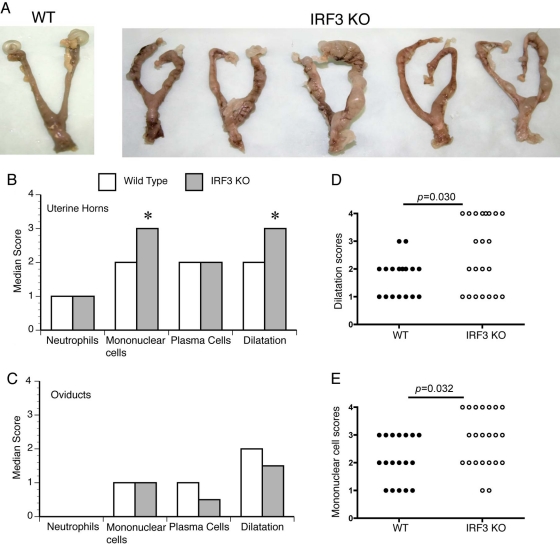
At day 45 postinfection, the genital tracts from infected WT and IRF3 KO mice were excised. The incidences of oviduct hydrosalpinx were not significantly different (P = 0.193) between the two study groups. However, IRF3 KO mice exhibited more-severe gross pathology in the uterine horns than WT mice, despite some variability within each of the two populations. This observation was visually manifested by cyst-like formations and increased to severe swelling in the horns of IRF3 KO mice (Fig. 2 A). Histopathological examination revealed a significantly higher median pathology score for dilatation (P = 0.030) and chronic inflammation as determined by the presence of mononuclear cells (lymphocytes and monocytes) (P = 0.032) in the IRF3 KO uterine horns (Fig. 2B) than in WT horns. There was no significant difference in the scores for acute inflammation in uterine horns, as measured by the presence of PMNs, in a comparison of KO and WT mice (Fig. 2B). Additionally, there were no significant differences between pathology scores for the endocervix (data not shown) or oviducts (Fig. 2C) of WT and IRF3 KO. Closer examination of the distribution of individual uterine horn dilatation scores demonstrated that the key difference between groups was a much greater probability for individual IRF3 KO horns to possess the most severe pathology score of 4 (10/24 horns) than for WT horns (0/18 horns) (Fig. 2D). This type of distribution was also apparent for the pathology scores for chronic inflammation (lymphocytes and monocytes) in the uterine horn (Fig. 2E). The increased uterine pathology was specific to IRF3 KO mice, since IRF7 KO mice exhibited uterine horn and oviduct pathologies similar to those of WT mice (data not shown).
Fig. 2.
IRF3 KO mice exhibit normal oviduct pathology but increased uterine horn pathology in comparison to WT controls. (A) Genital tracts from a representative WT mouse and five IRF3 KO mice at day 45 postinfection from one of three independent studies. Median pathology scores following histopathological examination of sections prepared from uterine horns (B) and oviducts (C) are graphed for each category. The distributions of individual uterine horn scores are shown for dilatation (D) and chronic cell inflammation (lymphocytes/monocytes) (E). An asterisk denotes a significant difference (P < 0.05) between WT and IRF3 KO groups, determined by the two-tailed Mann-Whitney test.
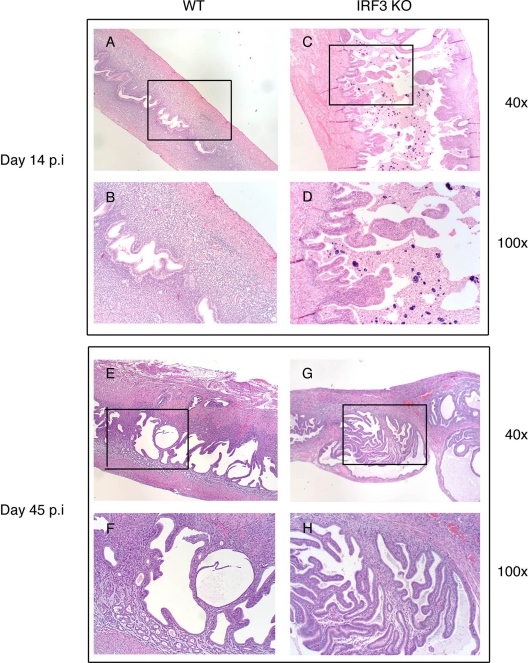
To further understand the differential pathology between WT and IRF3 KO uterine horns, sections were obtained from infected mice at day 14 postinfection for histopathology. At this time point, the IRF3 KO uterine horns are already starting to display swelling and lobed structures. Histopathological examination at this time point indicated that all mice developed granulomatous inflammation. Further, examination indicated an increased incidence of nonneoplastic, reactive hyperplasia due to proliferation and differentiation of uterine stromal cells in the IRF3 KO mice (6 of 8 uterine horns), compared to that in WT mice (1 of 10 uterine horns) (P < 0.01, using Fisher's exact test) (Fig. 3 A to D). These structures also remain evident at day 45 postinfection in some IRF3 KO horns (Fig. 3E to H). It is of importance to note that progesterone-treated uninfected IRF3 KO mice display no uterine horn pathology (data not shown), suggesting that the observed phenotype is infection driven.
Fig. 3.
IRF3 KO uterine horns display enhanced stromal nonneoplastic hyperplasia. WT control (n = 5) and IRF3 KO (n = 5) mice were infected with 1 × 106 IFU/ml of C. muridarum. Genital tracts were excised and processed for histopathology at day 14 (A to D) and day 45 postinfection (p.i.) (E to H). Representative images for WT and IRF3 KO mice are shown.
Genital tracts of IRF3 KO mice display reduced cellular apoptosis early in infection.
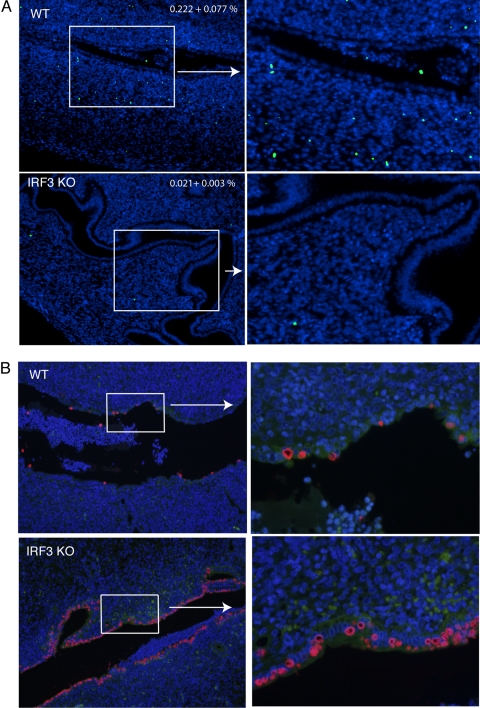
IRF3 has been suggested to play a major role in apoptosis (53). Independently, mice lacking the proapoptotic Bax protein display increased uterine horn pathology during C. muridarum genital infection (34). To test if there was a relationship between apoptosis and uterine horn pathology in IRF3 KO mice, TUNEL staining was performed on tissue sections obtained at day 4 postinfection. This time point was chosen because early events can set the stage for progressing pathology. TUNEL staining revealed significantly reduced numbers of apoptotic cells in the uterine stromata of IRF3 KO mice in comparison to WT mice (Fig. 4 A). The uterine epithelial cells did not show increased TUNEL staining in either WT or IRF3 KO mice following infection (Fig. 4A). The majority of TUNEL-positive cells were observed entrapped in the exudate in the lumen as expected (data not shown). These data demonstrate a clear reduction in the number of apoptotic cells in the uterine stromata of IRF3 KO compared to WT mice early in infection.
Fig. 4.
Reduced numbers of TUNEL-positive cells in the uterine horns but increased chlamydial infection in IRF3 KO mice in comparison to WT mice at day 4 postinfection. WT control (n = 5) and IRF3 KO (n = 5) mice were infected with 1 × 106 IFU/ml of C. muridarum. At day 4 postinfection, genital tracts were excised and processed for TUNEL as described in Materials and Methods. (A) Images acquired using a 10× objective and a magnified section from a representative horn from each group are shown. Blue indicates DAPI-positive cells, and green indicates nuclear DNA fragmentation by TUNEL staining. Numbers on the panel indicate the percentages of TUNEL-positive cells in the uterine horns as determined from 10 images for each mouse (5 mice per group ± standard error of the mean) (P = 0.019, unpaired t test.) (B) Staining for chlamydial antigen in adjacent sections shows inclusions in epithelial cells. Blue indicates DAPI-positive cells, and red indicates chlamydial staining with antichlamydial mouse serum followed by Texas Red-conjugated anti-mouse antibody. A representative section is shown from each group at two magnifications (×100 [left] and ×400 [right]).
To determine if there was a relationship between TUNEL staining and chlamydial infection, sections adjacent to the ones used for TUNEL were stained using antichlamydial mouse serum. As expected, chlamydial staining was restricted to epithelial cells in all tissues (Fig. 4B). However, unlike the WT sections, which showed intermittent staining for chlamydiae, IRF3 KO sections displayed continuous staining along the epithelia (Fig. 4B). These data are in agreement with the tissue IFU data indicating elevated colonization in the IRF3 KO mice at the same time point (Fig. 1B). Additionally, the integrity of the epithelial cells was well maintained in IRF3 KO mice but not in the WT controls. Together, these data suggest that increased early infection, along with decreased apoptosis in IRF3 KO mice, likely contributes to the increased hyperplasia of uterine stromal cells observed at day 14 postinfection (Fig. 3). Tissue sections obtained at day 14 postinfection showed a similar trend of reduced TUNEL staining in IRF3 KO uterine horns, but infection levels were comparable between WT and IRF3 KO uterine horns (data not shown).
IRF3 KO mice express less IFN-β and IRGs early during infection.
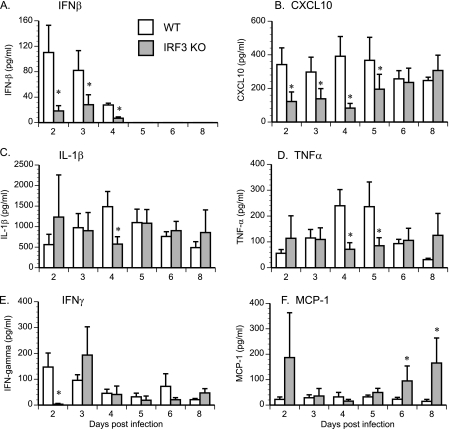
The two major phenotypes in IRF3 KO mice following chlamydial infection are the early increased bacterial load and the unusual uterine pathology. As a first step to explore this phenomenon further, we addressed the possible mechanism behind the increased bacterial load at day 4 postinfection. Since IRF3 is theorized to be essential for the initial IFN-β induction, but possibly not for later IFN-β expression due to compensation from IRF7, the most plausible early candidates are IFN-β and IFN response genes. Cytokine expression in genital tract secretions from infected animals was analyzed in order to test the hypothesis that expression of these genes was altered in the absence of IRF3. IFN-β was present in detectable quantities only at early time points (Fig. 5 A). Expression of both IFN-β (P = 0.018) and Cxcl10 (P = 0.026) was significantly decreased in the absence of IRF3 (Fig. 5A and B). Importantly, the role of IRF3 in cytokine expression was not universal. Levels of IL-1β (Fig. 5C) and TNF-α (Fig. 5D) in genital tract secretions were unaltered during the course of infection in the absence of IRF3, except at day 4 postinfection, when both were significantly decreased in the IRF3 KO mice. IFN-β expression can influence IFN-γ levels (43); therefore, IFN-γ levels were examined in genital secretions. IFN-γ levels were significantly reduced at day 2 postinfection in IRF3 KO mouse genital secretions but regained normal levels rapidly (Fig. 5E). In contrast, MCP-1 levels were found to be increased in IRF3 KO mouse genital secretions after day 6 postinfection (Fig. 5F). As IFN-β can impair chlamydial growth in tissue culture cells (42), decreased IFN-β combined with reduced early IFN-γ expression may have resulted in increased chlamydial burden early in infection.
Fig. 5.
Cytokine levels in genital secretions of IRF3 KO mice. WT control (n = 5) and IRF3 KO (n = 6) mice were infected with 1 × 106 IFU/ml of C. muridarum. IFN-β (A), Cxcl10 (B), IL-1β (C), TNF-α (D), IFN-γ (E), and MCP-1 (F) protein were quantified in genital tract secretions collected from infected mice at various times postinfection. Data for all panels represent the mean cytokine concentrations ± the standard errors of the means calculated from values obtained by assaying individual mouse secretions separately. An asterisk denotes a statistically significant difference (P < 0.05) on the particular day postinfection between values for WT and IRF3 KO mice.
IRF3 KO mice experience delayed recruitment of immune cells to the upper genital tract.
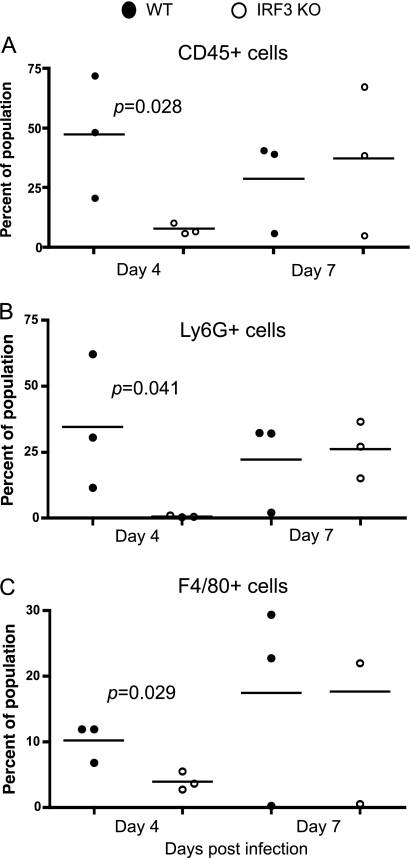
Differential expression of host chemokines and differential chlamydial burdens may alter the initial recruitment of immune cells to the site of infection. Additionally, to decipher the possible mechanism of uterine horn pathology, immune cell infiltration to the lower genital tract was analyzed by flow cytometry. Recruitment of CD45+ immune cells to the cervix at day 4 postinfection was normal in IRF3 KO mice (data not shown). In contrast, at this same time point, in the uterine horn, there was a significantly decreased percentage of infiltrating CD45+ cells (P = 0.0286), corresponding to fewer Ly6G+ neutrophils (P = 0.0411) and fewer F4/80+ macrophages (P = 0.0294) (Fig. 6 A to C) in the IRF3 KO mice than in WT mice. It is important to note that this impairment was only temporary, as normal recruitment to the same tissue site was observed at day 7 (Fig. 6) and day 14 (data not shown) postinfection. These data demonstrate that despite higher infection levels in IRF3 KO mice at day 4 postinfection, there was actually reduced infiltration of both macrophages and neutrophils. Alternatively, the increased infection at day 4 postinfection may have been a result of delayed recruitment of inflammatory cells.
Fig. 6.
Recruitment of immune cells to the uterine horns is delayed in IRF3 KO mice. WT control and IRF3 KO mice (three mice per group) were infected with 1 × 106 IFU/ml of C. muridarum. Genital tract tissues from individual mice were isolated and processed for flow cytometry as described in Materials and Methods at days 4 and 7 postinfection. The percentage of CD45+ cells (A), CD45+ Ly6G+ neutrophils (B), and CD45+ F4/80+ macrophages (C) were calculated for upper genital tract tissues by flow cytometric analysis. For all panels, dots represent data points calculated for tissue derived from individual mice. The line represents the mean. Statistically significant differences as measured by a two-tailed t test are indicated.
The absence of IRF3 leads to increased basal and antigen-specific proliferation in T cells derived from infected animals.
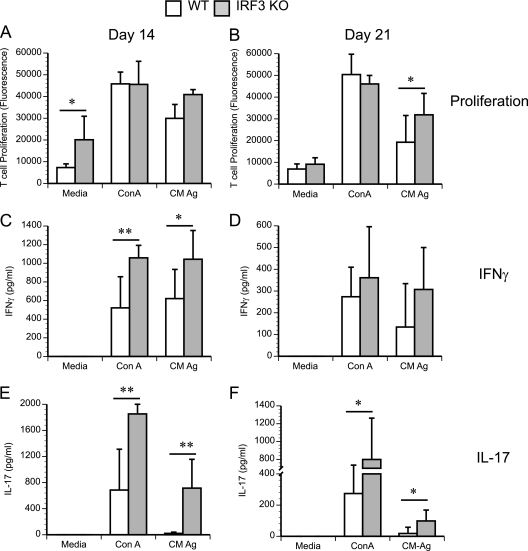
Despite increased bacterial colonization early on, IRF3 KO mice go on to clear infection at a normal rate. To understand the normal clearance of infection in IRF3 KO mice, we tested the Ag-specific T cell proliferation in the iliac lymph nodes of IRF3 KO mice. It should be noted that IFNAR KO mice infected with C. muridarum exhibit enhanced chlamydia-specific T cell proliferation compared to controls in response to chlamydial antigen (31). To determine if IRF3 contributes to this response, T cell proliferation was analyzed at days 14 and 21 postinfection (Fig. 7 A and B). As with the IFNΑR KO mice, T cells from infected IRF3 KO mice exhibited a trend toward increased proliferation in response to chlamydial antigen, especially at day 21 postinfection (Fig. 7B), when the difference became statistically significant between the two groups (P = 0.047). Additionally, the IRF3 KO T cells displayed an increased proliferation with no antigenic stimulation (medium control) at day 14 postinfection (P = 0.0308) (Fig. 7A) compared to WT control T cells. To determine the nature of the T cell response in IRF3 KO mice, the supernatants from T cell cultures stimulated ex vivo with chlamydial antigen were analyzed for IFN-γ and IL-17 secretion. Both IFN-γ (Fig. 7C and D) and IL-17 (Fig. 7E and F) levels were significantly increased in culture supernatants from IRF3 KO T cells compared to those from WT cells. This increase was observed in both ConA- and chlamydial-antigen-stimulated T cell supernatants, suggesting an overall increase in Th1 and Th17 responses in IRF3 KO mouse genital tracts. Together, these data suggest a temporal role for the IRF3-dependent IFN-β response. As a consequence, in IRF3 KO mice, the innate defect that leads to early increased bacterial colonization is compensated for by improved T cell function, leading to normal clearance.
Fig. 7.
IRF3 KO T cells display increased intrinsic and chlamydial-antigen-specific T cell responses during infection. WT control and IRF3 KO mice were infected with 1 × 106 IFU/ml of C. muridarum. Iliac lymph node cells were isolated from WT and IRF3 KO mice at days 7, 14, and 21 postinfection (four mice per group). Cells were stimulated in vitro with either medium, the T cell mitogen ConA, or C. muridarum antigen (CM Ag). Proliferation was quantitated using alamarBlue, and data for all panels represent the mean fluorescence ± the standard deviation calculated from values obtained by assaying T cell proliferation from cells derived from individual mice. Data from day 14 (A) and day 21 (B) postinfection are shown. T cell proliferation supernatants were diluted 1:10 for IFN-γ analysis (C and D) and assayed directly for IL-17 (E and F). *, P < 0.05; **, P < 0.01, as measured by a t test.
The in vivo effect of IRF3 deficiency is independent of IFN-β.
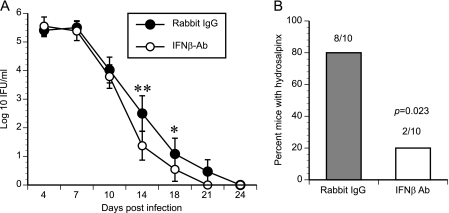
Overall, IRF3 KO mice resemble IFNAR KO mice with respect to their cytokine profile (low Cxcl10) and increased T cell proliferation but differ from the IFNAR KO mice with respect to infection course and pathology. To distinguish the effect of IRF3 from that of IFN-β signaling, WT mice were infected in the presence of either an IFN-β-neutralizing antiserum or preimmune serum. Mice receiving the neutralizing antibody resolved infection faster than the control group (P = 0.03) (Fig. 8 A) and exhibited a decreased incidence of hydrosalpinx (8/10 oviducts versus 2/10 oviducts; P = 0.023) (Fig. 8B). Overall, this reduction in pathology indicates that IFN-β is the major contributor to the phenotype of IFNAR KO mice. Further, IFN-β-depleted mice show no exacerbated uterine horn pathology, suggesting that IRF3, and not IFN-β, may play a special role in regulating uterine mucosal inflammatory responses during infection.
Fig. 8.
IFN-β neutralization leads to earlier clearance of C. muridarum. C57BL/6 mice (10 mice per group) were depleted of IFN-β by injection of IFN-β polyclonal antiserum (Fitzgerald) or preimmune serum before infection or on day 4 postinfection (p.i.), respectively. Infection courses were monitored by IFU quantitation (A). Data represent the mean log10 number of IFU/ml ± the standard error of the mean. Significance was determined by two-way ANOVA (P = 0.03). *, P < 0.05; **, P < 0.01, by one-way ANOVA. (B) Mice were sacrificed at day 45 postinfection, and the incidence of hydrosalpinx development was recorded. An asterisk represents a statistical difference as measured by the Fisher exact test (P = 0.023). Ab, antibody.
DISCUSSION
IFNAR KO mice experience a shortened course of infection and decreased pathology in the oviducts, suggesting that type I IFNs have a detrimental role in the mouse female genital tract during C. muridarum infection (31). IFN-β expression in macrophages infected in vitro with C. muridarum requires the transcription factor IRF3 (36), implying that IRF3 might also be a key player during infection in the genital tract. This inquiry led us to examine how IRF3 regulates the expression of IFN-β in vivo and how mice lacking IRF3 respond to infection with C. muridarum. Surprisingly, IRF3 KO mice did not resemble IFNAR KO mice in infection course and in genital tract pathology, suggesting an IFN-β-independent role for IRF3 in host protection during infection.
Expression of IFN-β was largely reliant on IRF3 during day 1 to day 4 of infection of the female genital tract (Fig. 5A). This lack of early IFN-β induction in IRF3 KO mice also results in decreased levels of the IRG Cxcl10 (Fig. 5B). The amount of Cxcl10 approaches wild-type levels in the absence of IRF3 as the infection progresses, possibly due to the contribution of IFN-γ, another known inducer of Cxcl10 (27). The diminished cytokine expression was specific to IFN-β and Cxcl10, as expression of genes such as those for IL-1β and TNF-α were largely unaltered in the IRF3 KO mice, except at day 4 postinfection (Fig. 5C and D), at which point inflammatory cell recruitment was also reduced (37). However, compensatory mechanisms seem to restore inflammatory cell recruitment to normal levels by day 7 postinfection, suggesting that the majority of the innate IRF3 effects are observed only early in infection.
In agreement with IFNAR KO mice, IRF3 KO mice exhibited increased chlamydial-antigen-specific T cell proliferation at later time points (Fig. 7B), which could be the reason for the normal clearance of chlamydial infection despite increased early bacterial burden. Additionally, increased secretion of IL-17 and IFN-γ were observed in IRF3 KO in comparison to WT mouse T cell supernatants, suggesting enhanced Th1 and Th17 responses. Recently, it was shown that IL-17 KO mice have a compromised Th1 response during C. muridarum infection (47), suggesting that elevated IL-17 may contribute to the enhanced Th1 response in IRF3 KO mice.
In contrast to IFNAR KO mice, IRF3 KO mice did not display a shortened course of infection. The presence of increased uterine pathology in the IRF3 KO mice also contradicted the observed phenotype of the IFNAR KO mice (31). It should be noted that this exacerbated pathology in IRF3 KO mice was observed in the uterine horns, while pathology in the oviducts was found to be similar to that of WT mice. Overall, the divergence in phenotype between the IRF3 KO and IFNAR KO mice suggests two possible interpretations. The first possibility is that IRF7 can compensate for the loss of IRF3, as was seen when IRF7 was overexpressed by the addition of exogenous IFN-β (36). However, the finding that IRF7 KO mice showed no difference from WT mice with respect to chlamydial clearance or the development of pathology does not support this interpretation. A second alternative is that IRF3 exerts IFN-β-independent roles during infection. IRF3 can upregulate immunologically relevant genes, such as those for IFN-λ, RANTES, ISG56, ISG54, TRAIL, Arginase II, and IL-27p28 (16, 17, 22, 26, 28, 50), independently of the actions of type I IFNs, while IRF7 can upregulate MxA, TRAIL, and Cxcl10 in an IFN-independent manner in plasmacytoid dendritic cells (DC) (11). An important IFN-independent effect of IRF3 in response to pathogens was similarly observed in a study using IRF3 KO mice infected with West Nile virus (8). The study showed impaired control of infection in the IRF3 KO mice despite normal levels of systemic type I IFN (8). Because IRF3 regulates many host genes in addition to IFN-β (2), the infection phenotype of the IRF3 KO mice includes the effect of losing all these other gene products in addition to IFN-β. A recent study identified 35 genes with known functions that were upregulated during infection with Newcastle disease virus in an IRF3-dependent but IFN-β-independent manner (2). Several more genes were similarly regulated but had no known function. Therefore, loss of a theoretically beneficial gene may counterbalance the effect of losing the detrimental type I IFNs.
Independently of its role in gene regulation, IRF3 has recently been directly linked to promoting cell death by binding to and activating the proapoptotic Bax protein, using its newly identified BH3 domain, resulting in Bax translocation to mitochondria and initiation of the intrinsic apoptotic pathway (7). It is intriguing to speculate that the early alteration in apoptosis might set the stage for the development of the increased pathology that was seen in the uterine horns of IRF3 KO mice. In that context, the genital tracts of infected IRF3 KO mice displayed significantly fewer TUNEL-positive cells than those of WT mice at day 4 postinfection. The TUNEL-positive cells were observed in the uterine stroma. There was no increased TUNEL staining in the epithelial cells, although it is quite possible that TUNEL-positive infected cells are no longer attached to the uterine stroma. Furthermore, at this time point, IRF3 KO mouse uterine horns, with strikingly healthy epithelial cells, were heavily infected with chlamydiae compared to WT uterine horns. The observation of hyperplasia in the uterine stromal cells in IRF3 KO mice by day 14 postinfection also suggests an inverse relationship between apoptosis and hyperplasia. The stromal cells play an important role in the replacement of infected epithelial cells that are shed following infection. It is likely that in the absence of normal apoptosis and with a lack of specific signaling from infected epithelial cells in IRF3 KO mice, an enhanced reaction of the stromal cells is observed during infection. Interestingly, mice lacking the proapoptotic protein Bax also exhibit worse pathology in their uterine horns (34), supporting the possibility that altered cell death along with increased infection could be the basis of exacerbated pathology at this site. It is important to note that it was recently discovered that the construction of IRF3 KO mice also resulted in the deletion of the gene of a BCL-2 family member with unknown function, BCL-2L12, which was carried in the other strand (33). Certain apoptotic phenotypes in cells derived from these mice were subsequently attributed to the loss of the latter gene and not that of IRF3 (33). It is also important to clarify that this finding does not pertain to the majority of studies showing that IRF3 plays a role in cell death, which did not use these KO mice. Therefore, either or both of the deleted genes may conceivably cause any cell death phenotype present in this mouse. In addition to having a proapoptotic role, IRF3 may play a direct role in the induction of specific chemokines necessary for selectively modulating recruitment of macrophages and PMNs, as observed during Pseudomonas aeruginosa lung infection (6). As a result, decreased or delayed recruitment of inflammatory cells may result in increased infection at early time points, setting the stage for uterine pathology. The subsequent development of an elevated Th1 response in IRF3 KO mice compensates for this initial impairment and results in normal infection clearance.
Antibody-mediated neutralization of IFN-β in WT mice was performed to further distinguish the effects of IFN-β and IRF3. Importantly, mice administered neutralizing antibody exhibited more-rapid clearance and decreased incidence of hydrosalpinx, mirroring the IFNAR KO phenotype (31). The faster clearance kinetics seems to run counter to the demonstrated antichlamydial role of IFN-β during growth in tissue culture (42). However, it hints that IFN-β may play distinct temporal roles during infection. Early production may aid in limiting chlamydial burden. This early production would help to explain the increased numbers of IFU seen at day 4 postinfection in the genital tract tissues of IRF3 KO mice. However, IFN-β produced later during infection that acts on T cells limits their activity. Ultimately, the infection course is determined by both the detrimental and beneficial roles of IFN-β. Overall, these IFN-β neutralization experiments suggest that IFN-β contributes more than the IFN-α genes to the IFNAR KO phenotype. Further, they support the interpretation that the delayed immune response and exacerbated pathology seen in the uterine horns of IRF3 KO mice are caused by an IFN-β-independent role that IRF3 plays in apoptosis. It remains to be determined whether the delayed immune response and exacerbated pathology occur by distinct mechanisms or whether they are intertwined.
ACKNOWLEDGMENTS
The project was supported by NIH grant AI067678 from the National Institute of Allergy and Infectious Diseases to U.M.N. and in part by the Arkansas Children's Hospital Research Institute, the Arkansas Biosciences Institute, and the UAMS DNA Damage and Toxicology Core. J. D. Sikes was partially supported by the Horace C. Cabe Foundation, Arkansas Children's Hospital.
We thank John Gregan and lab members for critically reading the manuscript. Special thanks go to Rosie Simmens (UAMS) for her evaluation of histopathology data.
Footnotes
Published ahead of print on 25 July 2011.
REFERENCES
- 1. Ackermann E. J., Taylor J. K., Narayana R., Bennett C. F. 1999. The role of antiapoptotic Bcl-2 family members in endothelial apoptosis elucidated with antisense oligonucleotides. J. Biol. Chem. 274:11245–11252 [DOI] [PubMed] [Google Scholar]
- 2. Andersen J., VanScoy S., Cheng T. F., Gomez D., Reich N. C. 2008. IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 9:168–175 [DOI] [PubMed] [Google Scholar]
- 3. Apostolov E. O., et al. 2009. DNase I is essential for DNA fragmentation induced by gamma radiation in mice. Radiat. Res. 172:481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buss C., et al. 2010. Essential role of mitochondrial antiviral signaling, IFN regulatory factor (IRF)3, and IRF7 in Chlamydophila pneumoniae-mediated IFN-beta response and control of bacterial replication in human endothelial cells. J. Immunol. 184:3072–3078 [DOI] [PubMed] [Google Scholar]
- 5. Caldwell H. D., Kromhout J., Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrigan S. O., et al. 2010. IFN regulatory factor 3 contributes to the host response during Pseudomonas aeruginosa lung infection in mice. J. Immunol. 185:3602–3609 [DOI] [PubMed] [Google Scholar]
- 7. Chattopadhyay S., et al. 2010. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 29:1762–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daffis S., Samuel M. A., Keller B. C., Gale M., Jr., Diamond M. S. 2007. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog. 3:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Darville T., et al. 1997. Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infect. Immun. 65:3065–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darville T., et al. 2003. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J. Immunol. 171:6187–6197 [DOI] [PubMed] [Google Scholar]
- 11. Di Domizio J., et al. 2009. TLR7 stimulation in human plasmacytoid dendritic cells leads to the induction of early IFN-inducible genes in the absence of type I IFN. Blood 114:1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Du W., Maniatis T. 1992. An ATF/CREB binding site is required for virus induction of the human interferon beta gene [corrected]. Proc. Natl. Acad. Sci. U. S. A. 89:2150–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duguay D., et al. 2002. In vivo interferon regulatory factor 3 tumor suppressor activity in B16 melanoma tumors. Cancer Res. 62:5148–5152 [PubMed] [Google Scholar]
- 14. Fu X. Y., Kessler D. S., Veals S. A., Levy D. E., Darnell J. E., Jr 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. U. S. A. 87:8555–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goubau D., et al. 2009. Transcriptional re-programming of primary macrophages reveals distinct apoptotic and anti-tumoral functions of IRF-3 and IRF-7. Eur. J. Immunol. 39:527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grandvaux N., et al. 2005. Regulation of arginase II by interferon regulatory factor 3 and the involvement of polyamines in the antiviral response. FEBS J. 272:3120–3131 [DOI] [PubMed] [Google Scholar]
- 17. Grandvaux N., et al. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heylbroeck C., et al. 2000. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J. Virol. 74:3781–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holm G. H., et al. 2007. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J. Biol. Chem. 282:21953–21961 [DOI] [PubMed] [Google Scholar]
- 20. Honda K., et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- 21. Kessler D. S., Levy D. E., Darnell J. E., Jr 1988. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. U. S. A. 85:8521–8525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirshner J. R., Karpova A. Y., Kops M., Howley P. M. 2005. Identification of TRAIL as an interferon regulatory factor 3 transcriptional target. J. Virol. 79:9320–9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kohl A., et al. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77:7999–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lad S. P., Fukuda E. Y., Li J., de la Maza L. M., Li E. 2005. Up-regulation of the JAK/STAT1 signal pathway during Chlamydia trachomatis infection. J. Immunol. 174:7186–7193 [DOI] [PubMed] [Google Scholar]
- 25. Lenardo M. J., Fan C. M., Maniatis T., Baltimore D. 1989. The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 57:287–294 [DOI] [PubMed] [Google Scholar]
- 26. Lin R., Heylbroeck C., Genin P., Pitha P. M., Hiscott J. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19:959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luster A. D., Unkeless J. C., Ravetch J. V. 1985. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 315:672–676 [DOI] [PubMed] [Google Scholar]
- 28. Molle C., et al. 2007. IL-27 synthesis induced by TLR ligation critically depends on IFN regulatory factor 3. J. Immunol. 178:7607–7615 [DOI] [PubMed] [Google Scholar]
- 29. Muller U., et al. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921 [DOI] [PubMed] [Google Scholar]
- 30. Nagarajan U. M., Ojcius D. M., Stahl L., Rank R. G., Darville T. 2005. Chlamydia trachomatis induces expression of IFN-gamma-inducible protein 10 and IFN-beta independent of TLR2 and TLR4, but largely dependent on MyD88. J. Immunol. 175:450–460 [DOI] [PubMed] [Google Scholar]
- 31. Nagarajan U. M., et al. 2008. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect. Immun. 76:4642–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagarajan U. M., et al. 2011. MyD88 deficiency leads to decreased NK cell gamma interferon production and T cell recruitment during Chlamydia muridarum genital tract infection, but a predominant Th1 response and enhanced monocytic inflammation are associated with infection resolution. Infect. Immun. 79:486–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakajima A., et al. 2009. Cell type-dependent proapoptotic role of Bcl2L12 revealed by a mutation concomitant with the disruption of the juxtaposed Irf3 gene. Proc. Natl. Acad. Sci. U. S. A. 106:12448–12452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perfettini J. L., et al. 2003. Role of proapoptotic BAX in propagation of Chlamydia muridarum (the mouse pneumonitis strain of Chlamydia trachomatis) and the host inflammatory response. J. Biol. Chem. 278:9496–9502 [DOI] [PubMed] [Google Scholar]
- 35. Peters K., Chattopadhyay S., Sen G. C. 2008. IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence. J. Virol. 82:3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prantner D., Darville T., Nagarajan U. M. 2010. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J. Immunol. 184:2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prantner D., et al. 2009. Critical role for interleukin-1β (IL-1β) during Chlamydia muridarum genital infection and bacterial replication-independent secretion of IL-1β in mouse macrophages. Infect. Immun. 77:5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qiu H., et al. 2008. Type I IFNs enhance susceptibility to Chlamydia muridarum lung infection by enhancing apoptosis of local macrophages. J. Immunol. 181:2092–2102 [DOI] [PubMed] [Google Scholar]
- 39. Rasmussen S. J., et al. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Invest. 99:77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ren Q., Robertson S. J., Howe D., Barrows L. F., Heinzen R. A. 2003. Comparative DNA microarray analysis of host cell transcriptional responses to infection by Coxiella burnetii or Chlamydia trachomatis. Ann. N. Y. Acad. Sci. 990:701–713 [DOI] [PubMed] [Google Scholar]
- 41. Romieu-Mourez R., et al. 2006. Distinct roles for IFN regulatory factor (IRF)-3 and IRF-7 in the activation of antitumor properties of human macrophages. Cancer Res. 66:10576–10585 [DOI] [PubMed] [Google Scholar]
- 42. Rothermel C. D., Byrne G. I., Havell E. A. 1983. Effect of interferon on the growth of Chlamydia trachomatis in mouse fibroblasts (L cells). Infect. Immun. 39:362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rothfuchs A. G., et al. 2001. IFN-alpha beta-dependent, IFN-gamma secretion by bone marrow-derived macrophages controls an intracellular bacterial infection. J. Immunol. 167:6453–6461 [DOI] [PubMed] [Google Scholar]
- 44. Sato M., et al. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106–110 [DOI] [PubMed] [Google Scholar]
- 45. Sato M., et al. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539–548 [DOI] [PubMed] [Google Scholar]
- 46. Savitsky D., Tamura T., Yanai H., Taniguchi T. 2010. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol. Immunother. 59:489–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scurlock A. M., et al. 2011. Interleukin-17 contributes to generation of Th1 immunity and neutrophil recruitment during Chlamydia muridarum genital tract infection but is not required for macrophage influx or normal resolution of infection. Infect. Immun. 79:1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Silvennoinen O., Ihle J. N., Schlessinger J., Levy D. E. 1993. Interferon-induced nuclear signalling by Jak protein tyrosine kinases. Nature 366:583–585 [DOI] [PubMed] [Google Scholar]
- 49. Stephens R. S. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 11:44–51 [DOI] [PubMed] [Google Scholar]
- 50. Thomson S. J., et al. 2009. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc. Natl. Acad. Sci. U. S. A. 106:11564–11569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trumstedt C., et al. 2007. Role of IRAK4 and IRF3 in the control of intracellular infection with Chlamydia pneumoniae. J. Leukoc. Biol. 81:1591–1598 [DOI] [PubMed] [Google Scholar]
- 52. Wathelet M. G., et al. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507–518 [DOI] [PubMed] [Google Scholar]
- 53. Weaver B. K., Ando O., Kumar K. P., Reich N. C. 2001. Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53. FASEB J. 15:501–515 [DOI] [PubMed] [Google Scholar]
- 54. Xia M., Bumgarner R. E., Lampe M. F., Stamm W. E. 2003. Chlamydia trachomatis infection alters host cell transcription in diverse cellular pathways. J. Infect. Dis. 187:424–434 [DOI] [PubMed] [Google Scholar]
- 55. Zhang P., Samuel C. E. 2008. Induction of protein kinase PKR-dependent activation of interferon regulatory factor 3 by vaccinia virus occurs through adapter IPS-1 signaling. J. Biol. Chem. 283:34580–34587 [DOI] [PMC free article] [PubMed] [Google Scholar]