Abstract
Nonribosomal peptide synthesis (NRPS) is a documented virulence factor for the opportunistic pathogen Aspergillus fumigatus and other fungi. Secreted or intracellularly located NRP products include the toxic molecule gliotoxin and the iron-chelating siderophores triacetylfusarinine C and ferricrocin. No structural or immunologically relevant NRP products have been identified in the organism. We investigated the function of the largest gene in A. fumigatus, which encodes the NRP synthetase Pes3 (AFUA_5G12730), by targeted gene deletion and extensive phenotypic analysis. It was observed that in contrast to other NRP synthetases, deletion of pes3 significantly increases the virulence of A. fumigatus, whereby the pes3 deletion strain (A. fumigatus Δpes3) exhibited heightened virulence (increased killing) in invertebrate (P < 0.001) and increased fungal burden (P = 0.008) in a corticosteroid model of murine pulmonary aspergillosis. Complementation restored the wild-type phenotype in the invertebrate model. Deletion of pes3 also resulted in increased susceptibility to the antifungal, voriconazole (P < 0.01), shorter germlings, and significantly reduced surface β-glucan (P = 0.0325). Extensive metabolite profiling revealed that Pes3 does not produce a secreted or intracellularly stored NRP in A. fumigatus. Macrophage infections and histological analysis of infected murine tissue indicate that Δpes3 heightened virulence appears to be mediated by aberrant innate immune recognition of the fungus. Proteome alterations in A. fumigatus Δpes3 strongly suggest impaired germination capacity. Uniquely, our data strongly indicate a structural role for the Pes3-encoded NRP, a finding that appears to be novel for an NRP synthetase.
INTRODUCTION
Aspergillus fumigatus is an opportunistic pathogen of immunocompromised individuals, exhibiting a 50 to 95% mortality rate, with the severity of disease depending on the immune status of the host (1, 17, 35). Nonribosomal peptide synthesis (NRPS) is a key mechanism for the biosynthesis of bioactive metabolites in bacteria and fungi (45, 50) and has been found to contribute to A. fumigatus virulence (1). Sequencing of the A. fumigatus genome has revealed the presence of at least 14 genes encoding putative nonribosomal peptide (NRP) synthetases (16, 50). NRP synthetases are modular or multimodular enzymes that direct the biosynthesis of NRPs independently of the ribosome (38, 66). Together with polyketides and terpenoids, NRPs constitute a class of structurally diverse secondary metabolites, which are generally considered to be components of the chemical arsenal necessary for adaptation to ecological niches (11, 22).
Each module within an NRP synthetase is responsible for the recognition, via an adenylation (A) domain, and incorporation of a single amino acid into the cognate peptide product (66); however, the basis of this recognition system is poorly understood. Furthermore, elucidating the specific NRP synthetase responsible for directing the biosynthesis of an NRP is nontrivial and defining the primary biochemical function of the NRP is challenging. Therefore, because few fungus-specific A domain substrates and NRP roles, particularly in A. fumigatus, have been established to date (20), it is imperative to assign functionality to the fungal NRP products. A few A. fumigatus NRP synthetases encoding secreted or intracellularly located peptides have been elucidated to date, mainly facilitated through targeted gene deletion and comparative metabolomic studies. Two NRP synthetases, SidC and SidD, were found to be essential for siderophore biosynthesis in A. fumigatus (58, 60). GliP, a bimodular NRP synthetase, was shown to be responsible for gliotoxin biosynthesis (5, 15); however, the contribution to organismal virulence remains to be definitely established. FtmA (brevianamide F synthetase) makes the diketopiperazine scaffold, brevianamide F, which is the precursor of the fumitremorgin family of tremorgenic secondary metabolites (39). A multimodular NRP synthetase pes1 was identified to be important for virulence and protection against oxidative stress (57). Importantly, where virulence studies have been undertaken, the disruption of a particular NRP synthetase has generally led to virulence attenuation, highlighting the importance of NRPS in the pathogenicity of A. fumigatus and other fungi (51, 57, 60, 68). To our knowledge, no structural role for an A. fumigatus NRP has been identified or proposed to date.
pes3 (CADRE locus identifier, AFUA_5G12730 [http://www.cadre-genomes.org.uk/index.html]; NRPS8 [16]), encoding a multimodular NRP synthetase (8,515 amino acids), is the largest gene (25 kb) in A. fumigatus, representing 0.01% of the genome (16, 65, 66). In addition, a recombinant Pes3 module was a target for posttranslational modification via solid-phase 4′-phosphopantetheinylation, thereby confirming the likelihood of in vivo functionalization via this process (65). BLAST searching of pes3 revealed no sequence orthologs in any organism for which the full genome sequence is available, making it difficult to predict the NRP produced by Pes3 by comparison to other organisms (16, 66). Pes3 contains six A domains and is predicted to encode a peptide with 6 amino acids, if each A domain exhibits unique substrate specificity. However, it has been proposed that the nonlinear modular and domain arrangement of Pes3 may suggest repetitive or nonlinear use of modules (16), confounding in silico prediction of cognate NRP structure, location, and function (45). Phylogenomic and expression analysis of all A. fumigatus NRP synthetase genes revealed that pes3 exhibited a unique pattern of expression compared to all other A. fumigatus NRP synthetase genes, whereby pes3 transcripts were most abundant in ungerminated spores, in contrast to all other NRP synthetase genes examined (16). This suggested that pes3 may be involved in germination; however, no further characterization of pes3 was carried out during that study.
We set out here to identify the role of the NRP synthetase Pes3 in A. fumigatus and to assess its contribution to virulence. Disruption of pes3 (resulting in a Δpes3 strain) resulted in an augmented virulence phenotype in invertebrate and hydrocortisone acetate immunosuppressed models for invasive aspergillosis (IA). Unexpectedly, this virulence appears to be mediated by an immunologically silenced phenotype for Δpes3. We propose that Pes3 biosynthesizes an NRP with a structural role within A. fumigatus conidia that is essential for effective innate immune recognition.
MATERIALS AND METHODS
Strains, growth conditions, and general DNA manipulation.
A. fumigatus strains were grown at 37°C in Aspergillus minimal medium (AMM), and fungal culturing was carried out as described in Schrettl et al. (61). Fungal strains used in the present study are listed in Table 1.
Table 1.
Fungal strains and plasmid constructs
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| ATCC 46645 | Aspergillus fumigatus wild-type strain | 24 |
| pes3 | Δpes3 deletion mutant (pes3::ptrA) | This study |
| pes3C | pes3 complemented strain | This study |
| Plasmids | ||
| pSK275 | Plasmid containing ptrA cassette conferring resistance to pyrithiamine | 30 |
| pAN 8.1 | Aspergillus marker cassette conferring resistance to phleomycin (PgpdA::ble/tk::trpC) |
PCRs for generation of DNA manipulation constructs were performed using the Expand long-range template kit (Roche). For general cloning procedures, the bacterial strain Escherichia coli DH5α was used (33); this strain was cultivated in LB (1% Bacto tryptone, 0.5% yeast extract, 1% NaCl [pH 7.5]) medium. A. fumigatus genomic DNA was purified using a ZR fungal/bacterial DNA kit (Zymoresearch).
Gene deletion and complementation of A. fumigatus pes3.
A. fumigatus transformation was carried out according to the method of Tilburn et al. (70). For the disruption of pes3 and generation of the Δpes3 mutant strain, a bipartite marker technique was used (49). The strategy designed for pes3 disruption in the present study led to the deletion of 1.5 kb of pes3 at the 5′ region of the gene. In A. fumigatus Δpes3, the deleted region comprises amino acids 688 to 1163 of pes3.
Briefly, A. fumigatus ATCC 46645 was transformed with two DNA constructs, each containing an incomplete fragment of a pyrithiamine resistance gene (ptrA) (31, 32), fused to 1.2 and 1.3 kb of the pes3 coding sequences, respectively, which flanked the region to be deleted. The marker fragments shared a 557-bp overlap within ptrA, serving as a potential recombination site during transformation. Two rounds of PCR generated each fragment. Table 2 provides a complete list of the primers used in the present study. First, each flanking region was amplified from ATCC 46645 genomic DNA by using the primer pair oAfpes3-1 and oAfpes3-4 for flanking region A (1.2 kb) and the primer pair oAfpes3-2 and oAfpes3-3 for flanking region B (1.3 kb). Subsequent to gel purification, the fragments were digested with EcolCRI and HindIII, respectively. The ptrA selection marker was released from plasmid pSK275 (kindly provided by Sven Krappmann, Goettingen, Germany) by digestion with EcolCRI and HindIII and ligated with the two flanking regions A and B described above. For transformation, two overlapping fragments were amplified from the ligation products by using the primers oAfpes3-5 and optrA-2 for fragment C (2.6 kb) and the primers oAfpes3-6 and optrA-1 for fragment D (2.3 kb). Subsequently, A. fumigatus ATCC 46645 was transformed simultaneously with the overlapping fragments C and D. In order to obtain homokaryotic transformants, colonies from single homokaryotic spores were picked, and single genomic integration was confirmed by PCR (data not shown) and Southern blot analysis.
Table 2.
Oligonucleotides used in this study
| Primer | Orientationa | Sequence (5′–3′) | PCR usage |
|---|---|---|---|
| oAfpes3-1 | F | GAGGCGGAACGTTGGAA | PCR 1, first-round PCR |
| oAfpes3-4 | R | CAGTGCTATGTTCCGCCAC | pes3 5′DIG probe |
| oAfpes3-3 | F | TTGGAGATGCGGTACTCG | |
| oAfpes3-2 | R | AAGCTGCGCTTCAACCTC | PCR 2, first-round PCR |
| oAfpes3-5b | F | GGAACCGATCACTCAAGAC | |
| optrA-2 | R | CATCGTGACCAGTGGTAC | PCR 3, second-round PCR |
| optrA-1 | F | GAGGACCTGGACAAGTAC | |
| oAfpes3-6b | R | CGACGTAGATGAGCGATC | PCR 4, second-round PCR |
| oAfpes3-5b | F | GGAACCGATCACTCAAGAC | |
| Afpes3Comp1 | R | CCAGAGATCGGCTAAGTGTCA | pes3 Comp 5′ |
| Afpes3Comp2 | F | CGGAGGTTTTATTAGACGCCTG | |
| oAfpes3-6b | R | CGACGTAGATGAGCGATC | pes3 Comp 3′ |
| Pes3RT-F | F | CTCTGGCACTCCTCCAAGTC | |
| Pes3RT-R | R | AGCAATCTATCCCACGGATG | pes3 RT-PCR |
| oAfpes3-5b | F | GGAACCGATCACTCAAGAC | |
| oAfpes3-6b | R | CGACGTAGATGAGCGATC | 3.7-kb pes3 replacement construct |
| calm F | F | CCGAGTACAAGGAAGCTTTCTC | calm RT-PCR |
| calm R | R | GAATCATCTCGTCGATTCGTCGTCTCAGT |
F, forward; R, reverse.
Some primers were used for multiple purposes as indicated.
For complementation of A. fumigatus Δpes3, a bipartite strategy was also used to precisely replace the deleted region in Δpes3, thus leading to pes3C. Two overlapping PCR fragments were amplified by using the primer pairs oAfpes3-5/Afpes3Comp1 (fragment 1) and Afpes3Comp2/oAfpes3-6 (fragment 2) (Table 2). These fragments overlapped by 405 bp, and recombination of these fragments should restore pes3, resulting in the removal of ptrA from the genome of pes3C. To achieve this, A. fumigatus Δpes3 was cotransformed with fragments 1 and 2 and a plasmid, Pan8.1, containing a phleomycin resistance gene (bler) for selection. Positive deletion and reconstituted strains were screened by Southern analysis, and digoxigenin-hybridization probes were generated by using the primers oAfpes3-5 and oAfpes3-4 (Table 2). Complementation of pes3 was further verified by restriction mapping part of the pes3 locus. PCR was carried out using the primer pair oAfpes3-5 and oAfpes3-6, and either wild-type, Δpes3 or pes3C genomic DNA as a template, leading to PCR products spanning the specific area of pes3 that was targeted for deletion (3,737 bp in wild-type and pes3C strains and 4,285 bp in the Δpes3 strain). These PCR products were restricted with AgeI to yield a range of expected fragment sizes as detailed in Table 3.
Table 3.
Restriction mapping expected fragments
| DNA type | Fragment sizes (bp) |
|---|---|
| Wild-type genomic DNA | 1,197, 1,053, 694, 569, 224 |
| Δpes3 genomic DNA | 2,197, 1,170, 694, 224 |
| Complemented pes3 strain (pes3C) | 1,197, 1,053, 694, 569, 224 |
RNA isolation and real-time PCR.
Fungal RNA was isolated and purified from A. fumigatus hyphae crushed in liquid nitrogen by using the RNeasy plant minikit (Qiagen). RNA was treated with DNase 1 (Invitrogen), and cDNA synthesis from mRNA (500 ng) was performed using a first-strand transcription cDNA synthesis kit (Roche) with oligo(dT) primers. The gene encoding calmodulin (calm) (AFUA_4G10050), which is constitutively expressed in A. fumigatus, served as a control in reverse transcription-PCR (RT-PCR) experiments (10).
Real-time PCR was performed using LightCycler 480 Sybr green 1 master mix (Roche) on a LightCycler 480 real-time PCR system. PCRs were carried out in 96-well plates in a reaction volume of 20 μl containing 5 μl of template cDNA. Standard curves were prepared for calm and pes3 by generating 5 orders of 10-fold serial dilutions of cDNA in H2O and performing five replicate PCRs on these dilutions. The primer pairs calm F/calm R or Pes3RT-F/Pes3RT-R were used for real-time PCR experiments (Table 2). Cycling conditions for PCRs were calculated using the recommendations from Roche, and 40 cycles of PCR were performed. Standard curves with a PCR efficiency of ≥1.8 and with an error value of <0.2 were accepted, and the cycling conditions were used for subsequent real-time PCR analysis by relative quantification using LightCycler 480 software. For real-time PCRs, a 1/10 dilution of cDNA from each sample was used as a template, and each reaction was performed in triplicate.
Metabolite extraction and analysis by reversed-phase HPLC.
Metabolite extraction was performed according to the micro extraction procedure of Smedsgaard (64). Plugs (0.6 cm2) were taken from petri dish cultures and extracted with either 1 ml of methanol-dichloromethane-ethyl acetate (1:2:3 [vol/vol/vol]) or 1 ml of 25% (vol/vol) CH3CN in water. These extracts were evaporated, and the residues were redissolved in 400 μl of methanol and stored at −20°C until analysis. Ultra-high-performance liquid chromatography-diode array detection (uHPLC-DAD) analyses were performed on a Dionex RSLC Ultimate 3000 (Dionex, Sunnyvale, CA). Separation was obtained on a Kinetex C18 column (150 by 2.1 mm, 2.6 μm; Phenomenex, Torrance, CA) maintained at 60°C using a linear gradient starting from 15% (vol/vol) CH3CN in water (containing 50 ppm of trifluoroacetic acid) increasing to 100% (vol/vol) CH3CN over 7 min at a flow rate of 0.8 ml/min. The injection volume was 1 μl. LC-DAD-mass spectrometry (MS) analysis was performed on an LCT oaTOF mass spectrometer (Micromass, Manchester, United Kingdom) as described by Nielsen et al. (47, 48). Chromatography was performed on a 5 cm, 3 μm, Luna C18 column (Phenomenex) (2) using a water-acetonitrile gradient from 15% (vol/vol) CH3CN to 100% (vol/vol) CH3CN in 20 min, with both solvents containing 20 mM formic acid.
Phenotypic analysis of A. fumigatus Δpes3.
For plate assays, conidia were harvested aseptically from 1-week-old malt extract agar plates and filtered through sterile Miracloth (Calbiochem) to remove mycelial matter. Conidia (104) were spotted onto a variety of test plates as shown in Table 4. Plates were incubated at 37°C unless otherwise stated. Colony diameter was measured periodically, and statistical analysis was carried out using two-way analysis of variance.
Table 4.
Phenotypic plate assay agents and observationsa
| Phenotypic test | Reagent(s) used | Concn(s) tested | Phenotype |
|---|---|---|---|
| Role of pes3 in siderophore biosynthesis | Iron stresses (low, high, and no iron) | 10 μM, 1.5 mM, and 200 μM BPS | N |
| Oxidative stress | Menadione | 20, 30, 40 μM | N |
| Diamide | 0.1, 0.2, 0.4, 1, and 2 mM | Y (Δpes3 more resistant to diamide [0.1 mM, P < 0.05] than the wild type) | |
| Hydrogen peroxide | 1, 2, and 3 mM | N | |
| Antifungal susceptibility | Voriconazole | 0.25, 0.5, 0.75, and 1.0 μg/ml | Y (Δpes3 more sensitive to voriconazole [0.5 μg/ml, P < 0.001] than the wild type) |
| Amphotericin B | 0.125, 0.25, 0.5, and 1.0 μg/ml | N | |
| Caspofungin | 0.2, 0.5, and 1.0 μg/ml | N | |
| Cell wall stress | Caffeine | 2 and 5 mM | N |
| Congo red | 5, 10, and 15 μg/ml | N | |
| Calcofluor white | 100 and 200 μg/ml | N | |
| High temp (48°C) | NA | N | |
| Membrane stress | SDS | 0.01 and 0.02% (wt/vol) | N |
BPS, bathophenanthroline sulfonate; SDS, sodium dodecyl sulfate; NA, not applicable; Y, yes; N, no.
Vegetative growth curves were performed according to the method of Reeves et al. (56). Freshly harvested conidia were used to inoculate AMM cultures (107 conidia/100 ml). Cultures were incubated for 24, 48, 72, or 96 h, and the mycelial biomass was collected. Mycelia were washed in sterile water and freeze-dried to remove all liquid. Dry weights were recorded, and statistical analysis was performed using a Student t test.
To determine germination rates, under shaking and static conditions, AMM cultures (10 ml) were inoculated with A. fumigatus conidia (107/ml) and incubated for at least 3 h to allow germination to commence. Subsequently, 500 μl of each culture was transferred into a 2-ml screw-cap tube containing a 0.2-ml scoop of 0.1-mm glass beads, and the tubes were vortexed for 10 s. To quantify germination, 10 μl of each specimen was placed on a hemocytometer, and conidia and germlings were counted up to 100 in total. Swollen conidia were not counted as germinated. Each specimen was counted in duplicate, and the assay was performed in triplicate. Germination was recorded every hour until germination rates became constant for all strains tested. Voriconazole (VFEND; Pfizer) MIC analysis was carried out according to EUCAST guidelines (54).
Galleria mellonella infection experiments.
G. mellonella larvae (n = 20) were inoculated into the hind proleg with 107 A. fumigatus conidia in 20 μl (per larva) as previously described (56). Mortality, defined by lack of movement in response to stimulation and discoloration (melanization) of the cuticle, was recorded at 72 h postinjection. Kaplan-Meier survival curves were analyzed by using a log-rank (Mantel-Cox) test for significance.
Murine infection experiments.
Outbred male mice (strain CD1, 20 to 28 g; Charles Rivers Breeders) were used for murine virulence analyses. For analysis of the fungal burden in neutropenic mice, Immunosuppression was carried out by subcutaneous injection of 112 mg of hydrocortisone acetate/kg on day −1 and by intraperitoneal injection of 150 mg of cyclophosphamide/kg on days −3, −1, and 2. For analysis of the fungal burden in corticosteroid-treated animals immunosuppression was induced with subcutaneous injections of hydrocortisone acetate (112 mg/kg on days −3, −1, and 2). On day 0, inocula containing 7.5 × 105 or 1.5 × 106 conidia were prepared for neutropenic and corticosteroid-treated animals, respectively, in 40 μl of saline. Spores were cultured on slants of solid ACM complete medium for 5 days at 37°C prior to harvest. Spores were filtered through Miracloth and washed twice with saline prior to enumeration. Mice were anesthetized by inhalation of isoflurane and infected by intranasal instillation. Bacterial infections were prevented by adding 1 g of tetracycline/liter and 64 mg of ciprofloxicin/liter to drinking water. The weights of the infected mice were monitored for 4 days, twice daily, before the animals were culled. An A. fumigatus para-aminobenzoic (PABA) acid auxotroph (referred to as A. fumigatus H515) that is unable to germinate in the murine lung (8) was used as a positive control for attenuated virulence. For histological analyses of ATCC 46645 and Δpes3 infections, corticosteroid-treated animals were immunosuppressed and infected with 107 spores as described above. The animals were sacrificed at 14 and 24 h postinfection, and the lungs were fixed in 4% (vol/vol) formalin prior to paraffin embedding. Lung sections (n = 4) were obtained at four independent vertical sectioning levels per sample and stained with Grocotts methanamine silver (GMS) for visualization of fungal elements and with hematoxylin and eosin (H&E) for visualization of the host cells.
Measurement of the fungal burden in murine lungs.
In order to assess fungal burden as a quantitative virulence criterion, quantitative PCR was performed on total genomic DNA extracted from homogenized whole lungs. The fungal burden was assessed by quantitative PCR amplification using the oligonucleotides BT-F (GAGCCCTTTTCCGACCTGAT) and BT-R (GGAACTCCTCCCGGATCTTG) to amplify the β-tubulin (AFUA_7G00250) locus from the A. fumigatus genome, and the oligonucleotides Actin-F (CGAGCACAGCTTCTTTGCAG) and Actin-R (CCCATGGTGTCCGTTCTGA) were used to amplify the murine actin locus (NM_007393). Quantification of both gene targets was performed in triplicate on groups of four to five mice. A modified mathematical model (incorporating adjustment for fungal inoculum) for relative quantification of the A. fumigatus actin and murine β-tubulin was adopted for each mouse sample using the formula ratio (R) = (Etarget)ΔCT(fungal actin − murine tubulin), where E is the efficiency of amplification of target gene and is equal to 2, based on the mean of eight independent murine infections (53). The R values were log10 transformed and corrected for inoculum size, and reciprocal values were plotted (1/log10 RC). The statistical significance of variances between fungal burdens was calculated by using a nonparametric Mann-Whitney t test. Genomic DNA, isolated from animals infected with attenuated A. fumigatus H515 (8), was used as a positive control for attenuation.
Fungal stimulation of BMMφ.
Immortalized murine bone marrow-derived macrophages (BMMφ) (62) were cultured in Dulbecco modified Eagle medium (Gibco) supplemented with 10% (vol/vol) fetal calf serum 1% (wt/vol) penicillin-streptomycin, and 0.5 ml of gentamicin solution. Cells were cultured in T175 flasks at 37°C with 5% CO2. For stimulation with A. fumigatus, BMMφ were washed in phosphate-buffered saline (PBS), counted, seeded onto 24-well plates containing Opti-MEM at a cell density of 106 cells per well, and left to adhere to 24-well plates overnight. A. fumigatus conidia (107) or germlings (5 × 106) were added to BMMφ, followed by incubation for 18 h at 37°C. Supernatants were subsequently collected for cytokine determination by enzyme-linked immunosorbent assay (ELISA). Germling formation was achieved as described above. Preparatory experiments indicated that between 90 and 100% of conidia had formed dichotomous germ tubes after 9 h of incubation. BMMφ were also exposed to filter sterilized culture supernatant (600 μl) from A. fumigatus strains. Aliquots of filter-sterilized Aspergillus culture supernatant were inoculated onto malt extract agar, and lack of fungal growth after 72 h confirmed that the supernatant was completely cell-free. Commercially available ELISA systems for tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), IL-10, and RANTES determination (Peprotech UK) were used according to the manufacturer's instructions.
SEM.
Dormant conidia and germlings (9 h) were Miracloth filtered and fixed using formaldehyde and glutaraldehyde in 100 mM sodium phosphate (pH 7.0) at room temperature (2 h) and 4°C (16 h). After being washed in 50 mM NH4Cl and deionized water, the specimens were analyzed by scanning electron microscopy (SEM) by commercial arrangement (CMA, Trinity College Dublin, Dublin, Ireland).
Confocal microscopy.
Germlings (9 h) were fixed with 4% (vol/vol) formaldehyde prior to two washes with PBS and blocking with bovine serum albumin (BSA; 50 mg/ml) in PBS. Incubation with murine monoclonal antibody to linear-(1,3)-β-glucan (Biosupplies; 1 μg/ml) for 1 h followed (13). The specimens were then washed twice with BSA (50 mg/ml) in PBS prior to staining with Alexa Fluor 488 goat anti-mouse IgG (H+L; Invitrogen) at 1/200 for 1 h. Germlings were washed twice with BSA (50 mg/ml) in PBS before image acquisition using an Olympus FluoView 1000 confocal microscope. Images were processed with FV10-ASW Software (v.01.07).
2D-PAGE and protein MS.
A. fumigatus ATCC 46645 and Δpes3 germlings (9 h) were harvested, washed, and disrupted in lysis buffer (1 ml/g germlings) (12) using bead beating (30 Hz; three times for 5 min each time on ice) in the presence of glass beads (0.4 g; 0.1 mm), followed by centrifugation (13,000 × g, 10 min), two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) fractionation (pH 4 to 7 Immobiline dry strips), and image analysis using Progenesis software (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom). Protein identification was carried out using a model 6340 ion trap LC mass spectrometer (Agilent Technologies, Santa Clara, CA) in which peptide separation was achieved by using a Nanoflow Agilent 1200 LC system and proteins were identified using Mascot MS/MS ion search (Matrix Science, London, United Kingdom).
RESULTS
Disruption and complementation of pes3 in A. fumigatus.
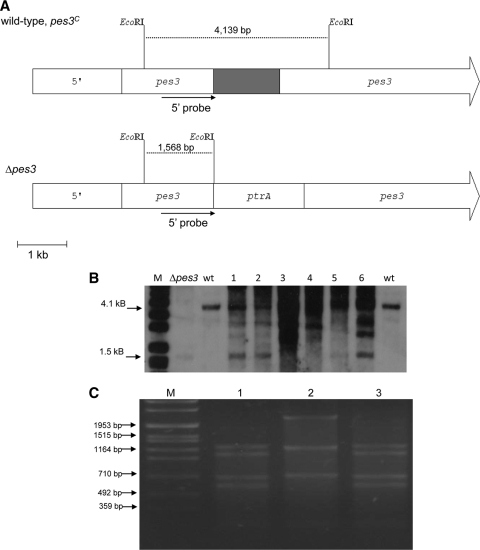
In order to gain insight into the biological role of Pes3, a pes3 mutant strain (Δpes3) was generated by homologous recombination by using the bipartite marker technique (49). Southern blot analysis was used to identify pes3 (negative) and ptrA (positive) colonies by probing for a 1,568-bp EcoRI restriction fragment in the Δpes3 strain and a 4,139-bp fragment in the wild-type strain (Fig. 1 A). Southern analysis of 14 isolates confirmed the disruption of pes3, and one representative clone was selected for further analysis. A complemented strain, pes3C, was generated by cotransformation of A. fumigatus Δpes3 with bipartite pes3 fragments and plasmid Pan8.1 using selection on phleomycin-containing media (Fig. 1B). Southern blot analysis of the complemented strains (pes3C) produced the expected wild-type 4,139-bp fragment due to restoration of the pes3 locus (Fig. 1A and B). Restriction mapping of part of the pes3 locus also confirmed the integrity of the pes3 locus following complementation (Fig. 1C). Two complemented strains (pes3C), termed pes3C1 and pes3C2, exhibited wild-type features in all subsequent experiments. RT-PCR and real-time PCR confirmed that pes3 transcripts were absent in the Δpes3 strain but present in wild-type and pes3C strains (Fig. 2). The culture conditions used for pes3 expression analysis were as described previously by Cramer et al. (16). pes3 transcripts were detected in wild-type and pes3C strains, but not in Δpes3 cultures, following 48 h of growth in RPMI medium (Fig. 2). The resulting set of strains was subjected to phenotypic analysis under various growth conditions (Table 4).
Fig. 1.
Disruption and complementation of A. fumigatus pes3. (A) Schematic representation of Southern blot and probe hybridization, displaying the pes3 loci in A. fumigatus wild-type, Δpes3, and pes3C strains. A 1.5-kb region close to the 5′ end of pes3 (gray) was deleted and replaced by ptrA as indicated. EcoRI-restricted DNA was probed with a digoxigenin (DIG)-labeled probe corresponding to a region at the 5′ end of pes3, leading to fragments of 4,139 bp in wild-type DNA and 1,568 bp in Δpes3 DNA, respectively. Probe hybridization region is indicated as black lines. The full pes3 coding sequence is 25.5 kb in length. For clarity, only a portion of pes3 is shown here, indicated by continuous arrows. (B) Southern blot analysis of A. fumigatus wild-type, Δpes3, and pes3C strains confirms pes3 disruption and subsequent complementation in A. fumigatus by the presence of the expected hybridization patterns as described in panel A. M, DIG-labeled marker VII (Roche); Δpes3, A. fumigatus Δpes3; wt, A. fumigatus wild type; lanes 3 and 4, pes3C1 and pes3C2. Loss of the Δpes3 fragment (1.5 kb) in lanes 3 and 4 indicates complementation of pes3. (C) AgeI restriction mapping of the region with pes3 deleted yields the expected restriction pattern (Table 3), confirming the integrity of pes3 locus following complementation. M, Roche VII marker; lanes 1 to 3, AgeI-digested PCR products (lane 1, A. fumigatus wild type; lane 2, Δpes3, lane 3, pes3C1).
Fig. 2.
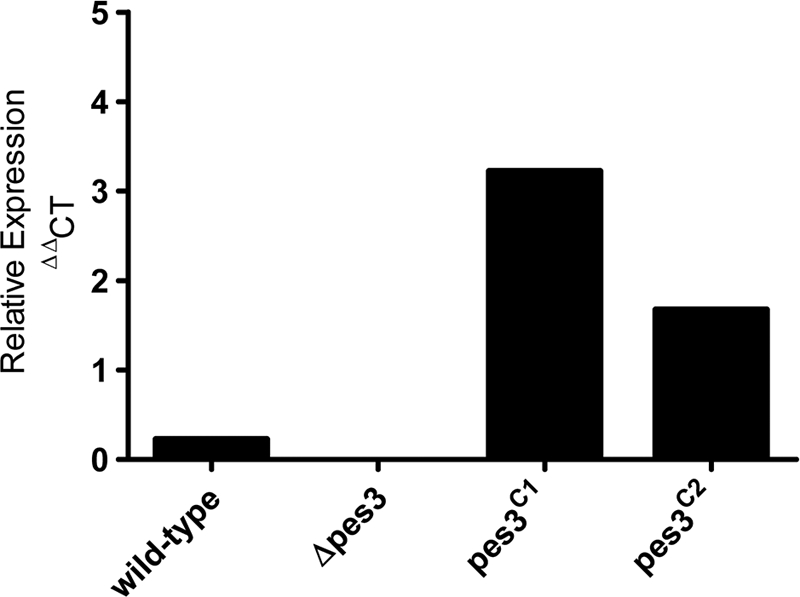
Real-time PCR analysis of pes3 expression. A. fumigatus wild-type, Δpes3, pes3C1, and pes3C2 strains were cultured for 48 h in RPMI medium, and real-time PCR analysis was undertaken on resultant cDNA samples. The relative abundances of pes3 and a housekeeping gene (calm) transcripts are given. The lack of pes3 expression (Δpes3) and the restoration of pes3 expression (pes3C1 and pes3C2) can be observed. The means of three replicate PCRs for each strain are shown.
Phenotypic analysis of A. fumigatus Δpes3 reveals voriconazole susceptibility and diamide resistance.
Antifungal susceptibility testing indicated that strain Δpes3 exhibited increased susceptibility to the azole, voriconazole. The addition of voriconazole (0.25 to 1.0 μg/ml) led to a significantly reduced growth phenotype of A. fumigatus Δpes3 compared to wild-type A. fumigatus, and growth was restored in pes3C. This increased sensitivity of Δpes3 toward voriconazole was most significant at 0.5 μg of voriconazole/ml at 62 and 72 h postinoculation (P < 0.001) (Fig. 3 A). The MICs of voriconazole were 0.125 and 0.0625 μg/ml for the A. fumigatus ATCC 46645 and Δpes3 strains, respectively. A. fumigatus Δpes3 grew at a rate identical to that of the wild type upon exposure to other antifungal agents (amphotericin B and caspofungin). With the exception of enhanced resistance to diamide (P < 0.05), A. fumigatus Δpes3 grew at a rate identical to that of the wild type under all other conditions tested (Table 4).
Fig. 3.
A. fumigatus Δpes3 is more sensitive to voriconazole compared to the wild type, but the growth rate is not affected in A. fumigatus Δpes3. (A) Conidia (104) of A. fumigatus wild-type and Δpes3 strains were spotted onto AMM plates in the presence or absence of voriconazole (0.25 to 1.0 μg/ml). Colony diameter was measured after 72 h of incubation at 37°C. The means ± the standard errors of three determinations are shown. ***, P < 0.001. (B) Germination rate analysis of A. fumigatus wild-type and Δpes3 strains indicates that both of them germinate at the same rate. Germination was first recorded in both strains at 3 h, and by 9 h ca. 90% of the conidia in both strains had germinated. The means ± the standard errors of three determinations are shown. (C) Analysis under static conditions confirmed that A. fumigatus Δpes3 germinates significantly slower than does the wild type (P < 0.01). Germination was first recorded in both strains at 6 h, and by 8 to 10 h only 10 to 20% of the mutant conidia had germinated. The means ± the standard errors of three determinations are shown. (D) Vegetative growth rates of A. fumigatus wild-type and Δpes3 strains over 96 h. No significant differences were observed between strains. The means ± the standard errors of three experiments are shown.
With the observation that pes3 transcripts were most abundant in ungerminated spores (16), germination and vegetative growth rates were compared for A. fumigatus wild-type and Δpes3 strains. Overall, A. fumigatus wild type and Δpes3 exhibited similar growth rates under shaking conditions (Fig. 3B). Conidial germination for both strains commenced after 3 h growth in liquid AMM, with ca. 5% germination for both strains by 5 h. By 6 h, germination had commenced for ca. 25 and 35% of the wild-type and Δpes3 conidia, respectively. Germination of A. fumigatus Δpes3 was significantly impeded under static conditions at 8 and 10 h, respectively (P < 0.01); moreover, only 10 to 20% of the conidia had germinated by 10 h (Fig. 3C). Vegetative growth determination by measuring the dry weights of the mycelial biomass for A. fumigatus wild-type and Δpes3 strains over a 96-h period indicated that both strains exhibited similar growth rates, with the exponential growth phase lasting until 48 h, and the growth rates remained stationary after this point (Fig. 3D).
pes3 does not encode a secreted peptide.
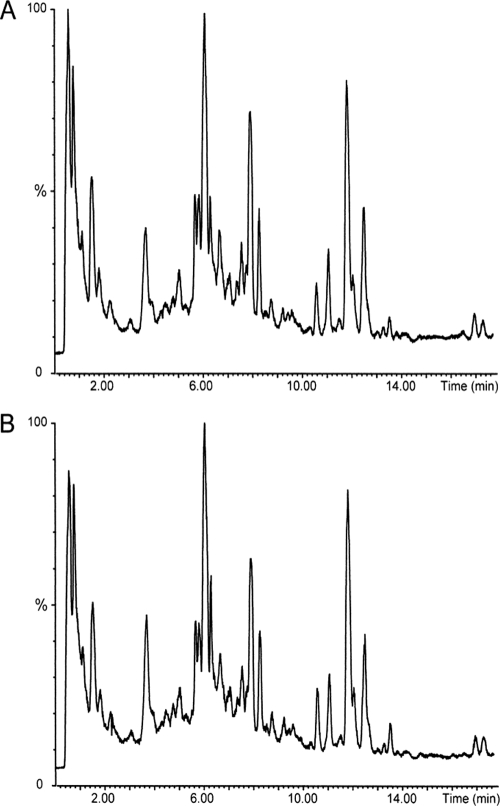
Availability of the A. fumigatus Δpes3 mutant facilitated comparative metabolite analysis to identify a Pes3-encoded NRP. To this end, A. fumigatus wild-type and Δpes3 were cultivated in a variety of conditions, including all conditions (Table 5) under which pes3 expression was detected. Comparative metabolite analysis, via LC-MS, of extracts from A. fumigatus wild-type and Δpes3 strains were identical and showed that there were no differences in the types or extents of metabolite production between both strains (Fig. 4). The fact that no products and/or peaks were missing in mutant samples, when analyzed using electrospray in the positive mode during MS, strongly indicates that pes3 is not responsible for the synthesis of a soluble NRP that is secreted by, or stored intracellularly within, A. fumigatus, since such a product should easily be protonated and thereby detected.
Table 5.
Conditions used for comparative metabolite analysis
| Condition | Description (all at 37°C) | Extraction solvent |
|---|---|---|
| A | RPMI liquid, 48 h of incubation | (i) Organic extraction of culture supernatant (SN) with SN-chloroform in a 1:1 ratio |
| B | Czapek liquid, 48 h of incubation | (ii) Neat culture supernatant injection |
| C | AMM liquid, 48 h of incubation | |
| D | Incubation on AMM agar for 6 days | Conidial extraction method (44) |
| E | Incubation on AMM supplemented with H2O2 (final concn, 2 mM) for 6 days | |
| F | Incubation on AMM agar for 6 days | Plug extraction method (64) using either 1 ml of methanol-dichloromethane-ethyl acetate (1:2:3 [vol/vol/vol]) or 1 ml of 25% (vol/vol) CH3CN in water |
| G | Incubation on Czapek agar for 6 days |
Fig. 4.
A. fumigatus wild-type and Δpes3 strains exhibit identical metabolite profiles, indicating that Pes3 does not encode a secreted peptide. Total ion chromatograms (0 to 15 min) representative of metabolites detected from A. fumigatus wild-type (A) and Δpes3 (B) strains after 6 days of growth on Czapek agar. Metabolites were extracted according to condition G (see Table 5) with 1 ml of 25% (vol/vol) CH3CN in water.
Disruption of pes3 leads to increased virulence in G. mellonella and a murine model of pulmonary aspergillosis.
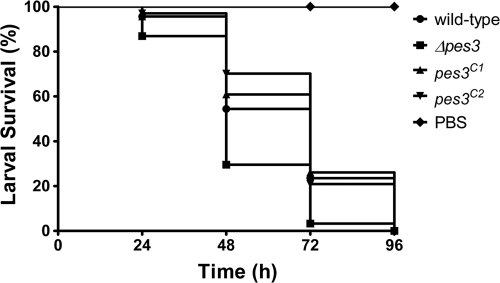
G. mellonella larvae infected with A. fumigatus wild type exhibited significantly lower mortality compared to those infected with A. fumigatus Δpes3 (P < 0.001), thereby revealing heightened virulence of the mutant strain. The percent larval survival is shown in Fig. 5. At 24 h after infection, ca. 95% of the larvae infected with the wild-type strain remained alive, whereas an 85% survival rate was observed for those infected with A. fumigatus Δpes3, indicating increased mortality associated with A. fumigatus Δpes3. At 48 h postinfection, 55% of the larvae infected with the wild type remained alive, in contrast to only 30% of larvae infected with Δpes3. By 72 h, only 3% of larvae infected with Δpes3 were viable versus 23% in the wild-type group. Larval survival levels associated with A. fumigatus wild-type were restored in both complemented strains (pes3C1 and pes3C2), and the overall differences in survival proportions between larvae infected with A. fumigatus wild type or Δpes3 is highly significant (P < 0.001).
Fig. 5.
Disruption of A. fumigatus Δpes3 leads to hypervirulence in an invertebrate model of invasive aspergillosis. The survival proportions of larvae (n = 20) infected with A. fumigatus wild-type, Δpes3, and pes3c strains are shown. Larval viability (%) was assessed at 24-h intervals after infection. A. fumigatus Δpes3 is more virulent than the wild type and is associated with reduced larval survival (P < 0.001) at all time points. Virulence is restored to wild-type levels in the two complemented strains (pes3C1 and pes3C1). PBS was used as an injection control, and all larvae in this group remained viable for the entire experiment.
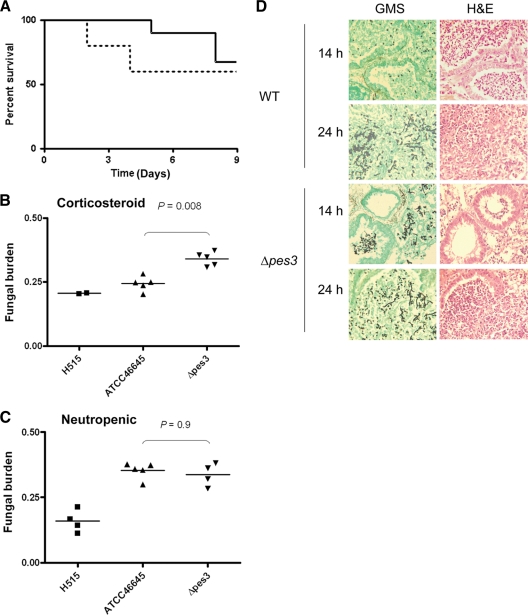
Two murine models of pulmonary aspergillosis were used to assess the comparative virulence of the A. fumigatus wild-type and Δpes3 strains by analysis of the fungal burden. Groups of five corticosteroid-treated or neutropenic animals were infected with 7.5 × 105 or 1.5 × 106 conidia, respectively. Comparative survival (A. fumigatus ATCC 46645 versus Δpes3) was assessed in a total of 10 mice for each strain (in two independent experiments). Although there was a tendency toward increased virulence of A. fumigatus Δpes3, the effect was not significant as determined by log-rank analysis (P = 0.32) (Fig. 6 A). However, corticosteroid-treated animals infected with Δpes3 demonstrated a rapid decline in weight relative to the day of infection; every animal infected with A. fumigatus Δpes3 exhibited weight loss over the course of the infection. Two A. fumigatus Δpes3-infected animals exhibited >20% weight loss by day 4 of the experiment, whereas animals infected with A. fumigatus wild type displayed moderate fluctuations in weight during infection, but none dropped below 20% of starting weight (see Fig. S1 in the supplemental material). The mean weights of animals at day 5 of the infection were significantly lower for the A. fumigatus Δpes3 infection group (93.00 ± 2.5% of the starting weight) than the wild-type counterpart (106.2% ± 1.62% of the starting weight) (P < 0.01) (see Fig. S1 in the supplemental material). Animals in the A. fumigatus H515 control group remained at a steady weight over the course of the experiment (see Fig. S1 in the supplemental material). The fungal burden was quantified, relative to murine β-tubulin, by quantitative PCR using genomic DNA isolated from whole, homogenized murine lung harvested from animals sacrificed on day 5. An attenuated p-amino benzoic acid auxotroph, H515 (8), was included as a control for attenuation. Infection of hydrocortisone acetate-treated animals with strain Δpes3 resulted in significantly heightened fungal burden (Fig. 6B, P = 0.008) relative to infection with ATCC 46645. In contrast, the fungal burdens following infection of neutropenic mice with Δpes3 and ATCC 46645 were comparable (Fig. 6C, P = 0.9).
Fig. 6.
Analysis of murine pulmonary aspergillosis after infection with A. fumigatus wild-type and Δpes3 conidia. (A) Survival of corticosteroid-treated mice (n = 10 per group) after infection with 106 A. fumigatus wild-type (solid line) or Δpes3 (dotted line) spores. (B and C) Fungal burden analysis corticosteroid (B) and neutropenic (C) models of pulmonary aspergillosis indicate a significantly heightened fungal burden in Δpes3-infected corticosteroid treated mice. The lungs of animals infected with Δpes3 exhibited greater fungal burden as observed by a lower threshold cycle (CT) value for A. fumigatus Δpes3 (P = 0.008). The fungal burden was quantified, relative to murine β-tubulin, by quantitative PCR using genomic DNA isolated from whole, homogenized murine lungs harvested from animals sacrificed on day 5. An attenuated p-amino benzoic acid auxotroph, H515 (8), was included as a control for attenuation. (D) Histological analysis of wild type- and Δpes3-infected mice demonstrates rapid germination of Δpes3 spores in the lungs of corticosteroid-treated animals. Animals were sacrificed at 14 and 24 h postinfection, and the lungs were fixed in 4% formalin prior to paraffin embedding. Lung sections (n = 4) were obtained at four independent vertical sectioning levels per sample and stained with GMS for visualization of the fungal elements and with H&E for visualization of the host cells.
In order to determine the basis of increased fungal burden in Δpes3-infected animals, we studied the histological progression of aspergillosis in the lungs of corticosteroid-treated animals at 14 and 24 h postinfection (Fig. 6D). The major difference between wild-type and Δpes3 infections was earlier germination of the Δpes3 spores. At 14 h postinfection, the majority of Δpes3 spores had germinated and produced germ tubes. Germinating spores were observable in the lung alveoli and in the bronchioles, and inflammatory cell infiltrates were observed in regions of spore deposition (Fig. 6D). In contrast, lungs infected with the wild-type isolate exclusively harbored ungerminated spores at 14 h postinfection. Large regions of inflammatory infiltration were observable in which the ultrastructure of the lung tissue was barely visible and spores were distributed throughout the lung parenchyma, including within the bronchioles that were heavily infiltrated with inflammatory cells. At 24 h postinfection, the observed disparate germination was limited to areas of high inflammation. Although a trend toward lessened inflammation in Δpes3-infected lungs was clearly discernible, the difference between wild type- and Δpes3-infected lungs was not dramatic, and quantitation of the difference was therefore not attempted from the lung sections. In summary, the heightened virulent phenotype observed for Δpes3 in the hydrocortisone acetate model is associated with a reduction in weight loss and an increase in fungal burden in the murine lung.
A. fumigatus Δpes3 exhibits an immunologically silent phenotype compared to the wild type.
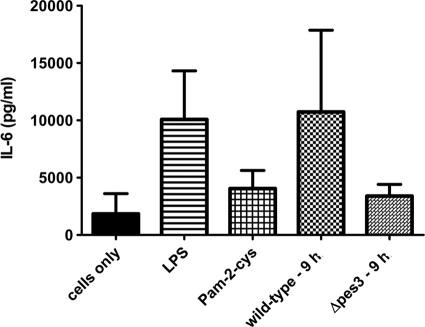
In order to test the hypothesis that increased virulence of A. fumigatus Δpes3 in both insect and murine models could be due to immunological silencing and a consequent attenuated immune response, we monitored cytokine production following coincubation of BMMφ with A. fumigatus 9-h germlings. This analysis revealed a reduction in four cytokines TNF-α (30% reduction; 2,592 ± 290 versus 3,686 ± 1,719 pg/ml [mean ± the standard deviation]), IL-6 (68% reduction), RANTES (25% reduction), and IL-10 (10% reduction) after coincubation of BMMφ with A. fumigatus Δpes3 compared to coincubation of BMMφ with the wild type (Fig. 7; see Fig. S2 in the supplemental material). The level of TNF-α produced in response to Δpes3 germlings resembled that of the response toward A. fumigatus conidia, either live or dead (2,000 to 2,500 pg/ml), and similar levels of TNF-α were produced in response to A. fumigatus wild-type and A. fumigatus Δpes3 conidia. Cytokines were not detected from BMMφ in response to A. fumigatus wild-type or Δpes3 culture supernatants (data not shown). These data suggest that the recognition of Δpes3 by pattern recognition receptors (PRRs) of the innate immune system may be less efficient than recognition of the wild type, leading to diminished cytokine secretion in BMMφ upon coincubation with A. fumigatus Δpes3, and this may, in part, explain the increased virulence and increased fungal burden compared to Δpes3.
Fig. 7.
Coincubation of murine macrophages with A. fumigatus Δpes3 leads to a reduction in IL-6 secretion compared to A. fumigatus wild type. BMMφ were stimulated with the TLR ligands lipopolysaccharide (LPS; TLR4 ligand) and Pam-2-cys (TLR2 ligand), as well as A. fumigatus wild-type and Δpes3 germlings/conidia for 9 h. Cytokine secretion was then assessed by ELISA. Less IL-6 (68% reduction) was detected from BMMφ after incubation with A. fumigatus Δpes3 (Δpes3 - 9 h) compared to wild-type germlings (wild-type - 9 h). The means ± the standard errors of two biological replicates are shown.
Loss of pes3 impairs germ tube formation.
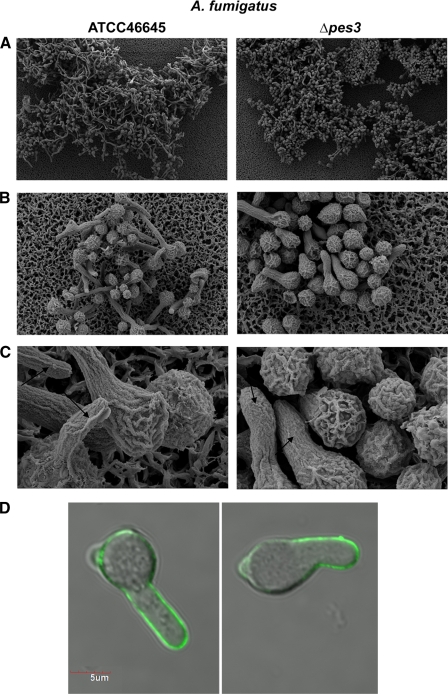
SEM pretreatment resulted in visible germling aggregation for A. fumigatus Δpes3. Comparative SEM analysis revealed impeded germination of A. fumigatus Δpes3 germlings. The hyphae of mutant germlings appeared smoother, compared to the wild type (Fig. 8 A), and the rod-like strands apparent on the hyphal surface of wild-type germinating conidia were effectively absent from the shorter hyphae of A. fumigatus Δpes3. No structural differences were apparent between ungerminated A. fumigatus wild-type and Δpes3 conidia (data not shown).
Fig. 8.
SEM and confocal microscopy analysis of A. fumigatus ATCC 46645 and Δpes3 germlings (9 h). (A) A. fumigatus wild-type germlings exhibit longer germ tubes and undergo a higher degree of germination compared to Δpes3 (×500 magnification). (B) A. fumigatus wild-type germlings exhibit narrower germ tubes than Δpes3 germlings (×2,000 to ×3,000 magnification) (C) The surface appearance of A. fumigatus Δpes3 is smoother than that of the wild type and lacks distinctive rod-like strands (×10,000 magnification). (D) Surface β-glucan expression on germlings is significantly different between A. fumigatus wild-type and Δpes3 strains; less β-glucan is present on mutant germlings (P = 0.0325). The difference in germling length is also clearly evident.
A. fumigatus Δpes3 exhibits proteome alteration.
2D-PAGE, quantitative image analysis, and tandem MS (MS/MS) confirmed the up- and downregulation of 15 and 10 proteins, respectively, in A. fumigatus Δpes3 compared to the wild type (Table 6). Upregulated proteins (1.6- to 4.1-fold) included the Rab11 GTPase and ribosomal, heat shock, and electron transport chain proteins, while those downregulated (1.6- to 3.1-fold) included the pentafunctional polypeptide AroM (AFUA_1G13740) and spermidine synthase (AFUA_1G13490).
Table 6.
Comparative proteomic analysis of A. fumigatus wild-type versus Δpes3 germlings
| Protein | No.a | Annotation name | Gene identity | Fold changeb | tpIc | tMrd | No. of peptides matched | % Sequence coverage |
|---|---|---|---|---|---|---|---|---|
| Upregulated in A. fumigatus Δpes3 | 328 | Glycine dehydrogenase | AFUA_4G03760 | 1.8 | 6.71 | 116,116 | 11 | 12 |
| 698 | t-Complex protein 1, zeta subunit | AFUA_3G09590 | 2.2 | 6.31 | 59,081 | 2 | 6 | |
| 1108 | NGG1 interacting factor Nif3 | AFUA_6G12480 | 1.7 | 6.08 | 42,126 | 7 | 23 | |
| 1291 | ATP synthase gamma chain, mitochondrial precursor | AFUA_1G03510 | 1.8 | 8.31 | 32,530 | 14 | 47 | |
| 1352 | Glucosamine-6-phosphate deaminase | AFUA_1G00480 | 3.2 | 8.35 | 40,181 | 1 | 4 | |
| 1478 | GTP-binding nuclear protein Ran | AFUA_6G13300 | 1.9 | 6 | 23 | |||
| 1497 | Heat shock protein 70 | AFUA_1G07440 | 2.6 | 5.09 | 69,792 | 3 | 2 | |
| 1512 | Heat shock protein Hsp30/Hsp42 | AFUA_3G14540 | 1.6 | 6.09 | 20,450 | 7 | 46 | |
| 1534 | Ubiquinol-cytochrome c reductase iron-sulfur subunit precursor | AFUA_5G10610 | 1.8 | 9.20 | 33,111 | 12 | 33 | |
| 1547 | Ubiquinol-cytochrome c reductase iron-sulfur subunit precursor | AFUA_5G10610 | 2.7 | 9.20 | 33,111 | 1 | 7 | |
| 1557 | Ras GTPase Rab11 | AFUA_1G02190 | 3.6 | 5.95 | 22,568 | 8 | 43 | |
| 1562 | 40S ribosomal protein S5 | AFUA_1G15020 | 3.2 | 9.35 | 25,886 | 1 | 6 | |
| 1566 | Mitochondrial ATPase subunit ATP4 | AFUA_8G05440 | 2.0 | 9.36 | 29,767 | 9 | 33 | |
| 1668 | 60S Ribosomal protein L5 | AFUA_1G12890 | 4.1 | 8.59 | 35,519 | 3 | 14 | |
| 1771 | Molecular chaperone Hsp70 | AFUA_1G07440 | 2.1 | 5.09 | 69,792 | 4 | 7 | |
| 1948 | Enolase/allergen AspF 22 | AFUA_6G06770 | 2.1 | 5.39 | 47,392 | 16 | 42 | |
| Downregulated in A. fumigatus Δpes3 | 128 | Pentafunctional polypeptide (AroM) | AFUA_1G13740 | 2.9 | 6.15 | 175,521 | 13 | 12 |
| 244 | Pyruvate carboxylase | AFUA_4G07710 | 2.2 | 6.23 | 132,003 | 16 | 16 | |
| 576 | Transketolase TktA | AFUA_1G13500 | 3.1 | 6.12 | 75,245 | 15 | 24 | |
| 585 | Transketolase TktA | AFUA_1G13500 | 2.5 | 6.12 | 75,245 | 22 | 31 | |
| 714 | Phosphoglucomutase | AFUA_3G03020 | 2.2 | 5.78 | 56,680 | 8 | 18 | |
| 1060 | Actin | AFUA_6G04740 | 2.1 | 5.37 | 43,397 | 17 | 52 | |
| 1242 | Spermidine synthase | AFUA_1G13490 | 1.8 | NAe | NA | 8 | 28 | |
| 1457 | Phosphoserine phosphatase | AFUA_4G12730 | 1.6 | 5.67 | 33,442 | 6 | 26 | |
| 1494 | Glycerol-3-phosphate phosphatase | AFUA_1G10570 | 2.5 | 5.48 | 25,765 | 7 | 34 | |
| 1773 | Nascent polypeptide-associated complex (NAC) subunit | AFUA_6G02750 | 2.0 | 5.32 | 20,591 | 7 | 27 | |
| 1923 | Aminopeptidase | AFUA_5G04330 | 2.0 | 6.26 | 109,277 | 16 | 22 |
No., spot number following 2D-PAGE.
The fold change upward is given for proteins that are upregulated in A. fumigatus Δpes3, and the fold change downward is given for proteins that are downregulated in A. fumigatus Δpes3.
tpI, theoretical isoelectric point.
tMr, theoretical molecular weight ratio.
NA, not applicable.
DISCUSSION
Sequencing of the A. fumigatus genome has revealed the presence of at least 14 genes encoding NRP synthetases (16, 50). However, specific data relating the majority of these NRP synthetases to an NRP product has not been forthcoming (66). The pes3 gene was disrupted and subsequently complemented in the A. fumigatus ATCC 46645 strain, with the aim of elucidating the peptide encoded by Pes3, a strategy used by others to identify the peptides encoded by NRP synthetases (15, 36, 39, 60). Unlike several other fungal NRP synthetases characterized to date, Pes3 is not primarily involved in the protection of A. fumigatus against oxidative stress and does not secrete an NRP product, as evidenced by the lack of peptide identification following exhaustive metabolite analysis. A. fumigatus Δpes3 displayed increased sensitivity toward the antifungal voriconazole compared to the wild type. A. fumigatus Δpes3 was hypervirulent in an invertebrate and showed heightened virulence in an HCA murine model of pulmonary aspergillosis, and stimulation of murine macrophages with A. fumigatus Δpes3 resulted in reduced cytokine production compared to stimulation with the wild type. We hypothesize that this is due to a structural alteration in A. fumigatus Δpes3, occurring in the vicinity of the cell wall which abrogates a pathogen associated molecular pattern (PAMP), thereby leading to defective recognition by innate immune receptors. With respect to the histology observed in infected animals, there are several plausible explanations for such a deficiency. Although we observed a slight reduction in inflammation associated with Δpes3 infection in corticosteroid-treated animals, recruitment of host inflammatory cells is evidently only marginally affected. This would imply that, rather than being “invisible” to host immunity (as, for instance, in the case of A. fumigatus ΔrodA [2]), the Δpes3 strain is resistant to killing by the host. This might feasibly result from either or both of (i) an altered local cytokine environment or (ii) the enhanced growth rate of Δpes3, which may outgrow macrophages and thereby avoid killing or possibly only transiently display a PAMP. In any case, the lungs of mice infected with A. fumigatus Δpes3 exhibited greater fungal burden compared to mice infected with the wild type, suggesting inefficient clearance of A. fumigatus Δpes3 by the immune system. Taken together, these data indicate a structural role for an NRP synthetase product, which represents a novel finding for an NRP synthetase in A. fumigatus.
Since relatively little is known about the biological role of NRP synthetases in A. fumigatus, a starting point for investigation was to probe pathways in which fungal NRP synthetase involvement has already been demonstrated. Phenotypic analyses eliminated a role for pes3 in siderophore biosynthesis and protection against oxidative stress, despite widely reported roles for A. fumigatus and other fungal NRP synthetases in these processes (36, 51, 57, 58, 60). Expression of pes3 was detected at low levels in liquid cultures in our study, in agreement with an earlier study by Cramer et al. (16). The high level of pes3 expression observed in ungerminated spores (16) hinted at a possible role for pes3 in germination or cell wall structure in A. fumigatus. Analysis of both vegetative and germination growth rates revealed that A. fumigatus Δpes3 grew at an rate identical to that of the wild type, ruling out an essential role for pes3 in conidial germination and mycelial growth. Exposure of A. fumigatus wild-type and Δpes3 strains to a range of cell wall-damaging agents in the present study according to a recently described protocol for identifying fungal cell wall mutants (55) revealed no altered phenotypes for Δpes3, therefore eliminating a major role for pes3 in modeling cell wall architectures affected by these agents. However, A. fumigatus Δpes3 did exhibit significantly increased sensitivity toward the antifungal voriconazole (P < 0.001), possibly caused by increased entry of voriconazole into conidia due to absence of the Pes3-encoded NRP. The increased voriconazole sensitivity of A. fumigatus Δpes3 may be related to the significantly reduced β-glucan presence on hyphal surfaces, if β-glucan acts to sequester antifungal drugs. Our observations are consistent with previous work which demonstrated that antifungal drug resistance in Candida albicans requires extracellular (1,3)-β-glucan, which acts to sequester and prevent fluconazole uptake into the cell (46). Thus, we hypothesize that altered β-glucan presence on hyphal surfaces, consequent to pes3 disruption, sensitizes A. fumigatus Δpes3 to voriconazole. Moreover, resistance to diamide may be due to decreased intracellular glutathione in A. fumigatus Δpes3.
Genes encoding secondary metabolites are often organized into clusters, and at least 22 such clusters have been identified in A. fumigatus (28, 50, 52). A master regulator of secondary metabolism, LaeA, has been identified in A. fumigatus (6), and this was found to transcriptionally regulate at least 13 secondary metabolite clusters in A. fumigatus (6, 52). Interestingly, pes3 was not found to be under LaeA regulation (52), which suggested it might not be responsible for the production of a secreted metabolite. Extensive comparative metabolite analysis between the wild-type and Δpes3 strains showed that this was in fact the case, with metabolite profiles for wild-type and Δpes3 identical under all of the conditions explored (Table 5). The metabolite extraction and analysis method used in the present study was sufficient to determine all known secreted secondary metabolites from 395 fungal isolates (64). Furthermore, the comparative analysis included conditions in which pes3 expression was confirmed in the present study and others, including analysis of ungerminated spores, where pes3 expression was most abundant (16). Taken together, these results unambiguously infer that Pes3 does not produce either a secreted or an intracellularly located metabolite in A. fumigatus.
NRP synthetases have been widely implicated in mediating fungal virulence. Surprisingly, in vivo virulence testing indicated that A. fumigatus Δpes3 exhibited increased virulence in both an invertebrate model, G. mellonella, and in an HCA immunosuppressed murine model of IA, demonstrating that deletion of pes3 augments the virulence of A. fumigatus. In order to explain the virulence of A. fumigatus Δpes3, several hypotheses were explored. A reduced growth rate of A. fumigatus Δpes3 compared to the wild type could result in augmented virulence of A. fumigatus Δpes3 by invoking an attenuated immune response due to reduced availability of fungal components for immune recognition. Alternatively, increased stimulation of the immune response could result from an accelerated growth rate, leading to greater inflammation, and/or greater production of toxic secondary metabolites. However, the identical growth rates observed between A. fumigatus wild-type and Δpes3 eliminate this hypothesis. Second, we found that Pes3 does not appear to encode a secreted metabolite, and the uniformity of metabolite profiles for wild-type and A. fumigatus Δpes3 makes it unlikely that an alteration in secondary metabolite biosynthesis is responsible for the increase in virulence of A. fumigatus Δpes3. A more plausible hypothesis is that A. fumigatus Δpes3 is immunologically silenced compared to the wild type, and support for this hypothesis was obtained from immune signaling data, whereby a reduction in cytokine induction (TNF-α, IL-6, RANTES, and IL-10) was observed in murine macrophages stimulated with A. fumigatus Δpes3 compared to those stimulated with the wild type. These results indicate that the NRP encoded by Pes3 may be necessary for efficient recognition of A. fumigatus by the innate immune system. No increase in virulence was observed between A. fumigatus wild-type and Δpes3 in a neutropenic model of IA, with similar weights and fungal burdens in the lungs of animals (n = 5) infected with either wild-type or Δpes3, indicating that neutrophil presence is required for the increased virulence of Δpes3, and that the role played by Pes3 in immune recognition leads to an immune response mediated largely by neutrophils.
Innate recognition of fungal pathogens involves specific recognition of invariant molecular components of pathogens, known as PAMPs, by PRRs (42, 72). Dectin-1, a type II transmembrane protein that belongs to the NK-like C-type lectin-like family, is a known receptor for fungal β-glucans (7). Since the initial discovery of dectin-1, this receptor has received significant attention for its essential role in the pulmonary innate immune response against A. fumigatus (25, 67, 75). The role of Toll-like receptors (TLRs) in the immunological recognition of A. fumigatus has also been studied, with TLR2 and TLR4 being implicated (40, 43, 74, 76). Several discrepancies have been documented between the role of TLR2 and TLR4 in recognition of A. fumigatus and, to date, no A. fumigatus PAMPs that are recognized by TLRs have been described (14), highlighting the importance of this research area. In fact, with the exception of C. albicans phospholipomannan, the fungal ligands stimulating TLR activation remain undefined (37). It has been suggested that the ligands activating TLRs in response to A. fumigatus may be of carbohydrate, lipid, or protein origin (25). Given the observations made during the present study, it is likely that this is the case and that Pes3 encodes a peptide with a structural role that is necessary for the recognition of A. fumigatus.
There is one other report describing a fungal NRP synthetase gene that is likely to encode an NRP with a structural role. AbNPS2 in the plant pathogen Alternaria brassicola was identified to encode a large multimodular NRP synthetase (29). Deletion of AbNPS2 resulted in several altered phenotypes in the resultant mutant; reduced virulence on cabbage leaves, lower conidial germination rates, reduced hydrophobicity of conidia, and an altered conidial surface. The conidial surface in the wild type was smooth and had a compact cell wall, whereas the surfaces of AbNPS2 mutant conidia were fluffy, and the cell wall layers were separated as determined by transmission electron microscopy (29). The authors of that study hypothesized that the AbNPS2-encoded peptide may be a component of, or facilitate linkage to, the outermost layer and the middle layer of the fungal conidial cell wall, a hypothesis that may also be relevant for the Pes3-encoded peptide. Dormant A. fumigatus conidia are covered by a hydrophobic rodlet layer (69). The rodlet layer is composed of hydrophobic proteins, and this layer maintains A. fumigatus conidia in an immunologically silenced state (2). Recently, it has been shown that one of the rodlet proteins, RodA, prevents A. fumigatus conidia from triggering NET formation by human neutrophils (9), indicating the importance of rodlet removal in leading to a neutrophil-mediated immune response. To our knowledge, the processes involved in linkage and removal of the rodlet layer to the cell wall have not yet been described, and we speculate that Pes3 may biosynthesize a peptide which is involved in removal of the rodlet layer from the surface of A. fumigatus conidia. Loss of a Pes3-encoded peptide could lead to incomplete or delayed removal of the rodlet layer during conidial germination, thereby maintaining A. fumigatus Δpes3 in an immunologically silenced state, leading to the phenotypes observed here. Alternatively, Pes3 might encode a peptide that functions in linking the rodlet layer to the cell wall. Loss of Pes3 could result in aberrant linking of the rodlet layer to the cell wall and may hinder the removal of the rodlet (e.g., RodAp) layer upon germination. Either hypothesis would result in a reduction in the exposure to fungal cell wall components and explain the reduced immune recognition of and observed virulence for A. fumigatus Δpes3. It has been proposed that RodA may be antiphagocytic, either via steric blocking and/or by impeding macrophage recognition of conidia (19).
Interestingly, the majority of A. fumigatus mutants for which hypervirulent phenotypes have been reported relate to a defect in the cell wall or conidial surface. For example, disruption of the glycosylphosphatidylinositol-anchored protein ECM33 resulted in hypervirulence in a murine model of IA, which was possibly due to a softer cell wall, allowing faster germination (59). Hypervirulent phenotypes were also observed following mutations in the α(1-3)-glucan synthase AGS3, the transcription factor ACE2, and the trehalose biosynthesis genes tpsA and tpsB, and in all cases defects could be detected at the cell walls of these mutants (3, 21, 41). The ACE2 mutant was hypervirulent in a non-neutropenic murine model of IA, but no difference in virulence was observed in a neutropenic model of IA (21), indicating that, as with A. fumigatus Δpes3, the presence of neutrophils was necessary for the increased virulence of the Δace2 strain. A. fumigatus ppoA to ppoC encode dioxygenases responsible for prostaglandin biosynthesis, and it has been shown that a triple A. fumigatus ppo mutant, generated using RNA interference, exhibited increased virulence in a murine model of invasive pulmonary aspergillosis (71). Like A. fumigatus Δpes3, this mutant indicated few phenotypes, however a ppoC deletion mutant exhibited an altered conidial size and retarded germination in static cultures compared to the wild type (18). In addition, A. fumigatus ΔppoC virulence was not significantly altered in a non-neutropenic mouse model, and yet it exhibited a greater susceptibility to macrophage killing than did the wild type. Together, these findings suggest an altered cell wall composition (18)—as we speculate for A. fumigatus Δpes3.
Proteome alterations in A. fumigatus Δpes3 (Table 6) reveal interesting insights into Pes3-encoded peptide functionality and the interplay between membrane-endosome trafficking and morphogenesis. Rab11 (AFUA_1G02190) shares up to 75% identity with mammalian orthologs, regulates plasma membrane-endosome trafficking in Drosophila melanogaster (4), and is upregulated (3.6-fold) in A. fumigatus Δpes3 (Table 6). This linkage between Pes3 loss and possible perturbation in endocytic trafficking is significant in the context of increased voriconazole sensitivity of this mutant (Fig. 3 and Table 4) and suggests a role for the Pes3-encoded peptide in membrane-cell wall interactions. Elevated expression of ribosomal, heat shock, and electron transport proteins in A. fumigatus Δpes3 (Table 6) suggest generalized cell stress and an increased energy requirement and is probably a consequence of impaired conidial germination. Downregulation of protein expression in A. fumigatus Δpes3 is correspondingly illuminating: decreased actin expression (2.1-fold) in A. fumigatus Δpes3 is in accordance with impaired germ tube length. The pentafunctional AroM protein interconverts metabolites of quinic acid in the shikimate pathway, to produce aromatic amino acids, by catalyzing the conversion of 3-deoxy-d-arabinoheptulosonic acid 7-phosphate into 5-enolpyruvylshikimate 3-phosphate (23). Previous work in A. nidulans showed that the constitutive overexpression of AroM inhibited conidial germination (34); thus, it is plausible that AroM downregulation in A. fumigatus Δpes3 (Table 6) represents a systems strategy to overcome the attenuated conidial germination due to Pes3 absence. Spermidine synthase in Ustilago maydis is required for both morphogenesis (yeast-to-mycelium transition) and fungal survival during the infection of plants (73); moreover, deletion mutants were auxotrophic for both lysine and spermidine. In addition, Jin et al. (26) demonstrated that spermidine synthase deletion in A. nidulans caused morphological defects and altered germination, as we observed for A. fumigatus Δpes3 (Fig. 8A). Given that spermidine synthase is downregulated in A. fumigatus Δpes3, we speculate that it may be downstream of, or affected by, Pes3 function; otherwise, increased expression would have been observed to counteract the inhibitory effects of Pes3 absence.
The exact nature of the Pes3-encoded peptide remains to be elucidated, as does that encoded by AbNPS2 (29), thereby highlighting the significant challenge that exists for relating NRP synthetases, particularly those with a structural role, to their respective NRP products. However, all of the data generated in the present study strongly suggest a structural role for the Pes3-encoded NRP in A. fumigatus, and elucidation of the exact nature of this peptide will be the focus of future studies. The agreement between the G. mellonella and the murine virulence data further consolidates the robustness of the larval model in discriminating between the virulence potential of different fungal isolates, a topic that has recently attracted much attention (27, 63).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by an Embark Ph.D. Fellowship to K.A.O. from the Irish Research Council for Science Engineering and Technology. T.C. was supported by a studentship awarded to the Section of Microbiology, Imperial College London, by the Biotechnology and Biosciences Research Council. D.S. and M.S. were recipients of HEA-PRTLI-3 funding and an EU Marie Curie Fellowship (MTKD-CT-2004-014436), respectively. Quantitative RT-PCR instrumentation was funded by Science Foundation Ireland (SFI; grant SFI/07/RFP/GEN/F571/ECO7). The LC-MS facilities were funded by the Danish Research Agency for Technology and Production (grant 09-064967) and the Irish Higher Education Authority.
We thank Ica Dix for assistance with confocal microscopy (SFI-funded instrumentation). We are also grateful to R. Cramer for very helpful advice on the germination assays.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 11 July 2011.
REFERENCES
- 1. Abad A., et al. 2010. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 27:155–182 [DOI] [PubMed] [Google Scholar]
- 2. Aimanianda V., et al. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1179 [DOI] [PubMed] [Google Scholar]
- 3. Al-Bader N., et al. 2010. Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect. Immun. 78:3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Assaker G., Ramel D., Wculek S. K., González-Gaitán M., Emery G. 2010. Spatial restriction of receptor tyrosine kinase activity through a polarized endocytic cycle controls border cell migration. Proc. Natl. Acad. Sci. U. S. A. 107:22558–22563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balibar C. J., Walsh C. T. 2006. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry 45:15029–15038 [DOI] [PubMed] [Google Scholar]
- 6. Bok J. W., Keller N. P. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown G. D., et al. 2002. Dectin-1 is a major β-glucan receptor on macrophages. J. Exp. Med. 196:407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown J. S., et al. 2000. Signature-tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol. Microbiol. 36:1371–1380 [DOI] [PubMed] [Google Scholar]
- 9. Bruns S., et al. 2010. Production of extracellular traps against Aspergillus fumigatus in vitro and in infected lung tissue is dependent on invading neutrophils and influenced by hydrophobin RodA. PLoS Pathog. 6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burns C., et al. 2005. Identification, cloning, and functional expression of three glutathione transferase genes from Aspergillus fumigatus. Fungal Genet. Biol. 42:319–327 [DOI] [PubMed] [Google Scholar]
- 11. Calvo A. M., Wilson R. A., Bok J. W., Keller N. P. 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66:447–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carberry S., Neville C. M., Kavanagh K. A., Doyle S. 2006. Analysis of major intracellular proteins of Aspergillus fumigatus by MALDI mass spectrometry: identification and characterization of an elongation factor 1B protein with glutathione transferase activity. Biochem. Biophys. Res. Commun. 314:1096–1104 [DOI] [PubMed] [Google Scholar]
- 13. Chamilos G., et al. 2010. Generation of IL-23 producing dendritic cells (DCs) by airborne fungi regulates fungal pathogenicity via the induction of T(H)-17 responses. PLoS One 5:e12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chignard M., Balloy V., Sallenave J. M., Si-Tahar M. 2007. Role of Toll-like receptors in lung innate defense against invasive aspergillosis: distinct impact in immunocompetent and immunocompromised hosts. Clin. Immunol. 124:238–243 [DOI] [PubMed] [Google Scholar]
- 15. Cramer R. A., et al. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cramer R. A., et al. 2006. Phylogenomic analysis of non-ribosomal peptide synthetases in the genus Aspergillus. Gene 383:24–32 [DOI] [PubMed] [Google Scholar]
- 17. Dagenais T. R. T., Keller N. P. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22:447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dagenais T. R., et al. 2008. Defects in conidiophore development and conidium-macrophage interactions in a dioxygenase mutant of Aspergillus fumigatus. Infect. Immun. 76:3214–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dagenais T. R., et al. 2010. Aspergillus fumigatus LaeA-mediated phagocytosis is associated with a decreased hydrophobin layer. Infect. Immun. 78:823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doyle S. 2009. Nonribosomal peptide synthesis: amino acids, peptides and proteins in organic chemistry, vol. 2 Wiley-VCH, Weinheim, Germany [Google Scholar]
- 21. Ejzykowicz D. E., et al. 2009. The Aspergillus fumigatus transcription factor Ace2 governs pigment production, conidiation, and virulence. Mol. Microbiol. 72:155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fox E. M., Howlett B. J. 2008. Secondary metabolism: regulation and role in fungal biology. Curr. Opin. Microbiol. 11:481–487 [DOI] [PubMed] [Google Scholar]
- 23. Hawkins A. R., Lamb H. K., Moore J. D., Charles I. G., Roberts C. F. 1993. The pre-chorismate (shikimate) and quinate pathways in filamentous fungi: theoretical and practical aspects. J. Gen. Microbiol. 139:2891–2899 [DOI] [PubMed] [Google Scholar]
- 24. Hearn V. M., Mackenzie D. W. R. 1980. Mycelial antigens from 2 strains of Aspergillus-Fumigatus: an analysis by two-dimensional immunoelectrophoresis. Mykosen 23:549–562 [PubMed] [Google Scholar]
- 25. Hohl T. M., et al. 2005. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. 1:232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin Y., Bok J. W., Guzman-de-Peña D., Keller N. P. 2002. Requirement of spermidine for developmental transitions in Aspergillus nidulans. Mol. Microbiol. 46:801–812 [DOI] [PubMed] [Google Scholar]
- 27. Kavanagh K., Fallon J. P. 2010. Galleria mellonella larvae as models for studying fungal virulence. Fungal Biol. Rev. 2010:1–5 [Google Scholar]
- 28. Keller N. P., Hohn T. M. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17–29 [PubMed] [Google Scholar]
- 29. Kim K. H., Cho Y., La Rota M., Cramer R. A., Lawrence C. B. 2007. Functional analysis of the Alternaria brassicicola non-ribosomal peptide synthetase gene AbNPS2 reveals a role in conidial cell wall construction. Mol. Plant Pathol. 8:23–39 [DOI] [PubMed] [Google Scholar]
- 30. Krappmann S., et al. 2006. The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis. Mol. Microbiol. 61:76–88 [DOI] [PubMed] [Google Scholar]
- 31. Kubodera T., Yamashita N., Nishimura A. 2000. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization, and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 64:1416–1421 [DOI] [PubMed] [Google Scholar]
- 32. Kubodera T., Yamashita N., Nishimura A. 2002. Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 66:404–406 [DOI] [PubMed] [Google Scholar]
- 33. Laboratories B. R. 1986. BRL pUC host: Escherichia coli DH5a competent cells. Focus 8:9. [Google Scholar]
- 34. Lamb H. K., Bagshaw C. R., Hawkins A. R. 1991. In vivo overproduction of the pentafunctional arom polypeptide in Aspergillus nidulans affects metabolic flux in the quinate pathway. Mol. Gen. Genet. 227:187–196 [DOI] [PubMed] [Google Scholar]
- 35. Latge J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee B. N., et al. 2005. Functional analysis of all nonribosomal peptide synthetases in Cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryot. Cell 4:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levitz S. M. 2010. Innate recognition of fungal cell walls. PLoS Pathog. 6:e1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lipmann F. 1971. Attempts to map a process evolution of peptide biosynthesis. Science 173:875–880 [DOI] [PubMed] [Google Scholar]
- 39. Maiya S., Grundmann A., Li S. M., Turner G. 2006. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. Chembiochem 7:1062–1069 [DOI] [PubMed] [Google Scholar]
- 40. Mambula S. S., Sau K., Henneke P., Golenbock D. T., Levitz S. M. 2002. Toll-like receptor (TLR) signaling in response to Aspergillus fumigatus. J. Biol. Chem. 277:39320–39326 [DOI] [PubMed] [Google Scholar]
- 41. Maubon D., et al. 2006. AGS3, an α(1-3)glucan synthase gene family member of Aspergillus fumigatus, modulates mycelium growth in the lung of experimentally infected mice. Fungal Genet. Biol. 43:366–375 [DOI] [PubMed] [Google Scholar]
- 42. Medzhitov R., Janeway C. A. 2000. How does the immune system distinguish self from nonself? Semin. Immunol. 12:185–188 [DOI] [PubMed] [Google Scholar]
- 43. Meier A., et al. 2003. Toll-like receptor (TLR) 2 and TLR4 are essential for Aspergillus-induced activation of murine macrophages. Cell. Microbiol. 5:561–570 [DOI] [PubMed] [Google Scholar]
- 44. Moon Y. S., et al. 2008. Agrobacterium-mediated disruption of a nonribosomal peptide synthetase gene in the invertebrate pathogen Metarhizium anisopliae reveals a peptide spore factor. Appl. Environ. Microbiol. 74:4366–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mootz H. D., Schwarzer D., Marahiel M. A. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochem 3:491–504 [DOI] [PubMed] [Google Scholar]
- 46. Nett J. E., Sanchez H., Cain M. T., Andes D. R. 2010. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J. Infect. Dis. 202:171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nielsen K. F., Mogensen J. M., Johansen M., Larsen T. O., Frisvad J. C. 2009. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Anal. Bioanal. Chem. 395:1225–1242 [DOI] [PubMed] [Google Scholar]
- 48. Nielsen K. F., Smedsgaard J. 2003. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardized liquid chromatography-UV-mass spectrometry methodology. J. Chromatogr. A 1002:111–136 [DOI] [PubMed] [Google Scholar]
- 49. Nielsen M. L., Albertsen L., Lettier G., Nielsen J. B., Mortensen U. H. 2006. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 43:54–64 [DOI] [PubMed] [Google Scholar]
- 50. Nierman W. C., et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 51. Oide S., et al. 2006. NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18:2836–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Perrin R. M., et al. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3:508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pfaller M., et al. 2011. Comparison of the broth microdilution methods of the European Committee on Antimicrobial Susceptibility Testing and the Clinical and Laboratory Standards Institute for testing itraconazole, posaconazole, and voriconazole against Aspergillus isolates. J. Clin. Microbiol. 49:1110–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ram A. F. J., Klis F. M. 2006. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 1:2253–2256 [DOI] [PubMed] [Google Scholar]
- 56. Reeves E. P., Messina C. G. M., Doyle S., Kavanagh K. 2004. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 158:73–79 [DOI] [PubMed] [Google Scholar]
- 57. Reeves E. P., et al. 2006. A nonribosomal peptide synthetase (Pes1) confers protection against oxidative stress in Aspergillus fumigatus. FEBS J. 273:3038–3053 [DOI] [PubMed] [Google Scholar]
- 58. Reiber K., et al. 2005. The expression of selected non-ribosomal peptide synthetases in Aspergillus fumigatus is controlled by the availability of free iron. FEMS Microbiol. Lett. 248:83–91 [DOI] [PubMed] [Google Scholar]
- 59. Romano J., et al. 2006. Disruption of the Aspergillus fumigatus ECM33 homologue results in rapid conidial germination, antifungal resistance, and hypervirulence. Microbiology 152:1919–1928 [DOI] [PubMed] [Google Scholar]
- 60. Schrettl M., et al. 2007. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 3:1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schrettl M., et al. 2010. Self-protection against gliotoxin: a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Siednienko J., Halle A., Nagpal K., Golenbock D. T., Miggin S. M. 2010. TLR3-mediated IFN-β gene induction is negatively regulated by the TLR adaptor MyD88 adaptor-like. Eur. J. Immunol. 40:3150–3160 [DOI] [PubMed] [Google Scholar]
- 63. Slater J. L., Gregson L., Denning D. W., Warn P. A. 2010. Pathogenicity of Aspergillus fumigatus mutants assessed in Galleria mellonella matches that in mice. Med. Mycol. doi:10.3109/13693786.2010.523852 [DOI] [PubMed] [Google Scholar]
- 64. Smedsgaard J. 1997. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J. Chromatogr. A 760:264–270 [DOI] [PubMed] [Google Scholar]
- 65. Stack D., Frizzell A., Tomkins K., Doyle S. 2009. Solid-phase 4′-phosphopantetheinylation: fungal thiolation domains are targets for chemoenzymatic modification. Bioconj. Chem. 20:1514–1522 [DOI] [PubMed] [Google Scholar]
- 66. Stack D., Neville C., Doyle S. 2007. Nonribosomal peptide synthesis in Aspergillus fumigatus and other fungi. Microbiology 153:1297–1306 [DOI] [PubMed] [Google Scholar]
- 67. Steele C., et al. 2005. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 1:323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sugui J. A., et al. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice Immunosuppressed with hydrocortisone. Eukaryot. Cell 6:1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thau N., et al. 1994. Rodletless mutants of Aspergillus-Fumigatus. Infect. Immun. 62:4380–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tilburn J., et al. 1983. Transformation in Aspergillus nidulans. Heredity 51:519–520 [Google Scholar]
- 71. Tsitsigiannis D. I., et al. 2005. Aspergillus cyclooxygenase-like enzymes are associated with prostaglandin production and virulence. Infect. Immun. 73:4548–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tsoni S. V., Brown G. D. 2008. β-Glucans and dectin-1, p. 45–60 In Year in immunology 2008, vol. 1143 Blackwell Publishing, Oxford, England: [DOI] [PubMed] [Google Scholar]
- 73. Valdés-Santiago L., Cervantes-Chávez J. A., Ruiz-Herrera J. 2009. Ustilago maydis spermidine synthase is encoded by a chimeric gene, required for morphogenesis, and indispensable for survival in the host. FEMS Yeast Res. 9:923–935 [DOI] [PubMed] [Google Scholar]
- 74. Wang J. E., et al. 2001. Involvement of CD14 and Toll-like receptors in activation of human monocytes by Aspergillus fumigatus hyphae. Infect. Immun. 69:2402–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Werner J. L., et al. 2009. Requisite role for the dectin-1 β-glucan receptor in pulmonary defense against Aspergillus fumigatus. J. Immunol. 182:4938–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhao J., Wu X. Y. 2008. Aspergillus fumigatus antigens activate immortalized human corneal epithelial cells via Toll-like receptors 2 and 4. Curr. Eye Res. 33:447–454 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.