Abstract
The 23-membered Esx protein family is involved in the host-pathogen interactions of Mycobacterium tuberculosis. These secreted proteins are among the most immunodominant antigens recognized by the human immune system and have thus been used to develop vaccines and immunodiagnostic tests for tuberculosis (TB). Gene pairs for 10 Esx proteins are contained in the ESX-1 to ESX-5 loci, encoding type VII secretion systems. A subset of Esx proteins can be further classified into the Mtb9.9, QILSS, and TB10.4 subfamilies. To survey genetic diversity in the Esx family and its potential for antigenic variation, we sequenced all esx genes from 108 clinical isolates of M. tuberculosis from different clades by using a targeted approach. A total of 109 unique single nucleotide polymorphisms (SNPs) were observed, and 59 of these were nonsynonymous. Some of the resultant amino acid substitutions affect known Esx epitopes, including two in the EsxB (CFP-10) and EsxH (TB10.4) antigens. Assessment of the SNP distribution across the Esx proteins revealed high genetic variability, especially in the Mtb9.9 and QILSS subfamilies, and more conservation in the ESX-1 to ESX-4 loci. Comparison of the DNA sequences of variable esx genes provided clear evidence for recombination events between different genes in the same strain, some of which are predicted to truncate the corresponding protein. Many of these polymorphisms escape detection by ultrahigh-throughput sequencing using short sequence reads, as such approaches cannot distinguish between closely related genes. The esx gene family is dynamic, and sequence changes likely lead to immune variation.
INTRODUCTION
Development of an effective tuberculosis (TB) control strategy requires detailed understanding of the biology of Mycobacterium tuberculosis and its interaction with human hosts. In humans, CD4+ and CD8+ T cells are known to mediate antigen-specific immune responses that are essential for conferring protective immunity against M. tuberculosis (7, 36). Several secreted proteins from mycobacteria have been shown to induce strong cellular immune responses due to the presence of short peptide fragments bearing epitopes that bind to major histocompatibility complex (MHC) molecules, which are recognized by T lymphocytes (7).
Two of the most frequently recognized T cell antigens from M. tuberculosis are the small secreted proteins EsxA or ESAT-6 (early secretory antigenic target of 6 kDa) and EsxB or CFP-10 (culture filtrate protein of 10 kDa), the prototypes of the Esx family (6). Genes encoding ESAT-6 (esxA) and CFP-10 (esxB) are located directly adjacent to each other and known to be cotranscribed (10). Analysis of the genome sequence of M. tuberculosis H37Rv revealed 11 pairs of tandem genes encoding paralogous Esx proteins located immediately downstream of the PE/PPE genes (15, 48). The esx family has 23 members (11 gene pairs and a singleton, esxQ) named esxA to esxW. Although the level of sequence identity varies between the Esx proteins (35% to 98%), all of them belong to the WXG100 family, which is characterized by a size of ∼100 amino acids and the presence of a Trp-Xaa-Gly (W-X-G) motif (37).
EsxA and EsxB interact to form a 1:1 heterodimer, which appears to be essential for their secretion (10, 43). Proteins encoded by two other paralogous gene pairs, EsxR-EsxS and EsxH-EsxG, also form 1:1 complexes, suggesting that this may be typical of all Esx protein couplets (8, 32). Five of the 11 tandem gene pairs are contained within conserved genetic loci ESX-1 to ESX-5, encoding components of a type VII secretory apparatus (Table 1) (2, 26, 48). The ESX-1 system, which is responsible for the secretion of EsxA and EsxB, has been extensively studied due to its important role in M. tuberculosis pathogenesis (11, 12, 27). Loss of the region of difference 1 (RD1) containing the ESX-1 locus contributes to the attenuation of the vaccine strains Mycobacterium bovis BCG and Mycobacterium microti (12, 40, 41). Of the other systems, ESX-5 is known to be necessary for the secretion of PE and PPE proteins in Mycobacterium marinum and for macrophage subversion (3, 4). ESX-3 is essential for in vitro growth and may be involved in iron/zinc homeostasis (44), while the functions of ESX-2 and ESX-4 remain unknown. Comparative genomic analysis suggested that the ESX loci in mycobacteria resulted from a series of duplication events, where ESX-4 was the progenitor (26).
Table 1.
Overview of the esx family of M. tuberculosisa
| Conserved ESX locus |
esxA (ESAT-6) paralogs |
esxB (CFP-10) paralogs |
||
|---|---|---|---|---|
| Inside the ESX locus | Outside the ESX locus | Inside the ESX locus | Outside the ESX locus | |
| ESX-1 | esxA (rv3875) | esxB (rv3874) | ||
| ESX-2 | esxC (rv3890c) | esxD (rv3891c) | ||
| ESX-3 | esxH (rv0288) | esxR (rv3019c), esxQb (rv3017c) | esxG (rv0287) | esxS (rv3020c) |
| ESX-4 | esxT (rv3444c) | esxU (rv3445c) | ||
| ESX-5 | esxN (rv1793) | esxI (rv1037c), esxL (rv1198), esxO (rv2346c), esxV (rv3619c) | esxM (rv1792) | esxJ (rv1038c), esxK(rv1197), esxP(rv2347c), esxW(rv3620c) |
| None | esxEc (rv3904c) | esxFc (rv3905c) | ||
Bold, TB10.4 subfamily; underlined, Mtb9.9 subfamily; bold and underlined, QILSS subfamily.
esxQ does not occur in tandem with a cfp-10 homologue.
esxE and esxF show no significant homology to any conserved ESX loci.
The 13 esx genes that are not part of the ESX-1 to ESX-5 loci seem to have arisen from singular duplication events (26). Consequently, the Esx family can be classified into distinct subfamilies based on high sequence identity between certain members (Table 1). The TB10.4 subfamily comprises esxH, which is a component of the ESX-3 locus, and two of its paralogs (45). The Mtb9.9 (31) and QILSS (48) subfamilies have 5 members each that include gene duplicates of esxN and esxM, respectively, both of which belong to the ESX-5 locus. These gene duplicates show a striking level of amino acid similarity (93 to 98%), indicating recent duplication from part of the ESX-5 locus (26). Comparative proteomics between M. tuberculosis H37Rv and the attenuated strain M. tuberculosis H37Ra revealed the absence of some EsxM and EsxN paralogs in M. tuberculosis H37Ra (28). Along with the ESX-1 system, two other loci, encoding paralogs of EsxM and EsxN, have been deleted from the vaccine strain M. bovis BCG: the RD5 and RD8 loci comprising esxP-esxO and esxW-esxV, respectively (9, 28).
The identification of EsxA and EsxB as potent T cell antigens prompted several immunological studies in different animal models and humans (1, 6, 21, 42, 49), and it has been shown that other Esx proteins also display antigenic qualities (12). ESAT-6 and CFP-10 have been utilized in the development of diagnostic tests that can distinguish between infected and vaccinated individuals (33, 49), while EsxH (TB10.4) has been used to obtain a fusion protein-based subunit vaccine (20). Furthermore, antigenic epitopes in several Esx proteins have been characterized experimentally. Investigation of TB10.4 paralogs revealed unique antigenic epitopes in the three proteins despite the high sequence similarity (∼75%) (45). Interestingly, the Mtb9.9 subfamily proteins, which display an even higher degree of amino acid similarity (≤93%), have also been shown to induce heterogeneous human T cell responses (5).
The success of M. tuberculosis as a pathogen has been due largely to its ability to survive in spite of a host immune response. In order to evade the host immune system, variants of pathogens emerge, with alterations in epitope regions recognized by critical host immune cells. Despite the high level of sequence conservation in the Esx family, there is heterogeneity in T cell responses to different Esx antigens. The aim of this study was to characterize sequence diversity in esx genes isolated from clinical M. tuberculosis samples in order to identify substitutions that may impact immunogenicity.
MATERIALS AND METHODS
TB patients.
The clinical strains used in this study were isolated from tuberculosis patients at Hôpital Ambroise Paré in Boulogne-Billancourt, France, over a period of 3 years from 2001 to 2004. A total of 103 patients were included, among whom 58 were male and 45 were female. The patient ages ranged from 12 to 96 years, with a median age of 38 years. Localization of tuberculosis was pulmonary in 84 cases and extrapulmonary in the remaining 19 cases (pleural TB, 3 cases; lymphatic TB, 8 cases; bone and joint TB, 3 cases; meningitis, 2 cases; miliary TB, 2 cases; digestive TB, 1 case). The HIV status was unknown for 31 patients, 67 patients tested HIV negative, and 5 patients tested HIV positive.
Strains.
With the exception of two patients, one single isolate per patient was included in this study. For one of the patients, 2 isolates were included, which had been isolated at a 3-week interval, while for another patient, 5 isolates were included, which had been isolated over a period of 3 years. The respiratory samples were decontaminated by the N-acetyl-cysteine-NaOH method. Routine laboratory procedures included microscopic examination after fluorescence staining and culture on solid Lowenstein-Jensen (Bio-Rad, Marnes-la Coquette, France) and liquid (MGIT; Becton Dickinson, Le Pont-de-Claix, France) media. Solid media were maintained at 37°C for 3 months and examined for growth once a week. The MGIT media were incubated in the Bactec MGIT 960 system and examined as soon as a positive signal was emitted. In the case of negativity, the liquid cultures were maintained in the MGIT system for 45 days. Species identification was done using the commercially available Accuprobe system (bioMérieux, Marcy l'Etoile, France), and antibiotic susceptibility testing was carried out with the Bactec MGIT 960 system.
Molecular typing.
For molecular typing, a loopful of Lowenstein-Jensen culture was suspended in 150 μl of Tris-EDTA (TE) buffer and heated at 95°C for 15 min. This crude lysate was used for PCRs. The M. tuberculosis H37Rv reference strain was included for control purposes. Amplification of the direct repeat (DR) regions and hybridization for spoligotyping were carried out as previously described (30), and hybridization profiles were expressed using the octal code and compared with the database, SpolDB4, to determine the corresponding spoligo international types (14). Amplification of the mycobacterial interspersed repetitive-unit (MIRU) loci for MIRU-variable-number tandem-repeat (VNTR) typing was carried out as described in the past (47). The fragment sizes of the amplification products were estimated by agarose gel electrophoresis in relation to molecular weight markers, and the number of MIRU copies was determined with reference to the MIRU-VNTR allele table (see Table S1 in the supplemental material).
PCR and sequencing.
Fragments bearing all esx genes were amplified using the sets of primers listed in Table S2A in the supplemental material. Amplification was performed using 25 μl of ReddyMix PCR Master Mix (Thermo Fisher Scientific, Inc.) and 1 μl of each primer (10 pmol). Dideoxy sequencing of the amplified gene fragments was carried out on both strands with the BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA), using the primers listed in Table S2B in the supplemental material, and with an ABI 3700 DNA analyzer.
SNP detection.
Sequences of esx genes from the clinical strains were compared with the corresponding sequences from the M. tuberculosis reference strain H37Rv. The positions of variant nucleotides were recorded as single nucleotide polymorphisms (SNPs) using the BLAST function on the TubercuList website (http://tuberculist.epfl.ch). By comparison of the amino acid resulting from the substitution with the reference amino acid sequence, these SNPs were further characterized as being synonymous (sSNPs; no change in amino acid) or nonsynonymous (nsSNPs; resulting in an amino acid change).
Epitope identification.
A total of 93 peptides belonging to the Esx proteins comprising human T cell epitopes and major histocompatibility complex (MHC)-binding peptides were obtained from the Immune Epitope Database and Analysis Resource (50) (see Table S3 in the supplemental material). The database was accessed on 20 July 2010 (http://www.immuneepitope.org/).
Phylogenetic analysis.
Comparison of the synonymous substitution rate per site (dS) to the nonsynonymous substitution rate per site (dN) via the ratio ω = dN/dS allows quantification of selection pressures on codon alignments. The number of SNPs observed in individual esx genes was not sufficient to determine a gene-specific dN/dS ratio from our data set. Instead, we concatenated all the codons containing SNPs to generate a single sequence for each isolate, which was used for subsequent analysis. Phylogenetic trees were obtained using the PHYLIP package (22) by implementing the neighbor-joining algorithm based on distances calculated under the K80 model. The neighbor-joining tree topologies and the sequence alignment were subjected to codon-based likelihood analysis using the CODEML (51) program in the PAML package (52). The M0 model was used to estimate a single ω ratio for all branches of the tree.
In order to screen for recombination, the genetic algorithm recombination detection (GARD) tool (38) was used, which is available on the web server of the HyPhy package (39) (http://www.datamonkey.org/). GARD searches for putative recombination breakpoints using a multiple-sequence alignment and generates the phylogenies for each nonrecombinant segment in order to assess a goodness of fit based on the Akaike information criterion (AIC) and AICc (AIC derived from a maximum likelihood model fit to each segment) (46). Information from all the fitted models is combined to assign a level of support to the placement of breakpoints and for different phylogenies inferred among nonrecombinant segments.
RESULTS
Diversity of bacterial strains.
A total of 108 clinical samples were included in this study to investigate the genetic diversity in the 23 genes constituting the esx family. Recent clinical isolates were used in order to assess the current situation and avoid possible sequence biases that could have been introduced in heavily passaged laboratory strains. The clinical isolates originated from different geographical locations in Europe, North Africa, America, and Asia and represented 75 different spoligo international types. MIRU-VNTR typing revealed the distribution of the isolates across different genotypic families, representing the main geographical lineages of M. tuberculosis (24). A subset of the isolates could be identified as members of the Beijing (6 isolates), Haarlem and Haarlem-like (23 isolates), East African Indian (EAI; 6 isolates), Latin American (LAM; 4 isolates), West African (7 isolates), and East African (4 isolates) families, representing the breadth of genomic diversity in the species (see Table S1 in the supplemental material). One of the clinical isolates was an M. bovis strain. The esx gene sequences obtained from the clinical samples and the M. tuberculosis reference strain H37Rv were compared in order to detect any variation at the nucleotide level. Gene sequences of esx from the attenuated strain H37Ra are identical to those of H37Rv (53).
Comparative esx genomics of clinical isolates.
In the entire clinical data set, a total of 797 substitutions were identified, corresponding to 109 unique SNPs across the 23 esx genes, each of which occurred in one or several isolates (see Table S4 in the supplemental material). Analysis of the 109 SNPs revealed 50 sSNPs and 59 nsSNPs. Based on the spoligotype and MIRU-VNTR information available for the clinical isolates, we investigated whether the highly prevalent SNPs were specific to any of the geographical lineages represented in our data set (see Table S1 and Fig. S1 in the supplemental material). Interestingly, three SNPs in esxV, including two nsSNPs, Q20L and S23L, and an sSNP in codon 57, occurred in 74 isolates which belonged to different lineages. The high prevalence of these SNPs in the clinical data set indicates that these positions in esxV are lineage markers. An E68K substitution in EsxB (CFP-10) was observed in 19 isolates, mostly belonging to the Haarlem, Haarlem-like, and LAM families, all of which represent the Euro-American lineage (24). On the other hand, there were also several SNPs with a very low prevalence that appeared to be specific to particular strains or lineages. The only M. bovis isolate in the data set showed two nsSNPs, M82V in EsxE and W58stop in EsxF, which were not observed in the other isolates. These were confirmed to be M. bovis specific upon comparison with the genome sequences of M. bovis AF2122/97 and M. bovis BCG Pasteur 1173P2 (13, 25).
SNPs involving stop codons can have a significant impact on the structure and function of a protein. Apart from EsxF, two other esx genes had substitutions that introduced stop codons. These include EsxL, containing a Q76stop, which was observed in three isolates, and EsxW, containing a Q59stop, which was observed in one isolate. In contrast to EsxW, a stop59Q substitution in EsxM was present in nine clinical isolates. Another nsSNP, A71S in EsxH, was seen in only two isolates, both of which represented West African lineages (I and II). Three out of six Beijing strains harbored an nsSNP, P63S in EsxU, that was not present in any of the other isolates.
SNP distribution across esx genes.
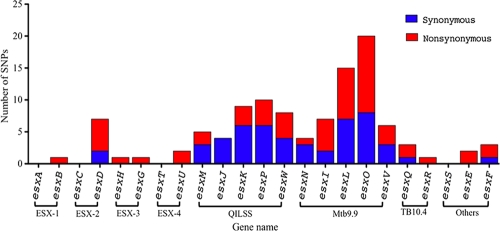
Analysis of 108 clinical isolates revealed SNPs in 19 of the 23 esx genes. Sequences for esxA, esxC, esxT, and esxS for all clinical isolates were invariant. The distribution of the SNPs was not uniform across the esx family members (Fig. 1). Grouping of the esx genes into their subfamilies revealed that the four secretion systems ESX-1 to ESX-4 displayed a low level of variation in general and a strikingly low level of synonymous substitutions. On the other hand, genes belonging to the Mtb9.9 and the QILSS subfamilies, including the ESX-5 system, accounted for the majority of the variation, displaying a large number of sSNPs as well as nsSNPs. On average, from whole-genome comparisons of two strains of M. tuberculosis (such as H37Rv and CDC1551 [23]), the SNP frequency is 1 per 4.1 kb. In contrast, in our esx data set, the frequency compared to H37Rv ranges from a median of 1 per kb to a maximum of 3 per kb depending on the clinical isolate examined.
Fig. 1.
Distribution of nonsynonymous (red) and synonymous (blue) SNPs across 23 esx genes. Brackets indicate genes encoded in the ESX-1 to ESX-4 loci and members of the Mtb9.9, QILSS, and TB10.4 subgroups.
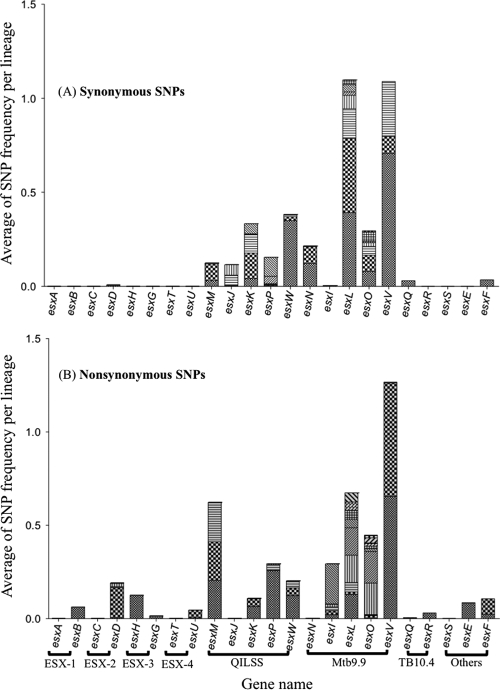
To estimate the incidence of SNPs across the clinical data set, while accounting for lineage-specific polymorphisms, we divided the clinical isolates into groups based on the lineage information. The isolates that could not be successfully genotyped were considered one separate group. The frequency of occurrence of an SNP was calculated separately for each lineage and represented as the average of SNP frequency per lineage. SNPs were grouped together based on the gene in which they occurred (Fig. 2). As seen with the number of polymorphisms, SNPs in the Mtb9.9 and QILSS genes were present in a large number of clinical isolates. In contrast, SNPs in genes encoded in the ESX-1 to ESX-4 loci and the TB10.4 paralogs were observed in very few isolates. Due to differences in the proportion of SNPs observed in different esx subfamilies, we carried out phylogenetic analysis separately on the two main classes of esx genes: the ESX-5 paralogs (Mtb9.9 and QILSS members) and components of the ESX-1 to ESX-4 loci.
Fig. 2.
Incidence of synonymous (A) and nonsynonymous (B) SNPs in 23 esx genes seen across 108 clinical isolates. Each pattern in the bars indicates a unique SNP, and the height of each bar correlates with the average frequencies per lineage.
Gene conversion in ESX-5 paralogs.
A particular feature of SNPs observed in esx genes belonging to the Mtb9.9 and QILSS subfamilies was the cooccurrence of two or more SNPs in neighboring codons. All the isolates containing an sSNP in codon 40 of esxJ also had an sSNP substitution in codon 41. The same was true for sSNPs in codons 6 and 8 of esxL. The highly prevalent Q20L and S23L SNPs in esxV occurred in over 70% of the clinical isolates, all of which harbored an sSNP in codon 57 of esxV. Reciprocal substitutions, Q59stop and stop59Q, were also seen in the esxM and esxW genes.
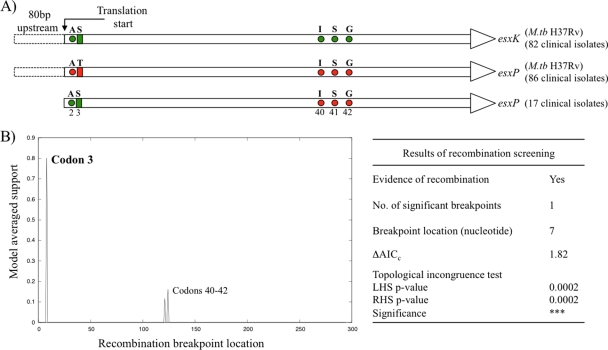
Sequences for esxP from 17 clinical isolates contained an sSNP in the second codon followed by an nsSNP (T3S) in the third codon (Fig. 3A). The resulting codons in the esxP clinical sequences were identical to the M. tuberculosis H37Rv sequences of the paralogous genes esxK, esxJ, and esxM from the QILSS subfamily. The nucleotide sequences in esxP, esxK, and esxJ are also identical for 108 bp downstream of the SNPs. Sequences up to 80 bp upstream of the SNPs, including the intergenic regions, are identical only between esxP and esxK (Fig. 3A). The esxK sequences of the 17 clinical isolates and M. tuberculosis H37Rv were identical, but note that in one other clinical isolate, codons 40, 41, and 42 all contain the same sSNPs as those present in esxP of M. tuberculosis H37Rv. We subjected the 17 esxK and esxP sequences to a recombination screen using the GARD tool. The results revealed the presence of one significant recombination breakpoint at the seventh position (codon 3) and two suggestive potential breakpoints between codons 40 and 42 (Fig. 3B).
Fig. 3.
Putative recombination between esxP and esxK genes. (A) Schematic representation of sequence variation in esxK and esxP from M. tuberculosis H37Rv and esxP sequences from clinical isolates. The M. tuberculosis H37Rv sequences for esxK and esxP are identical except at the positions indicated with circles (silent polymorphisms) and rectangles (amino acid differences). Positions of the SNPs observed in esxP from 17 clinical isolates are color coded depending on their similarity to the corresponding gene in M. tuberculosis H37Rv. Nucleotide sequences up to 80 bp upstream of the gene are also identical in esxP and esxK. (B) Plot and summary of GARD results showing the location of the putative recombination breakpoint. ΔAICc indicates improvement of the AICc score compared to that of the model with zero breakpoints. Significance of the difference in topologies between the partitions to the left and right of the breakpoint is indicated with P values for both sides.
Phylogenetic analysis.
In order to calculate the ω (dN/dS) ratio to quantify the selective pressure acting on the esx genes, we chose the random-site model, which accounts for variable selective pressures across codons and can detect amino acid residues under either positive or negative selection. Analysis of codon alignments, including SNPs observed in all esx genes, gave a low ω of 0.35, because the dS value of the ESX-5 paralogs is very high. Mtb9.9 and QILSS genes harbor 46 of the 50 sSNPs which are observed in a large proportion of the clinical isolates. However, the codon-based analysis assumes a single tree topology for the whole alignment and does not account for recombination. Therefore, we generated a codon alignment for SNPs in esx genes belonging to the ESX-1 to ESX-4 loci and reanalyzed them using CODEML. The estimated ω value of 1.66 (dN/dS > 1) indicated diversifying selection in this subset of esx genes.
SNPs occurring in Esx epitopes.
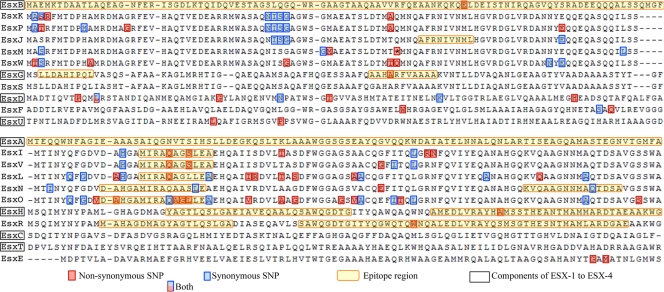
In order to identify whether any of the SNPs observed in our clinical data set could impact the immunogenicity of the Esx proteins, we searched the IEDB database for experimentally confirmed human T cell and MHC-binding peptides specific to the Esx family. A total of 93 peptides were obtained, 80 of which comprised overlapping peptides that belonged to EsxA and EsxB and spanned the entirety of these proteins. Sequence alignments of the EsxA (excluding EsxQ due to a low level of sequence homology to other EsxA paralogs) and EsxB paralogs (Fig. 4) highlight the antigenic epitopes and positions of all the SNPs observed in the clinical data set. There were 3 sSNP and 15 nsSNPs occurring in known epitope regions. Excluding the 80 epitopes from EsxA and EsxB, 9 of the 13 known epitopes in the other Esx proteins had at least one or more SNPs (Table 2). Mtb9.9 genes, which harbored 12 of the 18 SNPs, were overrepresented. The corresponding proteins share an MHC-binding peptide, MIRAQA[GA][SL]LEA, at positions 16 to 26 that is subject to sequence variation. Interestingly, three of the Mtb9.9 proteins, EsxI, EsxV, and EsxO, showed a Q20L polar-to-nonpolar substitution. One of the two variable positions in the MHC-binding peptide reflected an amino acid change, S23L in EsxI and EsxV, and a reverse L23S change in EsxO. In fact, we were able to identify several recurring positions both inside and outside known epitopes (Fig. 4) at which SNPs were observed in two or more Esx proteins.
Fig. 4.
Multiple sequence alignment of EsxB (CFP-10) and EsxA (ESAT-6) paralogs from M. tuberculosis H37Rv. Positions containing nonsynonymous (red boxes) and synonymous (blue boxes) changes have been annotated for each protein. Known immune epitope regions are highlighted in yellow.
Table 2.
SNPs affecting known epitope regions
| Protein | SNP | Type of change | Amino acid sequencea |
No. of isolates | |
|---|---|---|---|---|---|
| T cell epitope | MHC-II binding | ||||
| EsxB | E68K | Acidic to basic | KQELDEISTNIRQAG | 19 | |
| EsxG | A56T | Nonpolar to polar | AAHARFVAA | 1 | |
| EsxH | A71S | Nonpolar to polar | AMEDLVRAYHAMSSTHEA | 2 | |
| EsxQ | S90A | Polar to nonpolar | RCRRALRQIGVLERPVGDSS | 1 | |
| EsxQ | L76P | Nonpolar to nonpolar | RCRRALRQIGVLERPVGDSS | 1 | |
| EsxR | W58C | Nonpolar to polar | YQGWQTQWNQALEDLVRAYQ | 3 | |
| EsxI | S23L | Polar to nonpolar | MIRAQAGSL | 1 | |
| EsxI | Q20L | Polar to nonpolar | MIRAQAGSL | 13 | |
| EsxL | Q20L | Polar to nonpolar | MIRAQAGLL | 1 | |
| EsxN | 24 | Synonymous | DAHGAMIRAQAASLE | 4 | |
| EsxN | 83 | Synonymous | KVQAAGNNMAQTDSA | 1 | |
| EsxO | L23S | Nonpolar | MIRAQAGLL | 6 | |
| EsxO | G22A | Polar to nonpolar | MIRAQAGLL | 6 | |
| EsxO | A21P | Nonpolar to nonpolar | MIRAQAGLL | 1 | |
| EsxO | 20 | Synonymous | MIRAQAGLL | 1 | |
| EsxO | A12D | Nonpolar to acidic | DAHGAMIRAQAGLLE | 1 | |
| EsxV | S23L | Polar to nonpolar | MIRAQAGSL | 77 | |
| EsxV | Q20L | Polar to nonpolar | MIRAQAGSL | 74 | |
Obtained from the IEDB resource (49). Amino acids from codons containing nsSNPs are underlined, and sSNPs are italicized.
DISCUSSION
In this study, all 23 esx genes from 108 clinical samples were sequenced in order to identify substitutions that may have an impact on the immunogenicity or function of Esx proteins. A total of 797 substitutions corresponding to 109 distinct SNPs were observed in the entire clinical data set. Our analysis indicated that esx genes encoded within the ESX-1 to ESX-4 loci displayed less variation overall than the esx genes located outside these loci. EsxA and EsxB have been most extensively studied owing to their crucial role in M. tuberculosis pathogenesis and applications in the development of vaccines and diagnostics. A past study by Musser et al. involving sequencing of 24 important M. tuberculosis antigens from 16 clinical strains revealed no variation in EsxA and EsxB sequences (35). The commercially available gamma interferon (IFN-γ) release assays (IGRA) used for diagnosing TB, QuantiFERON gold test (33), and T-SPOT.TB (34) employ peptides from ESAT-6 (EsxA) and CFP-10 (EsxB). While no sequence variation has yet been observed in EsxA, there is an amino acid substitution (E68K) in EsxB present in 18 of the 108 strains representing diverse lineages. This substitution occurs within a known human T cell epitope, so it may influence responses to EsxB peptides in the IGRA. Although the strains containing the EsxB SNP represent different lineages, they also share three sSNPs in codons 6 and 8 of EsxL and codon 57 of EsxV.
A more recent study by Davila et al. to assess the genetic diversity of EsxA and EsxH genes among 88 clinical isolates revealed no variation in either of the two genes (17), and this is essentially in agreement with our findings (Table 2). Our results showed that three out of five EsxA paralogs encoded by the ESX-1 to ESX-5 loci were invariant across the entire clinical data set. There was also an absence of silent substitutions in the esx components of the ESX-1 to ESX-4 loci with the exception of esxD. Interestingly, the majority of the nsSNPs occur in regions of the gene coding for known epitopes. Our estimation of dN/dS (ω = 1.66) indicated that some of these epitopes might be under positive selection. These findings are in contrast with those in a recent publication by Comas et al. demonstrating the hyperconservation of human T cell epitopes among 21 strains representing the six main geographical lineages of M. tuberculosis (16). Their investigation uncovered more than 9,000 SNPs by whole-genome sequencing of 21 strains using the Illumina genome analyzer. Using the epitope information from the IEDB database, they estimated low ω values (dN/dS < 1) in antigens compared to those for essential and nonessential genes.
For technical reasons, we believe that Comas et al. (16) have underestimated the amount of variation in the Esx family. One of the limitations of the Illumina technology is sequence assembly and identification of SNPs in repetitive regions due to the short lengths of the sequence reads. This could mask any polymorphisms generated due to gene conversion events in duplicated paralogous genes, due to erroneous mapping of short reads. This includes genes in the Mtb9.9 and QILSS subfamilies. Our study involved traditional sequencing with primers specific for the different esx genes and significantly longer sequence reads than those generated by the Illumina technology and thus avoids this pitfall. Interestingly, one of the three outliers in the Comas et al. study was esxH, which displayed a high level of variation in epitope regions (dN/dS > 1).
Members of the Mtb9.9 and QILSS subfamilies show the highest levels (93 to 98%) of amino acid identity among the Esx family. Although several immunogenic epitopes were located in regions of 100% sequence identity, some of the major immunodominant epitopes were also identified in regions of sequence diversity in proteins from the Mtb9.9 and QILSS subfamilies. A recent study by Jones et al. confirmed that the Esx proteins, whose genes were not part of the ESX-1 to ESX-5 loci, showed similar levels of immunogenicity to the Esx proteins from the ESX-1 to ESX-5 loci (29). Strikingly, even single-residue differences in the epitope sequences altered the responder frequencies to these antigens. The amino acid residues critical for antigenicity include T58 for the QILSS subfamily and G22 and S23 for the Mtb9.9 subfamily. In the clinical data set, we observed an A58T mutation in EsxP and EsxK and a converse T58A substitution in EsxW. S23L substitutions in EsxI and EsxV and L23S and G22A substitutions in EsxO were also found in several isolates. A study using human CD4+ T cells performed by Alderson et al. demonstrated that T cell lines specific for EsxL (G22 and L23) failed to recognize peptides from EsxN (A22 and S23) and EsxV (G22 and S23) (5). On the other hand, peptide fragments from EsxV, which is absent in M. bovis, have been shown to induce IFN-γ responses in cattle infected with TB (29), suggesting cross-reactivity between highly similar epitopes in duplicated proteins. The conservation of silent as well as nonsynonymous SNPs between paralogs and orthologs of the Mtb9.9 family, as seen in esxP and esxM, respectively, suggests that even minor variation within the Mtb9.9 and QILSS families could significantly alter the length and expression of this protein subfamily. Based on the clinical SNPs, we were able to provide evidence for putative recombination between esxK and esxP. It is possible that gene conversion events may occur between other duplicated paralogous genes in the QILSS and Mtb9.9 subfamilies. Gene conversion has been described in members of the PE-PGRS gene family in M. bovis, where homogenotization has been reported for genes Mb1485 and Mb1487 (19). Frequent homologous recombination events in the highly repetitive PE/PPE multigene families with a potential role in antigenic variability have been described in M. tuberculosis (18), and the present work indicates that the esx gene family is also dynamic.
Amino acid substitutions encoded by duplicated genes may allow for genetic drift, by regulating expression of the functionally similar protein paralogs that differ in their immunodominant epitopes. Although in silico prediction of T cell binding epitopes is possible, we included only the experimentally confirmed epitope data in our analysis. With the exception of EsxA and EsxB, there is very little or no epitope data in IEDB for other members of the Esx family. As new Esx epitopes are identified in the future, we should be able to assess the role of the other nsSNPs observed in our clinical data set. In conclusion, our analysis of esx genes has revealed a number of previously unknown sequence polymorphisms in the highly immunogenic Esx family, including some in known epitope regions, which may affect the immunogenicity of the corresponding protein. This is the case with EsxB (CFP-10) and EsxH (TB10.4) and has implications for the fields of vaccines and diagnostics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Megan Murray for helpful discussions and comments on the manuscript.
This study was supported by funding from the European Community's Seventh Framework Program (FP7/2007-2013) under grant agreement no. 201762 and by SystemsX.ch and the Swiss National Science Foundation (31003A-125061).
S.U. and J.R. are affiliated with the Swiss Institute of Bioinformatics.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 1 August 2011.
REFERENCES
- 1. Aagaard C., Govaerts M., Meng Okkels L., Andersen P., Pollock J. M. 2003. Genomic approach to identification of Mycobacterium bovis diagnostic antigens in cattle. J. Clin. Microbiol. 41:3719–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdallah A. M., et al. 2007. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5:883–891 [DOI] [PubMed] [Google Scholar]
- 3. Abdallah A. M., et al. 2008. The ESX-5 secretion system of Mycobacterium marinum modulates the macrophage response. J. Immunol. 181:7166–7175 [DOI] [PubMed] [Google Scholar]
- 4. Abdallah A. M., et al. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 62:667–679 [DOI] [PubMed] [Google Scholar]
- 5. Alderson M. R., et al. 2000. Expression cloning of an immunodominant family of Mycobacterium tuberculosis antigens using human CD4(+) T cells. J. Exp. Med. 191:551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andersen P., Andersen A. B., Sørensen A. L., Nagai S. 1995. Recall of long-lived immunity to Mycobacterium tuberculosis infection in mice. J. Immunol. 154:3359–3372 [PubMed] [Google Scholar]
- 7. Andersen P., Askgaard D., Ljungqvist L., Bentzon M. W., Heron I. 1991. T-cell proliferative response to antigens secreted by Mycobacterium tuberculosis. Infect. Immun. 59:1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arbing M. A., et al. 2010. The crystal structure of the Mycobacterium tuberculosis Rv3019c-Rv3020c ESX complex reveals a domain-swapped heterotetramer. Protein Sci. 19:1692–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Behr M. A., et al. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520–1523 [DOI] [PubMed] [Google Scholar]
- 10. Berthet F. X., Rasmussen P. B., Rosenkrands I., Andersen P., Gicquel B. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144(Pt. 11):3195–3203 [DOI] [PubMed] [Google Scholar]
- 11. Brodin P., et al. 2005. Functional analysis of early secreted antigenic target-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex formation, virulence, and immunogenicity. J. Biol. Chem. 280:33953–33959 [DOI] [PubMed] [Google Scholar]
- 12. Brodin P., et al. 2004. Enhanced protection against tuberculosis by vaccination with recombinant Mycobacterium microti vaccine that induces T cell immunity against region of difference 1 antigens. J. Infect. Dis. 190:115–122 [DOI] [PubMed] [Google Scholar]
- 13. Brosch R., et al. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. U. S. A. 104:5596–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brudey K., et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cole S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 16. Comas I., et al. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42:498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davila J., Zhang L., Marrs C. F., Durmaz R., Yang Z. 2010. Assessment of the genetic diversity of Mycobacterium tuberculosis esxA, esxH, and fbpB genes among clinical isolates and its implication for the future immunization by new tuberculosis subunit vaccines Ag85B-ESAT-6 and Ag85B-TB10.4. J. Biomed. Biotechnol. 2010:208371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delogu G., Brennan M. J. 2001. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect. Immun. 69:5606–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delogu G., Cole S. T., Brosch R. 2008. The PE and PPE gene protein families of M. tuberculosis, p. 131–150In Kaufmann S. H. E., van Helden P., Rubin E., Britton W. J.(ed.), Handbook of tuberculosis. Wiley-VHC, Weinheim, Germany [Google Scholar]
- 20. Dietrich J., et al. 2005. Exchanging ESAT6 with TB10.4 in an Ag85B fusion molecule-based tuberculosis subunit vaccine: efficient protection and ESAT6-based sensitive monitoring of vaccine efficacy. J. Immunol. 174:6332–6339 [DOI] [PubMed] [Google Scholar]
- 21. Elhay M. J., Oettinger T., Andersen P. 1998. Delayed-type hypersensitivity responses to ESAT-6 and MPT64 from Mycobacterium tuberculosis in the guinea pig. Infect. Immun. 66:3454–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Felsenstein J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 23. Fleischmann R. D., et al. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gagneux S., Small P. M. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7:328–337 [DOI] [PubMed] [Google Scholar]
- 25. Garnier T., et al. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. U. S. A. 100:7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gey Van Pittius N. C., et al. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2(10):44.1–44.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harboe M., Oettinger T., Wiker H. G., Rosenkrands I., Andersen P. 1996. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect. Immun. 64:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He X. Y., Zhuang Y. H., Zhang X.-G., Li G. L. 2003. Comparative proteome analysis of culture supernatant proteins of Mycobacterium tuberculosis H37Rv and H37Ra. Microbes Infect. 5:851–856 [DOI] [PubMed] [Google Scholar]
- 29. Jones G. J., Gordon S. V., Hewinson R. G., Vordermeier H. M. 2010. Screening of predicted secreted antigens from Mycobacterium bovis reveals the immunodominance of the ESAT-6 protein family. Infect. Immun. 78:1326–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamerbeek J., et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Louise R., Skjøt V., Agger E. M., Andersen P. 2001. Antigen discovery and tuberculosis vaccine development in the postgenomic era. Scand. J. Infect. Dis. 33:643–647 [DOI] [PubMed] [Google Scholar]
- 32. Mahmood A., et al. 2011. Molecular characterization of secretory proteins Rv3619c and Rv3620c from Mycobacterium tuberculosis H37Rv. FEBS J. 278(2):341–353 [DOI] [PubMed] [Google Scholar]
- 33. Mazurek G. H., et al. 2001. Comparison of a whole-blood interferon gamma assay with tuberculin skin testing for detecting latent Mycobacterium tuberculosis infection. JAMA 286:1740–1747 [DOI] [PubMed] [Google Scholar]
- 34. Meier T., Eulenbruch H. P., Wrighton-Smith P., Enders G., Regnath T. 2005. Sensitivity of a new commercial enzyme-linked immunospot assay (T SPOT-TB) for diagnosis of tuberculosis in clinical practice. Eur. J. Clin. Microbiol. Infect. Dis. 24:529–536 [DOI] [PubMed] [Google Scholar]
- 35. Musser J. M., Amin A., Ramaswamy S. 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155:7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. North R. J., Jung Y. J. 2004. Immunity to tuberculosis. Annu. Rev. Immunol. 22:599–623 [DOI] [PubMed] [Google Scholar]
- 37. Pallen M. J. 2002. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol. 10:209–212 [DOI] [PubMed] [Google Scholar]
- 38. Pond S. L. K. 2005. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol. Biol. Evol. 22:478–485 [DOI] [PubMed] [Google Scholar]
- 39. Pond S. L. K., Frost S. D. W., Muse S. V. 2005. HyPhy: hypothesis testing using phylogenies. Stat. Methods Mol. Evol. 21(5):676–679 [DOI] [PubMed] [Google Scholar]
- 40. Pym A. S., Brodin P., Brosch R., Huerre M., Cole S. T. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709–717 [DOI] [PubMed] [Google Scholar]
- 41. Pym A. S., et al. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 9:533–539 [DOI] [PubMed] [Google Scholar]
- 42. Ravn P., et al. 1999. Human T cell responses to the ESAT-6 antigen from Mycobacterium tuberculosis. J. Infect. Dis. 179:637–645 [DOI] [PubMed] [Google Scholar]
- 43. Renshaw P. S., et al. 2005. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 24:2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siegrist M. S., et al. 2009. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. U. S. A. 106:18792–18797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Skjøt R. L. V., et al. 2002. Epitope mapping of the immunodominant antigen TB10.4 and the two homologous proteins TB10.3 and TB12.9, which constitute a subfamily of the ESAT-6 gene family. Infect. Immun. 70:5446–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sugiura N. 1978. Further analysis of the data by Akaike' s information criterion and the finite corrections. Commun. Stat. Theory Methods 7:13–26 [Google Scholar]
- 47. Supply P., et al. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762–771 [DOI] [PubMed] [Google Scholar]
- 48. Tekaia F., et al. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 79:329–342 [DOI] [PubMed] [Google Scholar]
- 49. van Pinxteren L. A., Ravn P., Agger E. M., Pollock J., Andersen P. 2000. Diagnosis of tuberculosis based on the two specific antigens ESAT-6 and CFP10. Clin. Diagn. Lab. Immunol. 7:155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vita R., et al. 2010. The immune epitope database 2.0. Nucleic Acids Res. 38:D854–D862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15:568–573 [DOI] [PubMed] [Google Scholar]
- 52. Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591 [DOI] [PubMed] [Google Scholar]
- 53. Zheng H., et al. 2008. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One 3:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.