Abstract
The intestine is inhabited by a large microbial community consisting primarily of anaerobes and, to a lesser extent, facultative anaerobes, such as Escherichia coli, which we have shown requires aerobic respiration to compete successfully in the mouse intestine (S. A. Jones et al., Infect. Immun. 75:4891-4899, 2007). If facultative anaerobes efficiently lower oxygen availability in the intestine, then their sustained growth must also depend on anaerobic metabolism. In support of this idea, mutants lacking nitrate reductase or fumarate reductase have extreme colonization defects. Here, we further explore the role of anaerobic respiration in colonization using the streptomycin-treated mouse model. We found that respiratory electron flow is primarily via the naphthoquinones, which pass electrons to cytochrome bd oxidase and the anaerobic terminal reductases. We found that E. coli uses nitrate and fumarate in the intestine, but not nitrite, dimethyl sulfoxide, or trimethylamine N-oxide. Competitive colonizations revealed that cytochrome bd oxidase is more advantageous than nitrate reductase or fumarate reductase. Strains lacking nitrate reductase outcompeted fumarate reductase mutants once the nitrate concentration in cecal mucus reached submillimolar levels, indicating that fumarate is the more important anaerobic electron acceptor in the intestine because nitrate is limiting. Since nitrate is highest in the absence of E. coli, we conclude that E. coli is the only bacterium in the streptomycin-treated mouse large intestine that respires nitrate. Lastly, we demonstrated that a mutant lacking the NarXL regulator (activator of the NarG system), but not a mutant lacking the NarP-NarQ regulator, has a colonization defect, consistent with the advantage provided by NarG. The emerging picture is one in which gene regulation is tuned to balance expression of the terminal reductases that E. coli uses to maximize its competitiveness and achieve the highest possible population in the intestine.
INTRODUCTION
The mouse colon is home to at least several hundred bacterial species and more than 100 billion bacteria per g of contents, a microbial community dominated by anaerobes with essentially no aerobes (2, 38, 57, 68). It is becoming increasingly clear that facultative anaerobes consume oxygen and thereby modify their host environment. For example, kidney infection caused by uropathogenic Escherichia coli results in local ischemia (47). The facultatively anaerobic enteric pathogen Shigella flexneri responds to oxygen in the gut by modulating virulence gene expression (45). It has been postulated that the importance of Salmonella motility for chemotaxis through the mucus layer (61) may be due to aerotaxis (44). That E. coli requires cytochrome bd oxidase to gain a competitive advantage implies that the colon is not the anaerobic environment that many consider it to be (30). Here, we explore the possibility that efficient oxygen scavenging by E. coli in the intestine causes it to depend also on anaerobic respiration.
To maximize their energy efficiency, enteric bacteria such as E. coli possess elaborate genetic regulatory networks for sensing oxygen (32), the redox state of the electron transport system (19, 43, 58), and the availability of alternative anaerobic electron acceptors (70). We previously showed that the anaerobic transcription factor Fnr, as well as the aerobic respiratory controller ArcA, is required for commensal and pathogenic E. coli strains to colonize the intestine in competition with wild-type E. coli parent strains (30). We further showed that mutants lacking ATPase (completely respiration deficient) and mutants lacking terminal reductases for low oxygen, nitrate, or fumarate each are at an extreme disadvantage in competition with their wild-type parents for colonization of the streptomycin-treated mouse intestine (30). On the basis of these results, we hypothesize that E. coli maximizes its cell yield from limited carbon sources by using the most energy-efficient respiratory electron acceptors that are available to it in the intestine.
There is clear evidence to indicate that oxygen is present in the gut (24, 37). We hypothesize that oxygen-scavenging facultative anaerobes such as E. coli create an anaerobic environment for the numerically dominant intestinal anaerobes, in analogy to the situation in anaerobic digesters (28). Our research supports this idea. Nevertheless, our experiments also indicate that reduction of the anaerobic electron acceptors nitrate and fumarate is essential for E. coli to maintain its competitive advantage in the intestine (30). We take this as evidence that the ability to efficiently respire oxygen and maintain an anaerobic environment (into which oxygen continuously diffuses) likewise requires maximum efficiency under anaerobic conditions. Here, we explore the possibility that additional alternative electron acceptors, i.e., nitrite, dimethyl sulfoxide (DMSO), and trimethylamine N-oxide (TMAO), support E. coli intestinal colonization. The results indicate that nitrate and fumarate are the only anaerobic electron acceptors used by commensal and pathogenic strains of E. coli to colonize the intestine. In addition, we present further evidence that anaerobic electron transport chain components, as well as the NarX-NarL two-component system that governs gene expression for anaerobic respiratory systems, are necessary for competitive fitness in the streptomycin-treated mouse intestine.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
In Table 1 we list the E. coli mutants used in this study; these were derived from E. coli MG1655 Strr (streptomycin resistant), a K-12 strain isolated from a human (4), and E. coli EDL933 Strr, the prototypical O157:H7 strain (53). For colonization experiments, the wild-type strains were E. coli MG1655 Strr Nalr (nalidixic acid resistant) (52) and E. coli EDL933 Strr Nalr (51); Nalr is used to distinguish the wild type from null allele mutants. Null alleles were constructed by using the allelic replacement method of Datsenko and Wanner (14) as described previously (8), such that target genes were deleted and replaced with kanamycin or chloramphenicol resistance cassettes (used as selectable markers in mouse colonization assays described below). The null allele strains are identified in the text by the genes that were deleted; single gene deletions began with the start codon and ended with the stop codon, and multiple gene deletions began with the start codon of the first gene deleted and ended with the stop codon of the last gene deleted. Strains containing multiple mutations were constructed by sequential allelic replacement; removal of the first inserted cassette with FLP recombinase (14) was followed by subsequent allelic replacement(s) and removal of the insertion as necessary, leaving the selectable marker in the last mutation made. It is possible that mutations were inadvertently introduced elsewhere on the genome during strain construction. However, we would argue that this was not the case because the allelic replacements described herein were obtained with a frequency that varied less than 1 order of magnitude. In addition, the results obtained in two different genetic backgrounds (EDL933 and MG1655) were essentially identical. The narXL and narP mutations were moved from strains VJS2458 and VJS4322, respectively, into the E. coli MG1655 Strr background by allelic replacement. Mutations were verified by phenotype and DNA sequence analyses.
Table 1.
Strains used in this study
| Strain | Relevant genotype, phenotype, or descriptiona | Source or reference |
|---|---|---|
| EDL933 | Wild-type O157:H7 | Alison O'Brien |
| EDL933 Strr | Spontaneous Strr | 51 |
| EDL933 Strr Nalr | Spontaneous Nalr | 51 |
| MG1655 | Wild type (CGSC 7740) | CGSCb |
| MG1655 Strr | Spontaneous Strr | 52 |
| MG1655 Strr Nalr | Spontaneous Nalr | 52 |
| VJS4322 | Δ(narP)253::Tn10d(cat) | Valley Stewart |
| VJS2458 | Δ(narXL)240::Knr | Valley Stewart |
| Derivatives of EDL933 Strr | ||
| cydAB mutant | Δ(cydAB)::cat | This study |
| cydDC mutant | Δ(cydDC)::cat | This study |
| dmsAB mutant | Δ(dmsAB)::cat | This study |
| frdA mutant | ΔfrdA::Knr | This study |
| napDA mutant | Δ(napDA)::cat | This study |
| narG mutant | ΔnarG::Knr | This study |
| narZ mutant | ΔnarZ::cat | This study |
| narG narZ mutant | ΔnarG ΔnarZ::cat | This study |
| narG narZ napDA mutant | ΔnarG ΔnarZ Δ(napDA)::cat | This study |
| ubiCA mutant | Δ(ubiCA)::cat | This study |
| ubiE mutant | ΔubiE::cat | This study |
| Derivatives of MG1655 Strr | ||
| cydAB mutant | Δ(cydAB)::cat | This study |
| cydDC mutant | Δ(cydDC)::cat | This study |
| dmsAB mutant | Δ(dmsAB)::cat | This study |
| dmsAB ynfEFGH mutant | Δ(dmsAB) Δ(ynfEH)::cat | This study |
| frdA mutant | ΔfrdA::Knr | This study |
| napDA mutant | Δ(napDA)::cat | This study |
| narG mutant | ΔnarG::Knr | This study |
| narZ mutant | ΔnarZ::cat | This study |
| narG narZ mutant | ΔnarG ΔnarZ::cat | This study |
| narG narZ napDA mutant | ΔnarG ΔnarZ Δ(napDA)::cat | This study |
| narP mutant | Δ(narP)253::Tn10d(cat) | This study |
| narXL mutant | Δ(narXL)240::Knr | This study |
| nirBD mutant | Δ(nirB-nirD)::cat | This study |
| nrfA mutant | ΔnirfA::cat | This study |
| nrfA nirBD mutant | ΔnirfA Δ(nirBD)::cat | This study |
| torCA mutant | Δ(torCA)::cat | This study |
| torYZ mutant | Δ(torYZ)::cat | This study |
| torCA torYZ mutant | Δ(torCA) Δ(torYZ)::cat | This study |
| torCA torYZ dmsAB mutant | Δ(torCA) Δ(torYZ) Δ(dmsAB)::cat | This study |
| ubiCA mutant | Δ(ubiCA)::cat | This study |
| ubiE mutant | ΔubiE::cat | This study |
Nalr, nalidixic acid resistance; Strr, streptomycin resistance; Knr, kanamycin resistance.
E. coli Genetic Stock Culture Collection, Yale University.
Phenotype analysis.
MOPS [3-(N-morpholino)propanesulfonic acid] defined medium was used for phenotype analysis, as described previously (8). Anaerobic cultures were grown in sealed Balch culture tubes filled to the top with N2-sparged medium. To test for nitrate or fumarate respiration, mutant strains were grown anaerobically overnight in MOPS medium with glycerol (1.6%) as the carbon source and either 50 mM nitrate or 50 mM fumarate as the electron acceptor. Cell growth was monitored spectrophotometrically at 600 nm (as the optical density at 600 nm [OD600]).
High-performance liquid chromatography (HPLC) analysis.
For anion analysis of mucus, we used a Dionex DX-500-Microbore system with a 10-μl injection volume, an IonPac AS11 column (at 30°C) with a 1 to 25 mM NaOH gradient as the eluent at a 0.5-ml/min flow rate, and a suppressed conductivity detector with the ASRS-ULTRA 2-mm AutoSuppression mode. The regression coefficient for the standard curve was used to demonstrate the linearity of the peak area with respect to concentration. The limit of detection of nitrate in this system was 3 μM. Mouse cecal mucus was isolated from the ceca of CD-1 male mice and lyophilized as described previously (72). Mucus samples from 5 mice were pooled and assayed in triplicate.
Mouse colonization experiments.
Streptomycin-treated mice have been used since 1954 to overcome the colonization resistance that prevents colonization of conventional animals by experimentally introduced enteric bacteria (7). We have made extensive use of the streptomycin-treated mouse model to study nutritional factors that govern colonization of the mouse large intestine by E. coli and Salmonella enterica serovar Typhimurium (3, 8, 16, 18, 30, 31, 34, 35, 48, 49, 51, 52). Treatment of mice with streptomycin selectively removes facultatively anaerobic E. coli bacteria, enterococci, streptococci, lactobacilli, and anaerobic lactobacilli and bifidobacteria without changing the overall populations of anaerobes, including Bacteroides and Eubacterium (25). Therefore, the streptomycin-treated mouse model allows colonization by experimentally introduced E. coli strains and competition with large numbers of strict anaerobes and, thus, is our model of choice for studying competition among E. coli strains in the intestine. Stable colonization requires that streptomycin treatment be continued for the duration of the experiments. For E. coli EDL933, we emphasize that streptomycin-treated mice are used to model colonization, not pathogenesis (13). Briefly, CD-1 male mice, 6 weeks of age, are given drinking water containing streptomycin sulfate (5 g/liter) for 24 h, which by removing the native facultative anaerobes opens the niche for E. coli to colonize (50). Following 18 h of starvation for food and water, the mice are fed 1 ml of 20% (wt/vol) sucrose containing 105 CFU of E. coli strains grown overnight in Luria broth. After the bacterial suspension is ingested, both food (Teklad mouse and rat diet; Harlan, Madison, WI) and streptomycin-water are returned to the mice, and 1-g samples of feces are collected after 5 and 24 h and on odd-numbered days thereafter. Mice are housed individually in cages without bedding and are placed in clean cages at 24-h intervals. Fecal samples (1 g) are therefore no older than 24 h. Each fecal sample is homogenized in 10 ml of 1% Bacto tryptone (Difco), diluted in the same medium, and plated onto MacConkey agar plates containing either streptomycin (100 μg/ml) and nalidixic acid (50 μg/ml) to count wild type CFU or streptomycin and kanamycin (40 μg/ml) or chloramphenicol (30 μg/ml) to count CFU of the null allele mutants. Streptomycin sulfate, chloramphenicol, and nalidixic acid were purchased from Sigma-Aldrich (St. Louis, MO). The limit of detection for fecal plate counts is 102 CFU/g feces. Each colonization experiment was replicated at least twice, with essentially identical results. Pooled data from at least 2 independent experiments (a total of 6 mice) are presented in the figures as the log of fecal plate counts.
RESULTS
Anaerobic quinones contribute to respiration in the intestine.
Previously, we showed that E. coli mutants lacking cytochrome bd oxidase, nitrate reductase (Nar), or fumarate reductase (Frd) are eliminated from the intestine by competition with the wild-type parent strains (30). The relative contributions of aerobic and anaerobic respiratory processes to colonization are not known. The respiratory dehydrogenases are linked to the terminal reductases by a pool of quinones comprising ubiqinone (UQ), menaquinone (MQ), and demethylmenaquinone (DMQ). Theoretically, this allows formation of branched respiratory chains in which most dehydrogenases can interact with most terminal reductases (70). However, the midpoint potential of UQ is best suited for aerobic respiration, whereas MQ and DMQ are involved primarily in anaerobic respiration, with DMQ having a midpoint potential between those of UQ and MQ (70). In general, anaerobic terminal reductases have higher activity with MQ and DMQ, cytochrome bo3 has the highest activity with UQ, and cytochrome bd apparently uses both UQ and MQ (23). Electron flow to fumarate, TMAO, and nitrite is via both MQ and DMQ, while electron flow to DMSO is primarily via MQ. The primary Nar enzyme (NarG) uses UQ and MQ but not DMQ (77). While all three quinones are present under both aerobic and anaerobic conditions, their relative concentrations vary with oxygen tension, with DMQ increasing rapidly and MQ increasing more slowly when oxygen is limited (6, 69, 75). Accordingly, while DMQ would be in the greatest abundance under the low oxygen tension in the mouse intestine (30), DMQ alone would not be sufficient for colonization since electron flow to nitrate and low oxygen would require MQ under these conditions. Thus, we predicted that respiratory electron flow via both MQ and DMQ supports colonization of the intestine by E. coli.
To confirm this hypothesis, we used mutants lacking one or more quinone biosynthetic pathways (46). Specifically, as shown in Table 2, ubiCA mutants cannot make UQ (33, 78, 80), ubiE mutants cannot make UQ or MQ (36, 77), and menA mutants cannot make MQ or DMQ (65, 79). The ubiCA, ubiE, and menA mutations were constructed in E. coli MG1655 and E. coli EDL933, the mutants were fed to streptomycin-treated mice together with the respective wild-type parent strains at 105 CFU of each strain, and the population of each strain was monitored by using fecal plate counts. The results of these experiments are shown in Table 2. The ubiCA mutants, lacking UQ, cocolonized with the wild type, which is evidence that MQ and DMQ are sufficient for colonization of the intestine by E. coli. The menA mutants, lacking both MQ and DMQ, were able to increase in population during the initiation stage of colonization (24 h), although not to the same extent as the wild type, but were completely eliminated (reaching levels of <102 CFU/g feces) by day 9 of the experiment. One interpretation of these results is that UQ helps support colonization initiation when MQ is unavailable. Given that the passage of electrons to oxygen is primarily via UQ when oxygen is not limiting, it may be that UQ is used during the initiation stage because oxygen levels are relatively high in animals that have been treated with streptomycin to remove the facultative anaerobes prior to colonization with E. coli. The ubiE mutants, which have only DMQ, failed to initiate colonization in competition with the wild type (in the first 24 h) and were eliminated from the mice (reaching levels of <102 CFU/g feces), which indicates that anaerobic electron flow via MQ in the intestine is more advantageous than that via DMQ and further supports the idea that UQ is used during colonization initiation. All four of the strains with competitive colonization defects (E. coli MG1655 ubiE, E. coli MG1655 menA, E. coli EDL933 ubiE, and E. coli EDL933 menA) colonized to yield between 107 and 108 CFU/g feces when fed alone to mice, indicating that the colonization defects were due to an inability to compete with the wild type rather than a general inability to grow in the intestine (data not shown). In summary, we conclude that respiratory electron flow via MQ and DMQ to cytochrome bd oxidase and anaerobic terminal reductases supports colonization of the intestine by E. coli.
Table 2.
Competitive colonization by E. coli quinone biosynthesis mutants and alternative anaerobic electron acceptor mutantsa
| Mutant genotype | Presence or absenceb of: |
Difference between E. coli EDL933 wild-type and mutant strains on: |
Difference between E. coli MG1655 wild-type and mutant strains on: |
||||
|---|---|---|---|---|---|---|---|
| UQ | MQ | DMQ | Day 1 | Day 9 | Day 1 | Day 9 | |
| Δ(ubiCA) | − | + | + | 0.21 ± 0.05 | 0.71 ± 0.14 | 0.78 ± 0.27 | 0.24 ± 0.22 |
| ΔubiE | − | − | + | 3.26 ± 0.13 | 5.80 ± 0.13 | 4.04 ± 0.09 | 5.03 ± 0.20 |
| ΔmenA | + | − | − | 1.23 ± 0.16 | 5.15 ± 0.19 | 1.00 ± 0.24 | 5.66 ± 0.11 |
| ΔdmsABC | + | + | + | 0.21 ± 0.11 | −0.31 ± 0.25 | 0.21 ± 0.18 | 0.21 ± 0.17 |
| ΔynfEFGH | + | + | + | NDc | ND | 0.16 ± 0.48 | 0.71 ± 0.25 |
| ΔdmsABC ΔynfEFGH | + | + | + | ND | ND | 0.11 ± 0.17 | 0.62 ± 0.12 |
| ΔtorCA | + | + | + | 0.52 ± 0.13 | 0.10 ± 0.11 | 0.18 ± 0.16 | 0.15 ± 0.26 |
| ΔtorYZ | + | + | + | ND | ND | 0.47 ± 0.37 | 0.51 ± 0.17 |
| ΔtorCA ΔtorYZ | + | + | + | ND | ND | 0.61 ± 0.54 | 0.15 ± 0.58 |
| ΔdmsABC ΔtorCA ΔtorYZ | + | + | + | ND | ND | 0.38 ± 0.19 | 0.26 ± 0.23 |
| ΔnirBD | + | + | + | 0.30 ± 0.24 | 0.49 ± 0.46 | 0.05 ± 0.16 | 0.64 ± 0.30 |
| ΔnrfA | + | + | + | ND | ND | 0.02 ± 0.25 | 0.11 ± 0.21 |
| ΔnirBD ΔnrfA | + | + | + | ND | ND | −0.21 ± 0.25 | 0.21 ± 0.11 |
Mice were fed 105 CFU each of the mutant and its wild-type parent. The average differences between the wild-type strain and its isogenic mutant (among 6 mice) are represented as the log number of wild-type CFU per gram of feces minus the log number of mutant CFU per gram of feces ± the standard error of the mean on days 1 and 9 of the experiments. All values shown in bold are statistically significant: P < 0.001 (Student's t test).
Presence (+) or absence (−) of UQ, MQ, or DMQ, in accordance with the indicated genotypes.
ND, not determined.
Alternative electron acceptors DMSO, TMAO, and nitrite do not support colonization.
In addition to its ability to respire nitrate and fumarate, E. coli possesses systems for reduction of DMSO, TMAO, and nitrite as terminal electron acceptors (11). To investigate these alternative respiratory pathways in the intestine, each of the redundant terminal reductase gene systems was knocked out in E. coli MG1655 by allelic replacement, the mutant strains were fed to mice together with their wild-type parents at 105 CFU of each strain, and the relative populations of the strains in fecal plate counts were determined. The DmsABC system reduces both DMSO and TMAO; a paralogous YnfEFGHI system for DMSO reduction (41) is now thought to reduce selenium (22). Mutants containing deletions of either or both of the corresponding gene systems were found to cocolonize with wild-type E. coli MG1655, indicating that reduction of DMSO does not provide a colonization advantage in the intestine (Table 2). The TorCAD and TorYZ systems, in addition to DmsABC, are responsible for TMAO reduction (9). Mutants lacking these TMAO reductase systems, either individually or collectively, cocolonized with the wild-type parent strain, indicating that reduction of TMAO does not contribute to the fitness of E. coli MG1655 in the intestine (Table 2). Lastly, to investigate the contribution of nitrite reductase to colonization fitness, we constructed mutants lacking either or both the cytoplasmic NirBCD and periplasmic NrfABCD nitrite reductase systems. We found that in competition experiments, each of these mutants cocolonized with wild-type E. coli MG1655 (Table 2), indicating that nitrite is not used in the intestine. Given that nitrate is used and that the product (nitrite) is potentially toxic (40), it is interesting that the loss of nitrite reductase does not result in a competitive disadvantage in the intestine. From these results we conclude that colonization of the intestine by E. coli is not supported by respiration of DMSO, TMAO, or nitrite, either because these electron acceptors are unavailable or because the terminal reductases are not functionally expressed. Together with results from the previous study (30), these new results lead us to conclude that respiration of nitrate and respiration of fumarate, in addition to fermentation, are the only anaerobic processes that contribute to the colonization fitness of E. coli in the mouse intestine.
Aerobic cytochrome bd is more advantageous than anaerobic respiration.
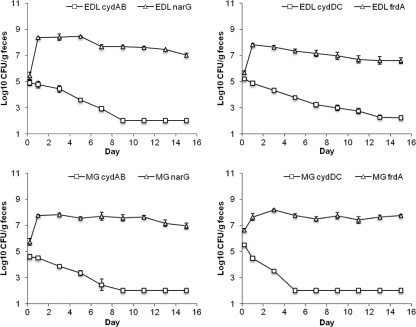
We sought to evaluate the relative contributions of aerobic and anaerobic respiration to colonization of the intestine. Previously, we showed that the low-affinity, high-oxygen-tension cytochrome bo3 oxidase does not contribute to colonization but that the high-affinity, low-oxygen cytochrome bd oxidase is essential for competition with wild-type E. coli in the intestine (30). The E. coli EDL933 and E. coli MG1655 cydAB mutants were competed against E. coli EDL933 and E. coli MG1655 narG mutants, respectively, by feeding the two strains together to mice at 105 CFU of each strain. In both experiments the cydAB mutants were eliminated from the intestine (Fig. 1). Likewise, the E. coli EDL933 and E. coli MG1655 cydDC mutants were eliminated from the intestine during competition against E. coli EDL933 and E. coli MG1655 frdA mutants, respectively (Fig. 1). We showed previously that E. coli cydDC mutants that cannot assemble cytochrome bd oxidase in the membrane (54) have colonization defects similar to those of cydAB mutants (30). These results indicate that aerobic respiration via cytochrome bd oxidase provides an advantage over anaerobic respiration of nitrate or fumarate in the intestine.
Fig. 1.
Competition between mutants lacking cytochrome bd and mutants lacking either nitrate reductase or fumarate reductase. Upper left, E. coli EDL933 ΔcydAB (EDL cydAB) versus E. coli EDL933 ΔnarG (EDL narG); upper right, E. coli EDL933 ΔcydDC (EDL cydDC) versus E. coli EDL933 ΔfrdA (EDL frdA); lower left, E. coli MG1655 ΔcydAB (MG cydAB) versus E. coli MG1655 ΔnarG (MG narG); lower right, E. coli MG1655 ΔcydDC (MG cydDC) versus E. coli MG1655 ΔfrdA (MG frdA).
The primary Nar enzyme contributes a major colonization advantage.
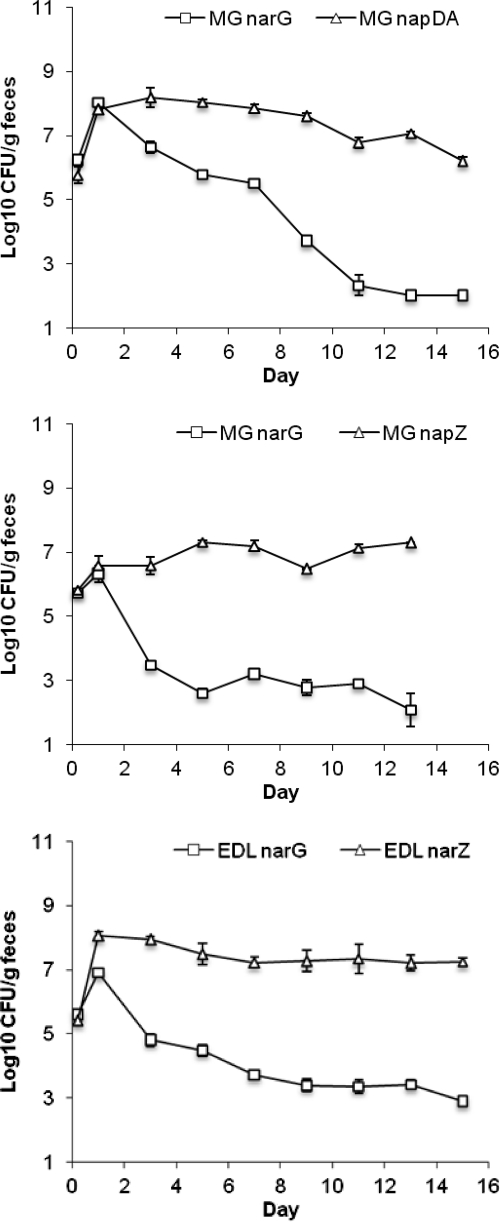
There are three nitrate reductase systems in E. coli (70). Previously, we showed that strains lacking the primary enzyme, NarG, have a significant colonization defect in competition with the wild type but that strains lacking the secondary cytoplasmic enzyme NarZ or Nap are as competitive as the wild type, which suggests that NarG is the major contributor to nitrate respiration in the intestine (30). Strains lacking all three systems had colonization defects up to 1 log more severe than the strain lacking NarG alone, which suggests that the other systems may contribute modestly to colonization in the absence of NarG (30). To further evaluate the relative importance of the Nar systems, we competed narG mutants individually against narZ and napDA mutants (Fig. 2). In competition with the narZ mutant, the narG mutants had a 4-log colonization defect. In competition with the napDA mutant, the narG mutant initially reached the same high population (108 CFU/g feces) before showing a 2-log defect at day 7 and then a more rapid decline to an undetectable number (102 CFU/g feces) by day 13 of the experiment. These results confirm that electron flow to nitrate occurs primarily via NarG.
Fig. 2.
Competition between nitrate reductase mutants. Top, E. coli MG1655 ΔnapDA versus E. coli MG1655 ΔnarG; middle, E. coli MG1655 ΔnarZ versus E. coli MG1655 ΔnarG; bottom, E. coli EDL933 ΔnarZ versus E. coli EDL933 ΔnarG.
Nitrate availability dictates competition for anaerobic electron acceptors in the intestine.
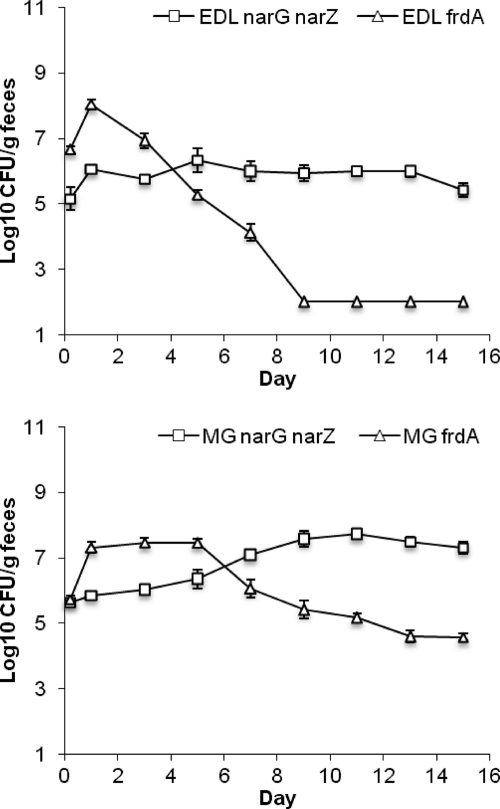
To determine which of the two anaerobic electron acceptors used by E. coli in the intestine provides the superior colonization advantage, we competed E. coli EDL933 narG narZ with E. coli EDL933 frdA and E. coli MG1655 narG narZ with E. coli MG1655 frdA (Fig. 3). In both experiments the frdA mutants, which can respire nitrate but not fumarate, initially (at day 3 or day 5) had a colonization advantage but subsequently declined in numbers relative to the narG narZ mutants, which persisted at relatively high populations. Apparently, nitrate respiration provides an advantage during the initial stage of colonization, when the level of nitrate is expected to be high if no other members of the microbial community consume it. However, we conclude that the ability to respire fumarate provides a more significant colonization advantage than the ability to respire nitrate for long-term persistence of established populations of E. coli.
Fig. 3.
Competition between nitrate reductase and fumarate reductase mutants. Top, E. coli EDL933 ΔnarG ΔnarZ versus E. coli EDL933 ΔfrdA; bottom, E. coli MG1655 ΔnarG ΔnarZ versus E. coli MG1655 ΔfrdA.
To determine the role of nitrate availability in the competition between Nar and Frd mutants, we measured the nitrate concentration in cecal mucus after association with 105 CFU each of E. coli EDL933 narG narZ napDA and E. coli EDL933 frdA (Table 3). In control experiments, mice colonized with E. coli EDL933 Strr (the wild type) had undetectable levels of nitrate (<3 μM) in their cecal mucus on days 1, 5, and 11 following association. Mice colonized with E. coli EDL933 narG narZ napDA alone had an average of 2.8 mM nitrate. Following association with E. coli EDL933 narG narZ napDA and E. coli EDL933 frdA, nitrate in cecal mucus declined to a mean ± standard error of the mean of 0.77 ± 0.31 mM on day 1, reached 0.31 ± 0.25 mM on day 5, and was undetectable (<3 μM) on day 11 of the experiment (Table 3). In the colonization experiment (Fig. 3), the Nar+ Frd− strain increased in population at 24 h, consistent with the availability of nitrate, whereas the Nar− Frd+ strain was unable to increase in population and remained at 6 log CFU/g feces. By day 3 of the experiment, as the nitrate concentration decreased (presumably through respiratory metabolism by the competing Nar+ strain), the Nar+ Frd− strain declined in population whereas the Nar− Frd+ strain persisted. Thus, the population of the Nar+ Frd− strain declined as nitrate diminished, indicating that respiration of nitrate by E. coli in the intestine is limited by nitrate availability but that fumarate is more readily available and therefore more important for persistence in the intestine.
Table 3.
HPLC analysis of nitrate in mouse cecal mucus
| Strain | NO3− concna (mM) in mucus | Day of colonization |
|---|---|---|
| EDL933 Strr | ND | 1 |
| ND | 5 | |
| ND | 11 | |
| narG narZ napDA mutant | 2.76 ± 0.27 | 1 |
| 2.89 ± 0.33 | 5 | |
| 2.65 ± 0.49 | 11 | |
| narG narZ napDA frdA mutant | 0.77 ± 0.31 | 1 |
| 0.31 ± 0.25 | 5 | |
| ND | 11 |
Nitrate was measured in samples of cecal mucus prepared from mice colonized with the indicated E. coli strains. Values are means ± standard errors of the means for samples from 5 mice. ND, not detected (<3 μM).
The NarX-NarL regulatory circuit contributes to colonization.
Regulation of anaerobic respiration is thought to contribute to the success of E. coli in changing environments (10). Thus, it was of interest to determine the relative contributions to colonization of the two two-component systems that control anaerobic gene systems involved in nitrate respiration. NarG is activated by the NarX-NarL two-component system when nitrate is abundant, and NarX-NarL represses Nap, which is activated by the NarQ-NarP system when nitrate is 3 orders of magnitude lower (74, 76). To determine the relative contributions of the NarX-NarL and NarQ-NarP systems to colonization, mice were fed E. coli MG1655 narXL or E. coli MG1655 narP together with the wild-type parent at 105 CFU of each strain and the relative populations of the strains were determined (data not shown). The narXL mutant had a 1-log colonization defect (P < 0.05) in competition with the wild type, and the narP mutant cocolonized with the wild type. As discussed further below, it makes sense that the structural gene (narG) colonization defect is more severe than the regulatory gene (narXL) defect. These results confirm the importance of NarG for colonization, since the main function of NarX-NarL is to activate NarG.
Aerobic respiratory control is more important than aerobic respiration.
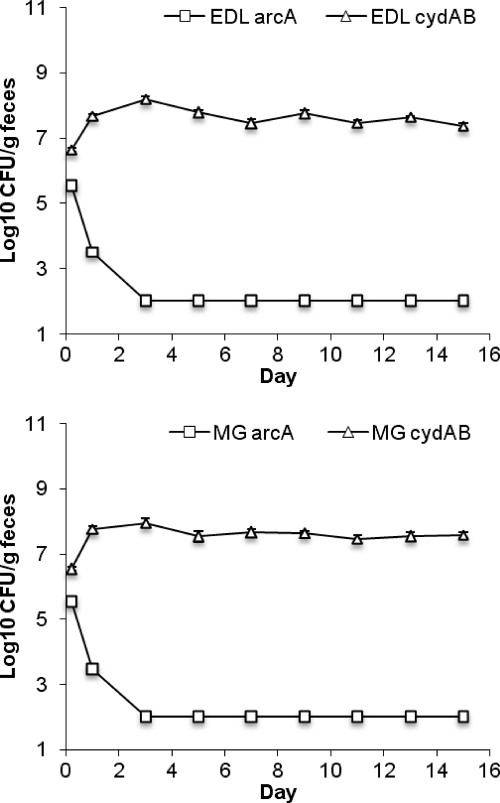
Previously, we showed that Fnr and ArcA mutants are eliminated from the intestine in competition with the wild type (30). Since mutants lacking the high-affinity cytochrome bd oxidase have a colonization defect, we concluded that E. coli experiences microaerobic or alternately aerobic and anaerobic conditions in the intestine (30). Interestingly, the two-component ArcAB system is most active at low oxygen tension (1), i.e., at oxygen levels present in the large intestine (24). Thus, we considered the possibility that ArcAB contributes to colonization fitness because it activates transcription of the cytochrome bd oxidase genes (24). To test whether the loss of ArcAB activation of cytochrome bd was the sole reason for the previously observed colonization defect of the ΔarcA mutant in competition with the wild type (30), ΔarcA::kan and Δ(cydAB)::cat mutants of E. coli EDL933 and MG1655 were fed together to mice at a level of 105 CFU of each strain/mouse. The arcA mutants with either genetic background were quickly eliminated during competition with the cydAB mutants (Fig. 4). These results indicate that failure to activate cydAB under microaerobic conditions is not the sole reason for the colonization defect of the arcA mutant and imply that other aspects of ArcAB control, such as repression of cytochrome bo3 oxidase genes and other genes that participate in aerobic processes (e.g., the tricarboxylic acid [TCA] cycle), are also important in the intestine.
Fig. 4.
Competition between E. coli EDL933 arcA and E. coli EDL933 cydAB (top) and E. coli MG1655 arcA and E. coli MG1655 cydAB (bottom).
DISCUSSION
Commensal E. coli MG1655 and pathogenic E. coli EDL933 must respire oxygen, nitrate, and fumarate to be competitive in the intestine (30). Analysis of a series of quinone biosynthesis mutants (Table 2) demonstrated that electron flow in colonized E. coli is primarily via the naphthoquinone pool (MQ and DMQ), which is used by cytochrome bd oxidase and the anaerobic terminal reductases that use nitrate and fumarate. Competition experiments revealed that respiration of oxygen provides a greater colonization advantage than anaerobic respiration (Fig. 1). NarG is the most important of the three systems for nitrate respiration (Fig. 2). Similarly, a mutant lacking the NarX-NarL regulatory system, which activates NarG synthesis, had a colonization defect. Competition of mutants lacking nitrate reductase with mutants lacking Frd revealed an interesting interplay between the two strains, which can be explained by nitrate limitation in the cecum (Fig. 3 and Table 3). Lastly, competition between a mutant lacking the aerobic respiratory control system and a mutant lacking cytochrome bd oxidase indicated that the role of ArcA in colonization extends beyond simple activation of cydAB (Fig. 4).
The emerging picture is one in which E. coli generates the energy it requires to colonize the mammalian intestine by respiring oxygen, nitrate, and fumarate. As would be expected when considering the potential of these processes to generate energy (11), oxygen is favored over nitrate or fumarate. However, it is clear from published studies (24, 37) that oxygen is limiting in the intestine, consistent with our observations that anaerobic respiration also contributes to colonization. The studies described herein support the conclusion that nitrate, too, is limiting in the intestine. In this regard, E. coli can be thought of as a scavenger of available electron acceptors, using whichever one is available to maximize its competitiveness and achieve the highest possible population.
Availability and use of electron acceptors.
Oxygen diffuses from the intestinal epithelium into the mucus layer (59). Fumarate is generated endogenously from carbohydrates via glycolysis and the reductive branch of the TCA cycle and also from available C4-dicarboxylates and related compounds such as aspartate via metabolism (29). In humans, dietary nitrate, mostly from vegetables (42), as well as nitrate biosynthesis (21), contributes to the available nitrate in the body. While it is known that the availability of nitrate in the ferret intestine increases with antibiotic treatment to remove the microbiota (15), microbial nitrate metabolism in animals and humans remains largely unexplored.
We measured 2.8 mM nitrate in cecal mucus prepared from mice that were treated with streptomycin to remove native E. coli and subsequently colonized with an E. coli EDL933 mutant that is completely Nar deficient. Upon colonization of the streptomycin-treated mouse intestine with wild-type E. coli EDL933, the nitrate concentration was undetectable 24 h postassociation. This suggests, first, that E. coli is the only microorganism that consumes nitrate in the intestines of streptomycin-treated mice and, second, that at least in streptomycin-treated mice fed a controlled diet, intestinal nitrate is consumed as fast as it becomes available. Together with our previous results, our present results point to the intestinal environment as one containing limiting oxygen and limiting nitrate as the only available exogenous electron acceptors.
Terminal reductase synthesis is subject to hierarchical regulation such that electron acceptors with higher midpoint potentials are used preferentially (11, 26, 39, 60, 63, 70). The streptomycin-treated mouse model afforded the opportunity to test the hypothesis that this paradigm also operates in vivo. Our competition assays with streptomycin-treated mice revealed that mutants lacking cytochrome bd oxidase are outcompeted by mutants lacking either Nar or Frd, which is in keeping with the observed preference for oxygen over nitrate and for nitrate over fumarate. Given that nitrate has the potential to yield more energy than fumarate respiration, we expected a mutant lacking Nar to be outcompeted by a mutant lacking Frd. Instead, we found that Nar+ Frd− strains had an initial advantage but that after several days Nar− Frd+ strains gained an advantage. The population of Nar+ Frd− strains correlated with the availability of nitrate, i.e., the nitrate concentration in cecal mucus was reduced to submillimolar concentrations at the time when the Nar− Frd+ strains became dominant, presumably because they can use fumarate. Just as in a nitrate-limited chemostat (74), it appears that in the intestine the respiratory activity of E. coli decreases the low initial nitrate concentration to an undetectable level. Thus, in the streptomycin-treated mouse intestine it appears that nitrate is limiting and that fumarate, because it is generated from exogenous carbon sources, is more abundant. The observed competition between the Nar+ Frd− and Nar− Frd+ strains is what one would predict for an ecological succession based on competition for a single nutrient that becomes limiting (66).
Role of nitrate respiration and its regulation.
Evidence suggests that NarG and NapA nitrate reductases function optimally at relatively high and low nitrate concentrations, respectively (55, 56). This predicts that the NapA enzyme would be more important than the NarG enzyme for intestinal colonization (11). However, our results support the opposite conclusion. As noted above, these results may reflect nitrate dynamics that are particular to the streptomycin-treated mouse intestine and colonization protocols employed in our studies. Certainly, the relative roles for these two forms of nitrate reductase in different natural environments deserve further study.
Dual nitrate-responsive two-component systems, NarX-NarL and NarQ-NarP, control terminal reductase synthesis. The NarX-NarL system activates synthesis of the NarG enzyme and represses synthesis of other terminal reductases, including NapA and FrdA (62). The NarQ-NarP system plays a more restricted role and is involved primarily in activating NapA and NrfA synthesis (12, 62). We found a colonization defect for the Δ(narXL) but not the ΔnarP strain, congruent with the colonization defect of the ΔnarG but not the Δ(napDA) strain.
However, whereas the ΔnarG strain exhibited an approximately 5-log defect, the Δ(narXL) strain colonization defect was near the detection limit, approximately 1 log. Similarly, work with Pseudomonas aeruginosa demonstrated that a Δ(narGH) mutant is avirulent in Caenorhabditis elegans infection but that a Δ(narXL) mutant exhibits nearly wild-type virulence (71). We offer two hypotheses for why the ΔnarG and Δ(narXL) mutant phenotypes differed in the streptomycin-treated mouse model. First, the ΔnarG strain is devoid of the corresponding activity whereas the narG operon is expressed at low but detectable levels in the Δ(narXL) strain (64). Perhaps this low-level expression is sufficient to allow significant nitrate-dependent growth in this environment. Second, during growth with nitrate, the NarX-NarL system represses synthesis of enzymes for the manufacture and respiration of fumarate (12, 20, 27). Thus, the ΔnarG mutant is defective not only for nitrate respiration but also for fumarate respiration, because of NarX-NarL-mediated repression. In contrast, the Δ(narXL) mutant is able to manufacture and respire fumarate even in the presence of nitrate. These hypotheses are not mutually exclusive, and further analysis will determine if either or both are likely to be correct.
In conclusion, our studies support the idea that E. coli in the intestine is poised for respiration of oxygen, nitrate, and fumarate as the availability of these electron acceptors dictates. Regulation of these respiration systems under low oxygen tension would allow simultaneous production of cytochrome bd oxidase and nitrate reductase (1, 5, 67). The low concentration of available nitrate (and oxygen) would also allow production of Frd (74, 76). Lastly, the flexibility of the quinone pool facilitates electron transfer from NADH dehydrogenase to whichever terminal reductase has a substrate available. It appears that the regulatory interplay of the ArcA, Fnr, and NarX-NarL (and perhaps other) systems is tuned to balance expression of the three terminal reductases that E. coli uses to colonize the intestine.
ACKNOWLEDGMENTS
We thank the following undergraduate students for assistance with animal colonization experiments: Amanda L. Rinkenbaugh, Darrel M. Schreiner, Kathryn E. Watkins, Alyson Crawford, Amanda Laughlin, and Anetra House. We thank Andrew Fabich for confirmatory colonizations.
This work was supported by grants AI48945 and GM095370 from the National Institutes of Health.
Footnotes
Published ahead of print on 8 August 2011.
REFERENCES
- 1. Alexeeva S., Hellingwerf K. J., Teixeira de Mattos M. J. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185:204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antonopoulos D. A., et al. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Autieri S. M., et al. 2007. l-Fucose stimulates utilization of d-ribose by Escherichia coli MG1655 ΔfucAO and E. coli Nissle 1917 ΔfucAO mutants in the mouse intestine and in M9 minimal medium. Infect. Immun. 75:5465–5475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachmann B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460–2488 In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 5. Becker S., Holighaus G., Gabrielczyk T., Unden G. 1996. O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J. Bacteriol. 178:4515–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bekker M., et al. 2007. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153:1974–1980 [DOI] [PubMed] [Google Scholar]
- 7. Bohnhoff M., Drake B. L., Miller C. P. 1954. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc. Soc. Exp. Biol. Med. 86:132–137 [DOI] [PubMed] [Google Scholar]
- 8. Chang D. E., et al. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U. S. A. 101:7427–7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng V. W. T., Weiner J. H. 31 August 2007, posting dateChapter 3.2.8, S- and N-oxide reductases. In Böck A., et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: doi:10.1128/ecosal.3.2.8 [Google Scholar]
- 10. Cole J. 1996. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol. Lett. 136:1–11 [DOI] [PubMed] [Google Scholar]
- 11. Cole J. A., Richardson D. J. 18 August 2008, posting date Chapter 3.2.5, Respiration of nitrate and nitrite. In Böck A., et al. (ed.), EcoSal-Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: doi:10.1128/ecosal.3.2.5 [Google Scholar]
- 12. Constantinidou C., et al. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281:4802–4815 [DOI] [PubMed] [Google Scholar]
- 13. Conway T., Cohen P. S. 2007. Escherichia coli at the intestinal mucosal surface, p. 175–196 In Brogden K. A., et al. (ed.), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC [Google Scholar]
- 14. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dull B. J., Hotchkiss J. H. 1984. Nitrate balance and biosynthesis in the ferret. Toxicol. Lett. 23:79–89 [DOI] [PubMed] [Google Scholar]
- 16. Fabich A. J., et al. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76:1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. Gauger E. J., et al. 2007. Role of motility and the flhDC operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect. Immun. 75:3315–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Georgellis D., Kwon O., Lin E. C. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314–2316 [DOI] [PubMed] [Google Scholar]
- 20. Goh E. B., et al. 2005. Hierarchical control of anaerobic gene expression in Escherichia coli K-12: the nitrate-responsive NarX-NarL regulatory system represses synthesis of the fumarate-responsive DcuS-DcuR regulatory system. J. Bacteriol. 187:4890–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Green L. C., et al. 1981. Nitrate biosynthesis in man. Proc. Natl. Acad. Sci. U. S. A. 78:7764–7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guymer D., Maillard J., Sargent F. 2009. A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 191:519–528 [DOI] [PubMed] [Google Scholar]
- 23. Hastings S. F., et al. 1998. Identification of a stable semiquinone intermediate in the purified and membrane bound ubiquinol oxidase-cytochrome bd from Escherichia coli. Eur. J. Biochem. 255:317–323 [DOI] [PubMed] [Google Scholar]
- 24. He G., et al. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. U. S. A. 96:4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hentges D. J., Que J. U., Casey S. W., Stein A. J. 1984. The influence of streptomycin on colonization in mice. Microecol. Theor. 14:53–62 [Google Scholar]
- 26. Ingledew W. J., Poole R. K. 1984. The respiratory chains of Escherichia coli. Microbiol. Rev. 48:222–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iuchi S., Lin E. C. 1987. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 84:3901–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ivanov V., Nejad S. R., Yi S., Wang X. H. 2008. Physiological heterogeneity of suspended microbial aggregates. Water Sci. Technol. 58:2435–2441 [DOI] [PubMed] [Google Scholar]
- 29. Janausch I. G., Zientz E., Tran Q. H., Kroger A., Unden G. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39–56 [DOI] [PubMed] [Google Scholar]
- 30. Jones S. A., et al. 2007. Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75:4891–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones S. A., et al. 2008. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect. Immun. 76:2531–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kiley P. J., Beinert H. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341–352 [DOI] [PubMed] [Google Scholar]
- 33. Lawrence J., Cox G. B., Gibson F. 1974. Biosynthesis of ubiquinone in Escherichia coli K-12: biochemical and genetic characterization of a mutant unable to convert chorismate into 4-hydroxybenzoate. J. Bacteriol. 118:41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leatham M. P., et al. 2009. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect. Immun. 77:2876–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leatham M. P., et al. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039–8049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee P. T., Hsu A. Y., Ha H. T., Clarke C. F. 1997. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179:1748–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levitt M. D. 1970. Oxygen tension in the gut. N. Engl. J. Med. 282:1039–1040 [DOI] [PubMed] [Google Scholar]
- 38. Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023 [DOI] [PubMed] [Google Scholar]
- 39. Lin E. C., Iuchi S. 1991. Regulation of gene expression in fermentative and respiratory systems in Escherichia coli and related bacteria. Annu. Rev. Genet. 25:361–387 [DOI] [PubMed] [Google Scholar]
- 40. Lin H. Y., Bledsoe P. J., Stewart V. 2007. Activation of yeaR-yoaG operon transcription by the nitrate-responsive regulator NarL is independent of oxygen-responsive regulator Fnr in Escherichia coli K-12. J. Bacteriol. 189:7539–7548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lubitz S. P., Weiner J. H. 2003. The Escherichia coli ynfEFGHI operon encodes polypeptides which are paralogues of dimethyl sulfoxide reductase (DmsABC). Arch. Biochem. Biophys. 418:205–216 [DOI] [PubMed] [Google Scholar]
- 42. Lundberg J. O., Weitzberg E., Cole J. A., Benjamin N. 2004. Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2:593–602 [DOI] [PubMed] [Google Scholar]
- 43. Lynch A. S., Lin E. C. C. 1996. Responses to molecular oxygen, p. 1526–1538 In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 44. Marteyn B., Scorza F. B., Sansonetti P. J., Tang C. 2011. Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell. Microbiol. 13:171–176 [DOI] [PubMed] [Google Scholar]
- 45. Marteyn B., et al. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meganathan R. 1996. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q), p. 642–655 In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 47. Melican K., et al. 2008. Bacterial infection-mediated mucosal signalling induces local renal ischaemia as a defence against sepsis. Cell. Microbiol. 10:1987–1998 [DOI] [PubMed] [Google Scholar]
- 48. Mercado-Lubo R., Gauger E. J., Leatham M. P., Conway T., Cohen P. S. 2008. A Salmonella enterica serovar Typhimurium succinate dehydrogenase/fumarate reductase double mutant is avirulent and immunogenic in BALB/c mice. Infect. Immun. 76:1128–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mercado-Lubo R., Leatham M. P., Conway T., Cohen P. S. 2009. Salmonella enterica serovar Typhimurium mutants unable to convert malate to pyruvate and oxaloacetate are avirulent and immunogenic in BALB/c mice. Infect. Immun. 77:1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller C. P., Bohnhoff M. 1963. Changes in the mouse's enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin treatment. J. Infect. Dis. 113:59–66 [DOI] [PubMed] [Google Scholar]
- 51. Miranda R. L., et al. 2004. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 72:1666–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moller A. K., et al. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71:2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perna N. T., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 54. Poole R. K., Gibson F., Wu G. 1994. The cydD gene product, component of a heterodimeric ABC transporter, is required for assembly of periplasmic cytochrome c and of cytochrome bd in Escherichia coli. FEMS Microbiol. Lett. 117:217–223 [DOI] [PubMed] [Google Scholar]
- 55. Potter L., Angove H., Richardson D., Cole J. 2001. Nitrate reduction in the periplasm of gram-negative bacteria. Adv. Microb. Physiol. 45:51–112 [DOI] [PubMed] [Google Scholar]
- 56. Potter L. C., Millington P., Griffiths L., Thomas G. H., Cole J. A. 1999. Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem. J. 344(Pt. 1):77–84 [PMC free article] [PubMed] [Google Scholar]
- 57. Robinson C. J., Bohannan B. J., Young V. B. 2010. From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74:453–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rolfe M. D., et al. 2011. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J. Biol. Chem. 286:10147–10154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saldena T. A., Saravi F. D., Hwang H. J., Cincunegui L. M., Carra G. E. 2000. Oxygen diffusive barriers of rat distal colon: role of subepithelial tissue, mucosa, and mucus gel layer. Dig. Dis. Sci. 45:2108–2114 [DOI] [PubMed] [Google Scholar]
- 60. Spiro S., Guest J. R. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6:399–428 [DOI] [PubMed] [Google Scholar]
- 61. Stecher B., et al. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell. Microbiol. 10:1166–1180 [DOI] [PubMed] [Google Scholar]
- 62. Stewart V. 2003. Biochemical Society Special Lecture. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 31:1–10 [DOI] [PubMed] [Google Scholar]
- 63. Stewart V. 1988. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol. Rev. 52:190–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stewart V. 1982. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J. Bacteriol. 151:1320–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Suvarna K., Stevenson D., Meganathan R., Hudspeth M. E. 1998. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J. Bacteriol. 180:2782–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tilman D. 1982. Resource competition and community structure. Monogr. Popul. Biol. 17:1–296 [PubMed] [Google Scholar]
- 67. Tseng C. P., Albrecht J., Gunsalus R. P. 1996. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 178:1094–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Turnbaugh P. J., et al. 2010. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. U. S. A. 107:7503–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Unden G. 1988. Differential roles for menaquinone and demethylmenaquinone in anaerobic electron transport of E. coli and their Fnr-independent expression. Arch. Microbiol. 150:499–503 [DOI] [PubMed] [Google Scholar]
- 70. Unden G., Dunnwald P. 11 March 2008, posting date Chapter 3.2.2, The aerobic and anaerobic respiratory chain of Escherichia coli and Salmonella enterica: enzymes and energetics. In Böck A., et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: doi:10.1128/ecosal.3.2.2 [DOI] [PubMed] [Google Scholar]
- 71. Van Alst N. E., Picardo K. F., Iglewski B. H., Haidaris C. G. 2007. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect. Immun. 75:3780–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wadolkowski E. A., Laux D. C., Cohen P. S. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect. Immun. 56:1030–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reference deleted.
- 74. Wang H., Tseng C. P., Gunsalus R. P. 1999. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J. Bacteriol. 181:5303–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Whistance G. R., Threlfall D. R. 1968. Effect of anaerobiosis on the concentrations of demethylmenaquinone, menaquinone and ubiquinone in Escherichia freundii, Proteus mirabilis and Aeromonas punctata. Biochem. J. 108:505–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Williams S. B., Stewart V. 1997. Nitrate- and nitrite-sensing protein NarX of Escherichia coli K-12: mutational analysis of the amino-terminal tail and first transmembrane segment. J. Bacteriol. 179:721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wissenbach U., Ternes D., Unden G. 1992. An Escherichia coli mutant containing only demethylmenaquinone, but no menaquinone: effects on fumarate, dimethylsulfoxide, trimethylamine N-oxide and nitrate respiration. Arch. Microbiol. 158:68–73 [DOI] [PubMed] [Google Scholar]
- 78. Wu G., Williams H. D., Gibson F., Poole R. K. 1993. Mutants of Escherichia coli affected in respiration: the cloning and nucleotide sequence of ubiA, encoding the membrane-bound p-hydroxybenzoate:octaprenyltransferase. J. Gen. Microbiol. 139:1795–1805 [DOI] [PubMed] [Google Scholar]
- 79. Young I. G. 1975. Biosynthesis of bacterial menaquinones. Menaquinone mutants of Escherichia coli. Biochemistry 14:399–406 [DOI] [PubMed] [Google Scholar]
- 80. Young I. G., Leppik R. A., Hamilton J. A., Gibson F. 1972. Biochemical and genetic studies on ubiquinone biosynthesis in Escherichia coli K-12: 4-hydroxybenzoate octaprenyltransferase. J. Bacteriol. 110:18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]