Abstract
Filifactor alocis, a Gram-positive anaerobic rod, is one of the most abundant bacteria identified in the periodontal pockets of periodontitis patients. There is a gap in our understanding of its pathogenicity and ability to interact with other periodontal pathogens. To evaluate the virulence potential of F. alocis and its ability to interact with Porphyromonas gingivalis W83, several clinical isolates of F. alocis were characterized. F. alocis showed nongingipain protease and sialidase activities. In silico analysis revealed the molecular relatedness of several virulence factors from F. alocis and P. gingivalis. In contrast to P. gingivalis, F. alocis was relatively resistant to oxidative stress and its growth was stimulated under those conditions. Biofilm formation was significantly increased in coculture. There was an increase in adherence and invasion of epithelial cells in coculture compared with P. gingivalis or F. alocis monocultures. In those epithelial cells, endocytic vesicle-mediated internalization was observed only during coculture. The F. alocis clinical isolate had an increased invasive capacity in coculture with P. gingivalis compared to the ATCC 35896 strain. In addition, there was variation in the proteomes of the clinical isolates compared to the ATCC 35896 strain. Hypothetical proteins and those known to be important virulence factors in other bacteria were identified. These results indicate that F. alocis has virulence properties that may enhance its ability to survive and persist in the periodontal pocket and may play an important role in infection-induced periodontal disease.
INTRODUCTION
Bacteria in the periodontal pocket can develop complex sessile communities that play a significant role in infection-induced periodontal disease. Assembly of these bacterial communities involves inter- and intrageneric attachment and coaggregation among initial, early, and late colonizers (44). In addition, the temporal and spatial development is modulated by specific interbacterial signaling, which may cause physiologically compatible organisms to accumulate in mutualistic groupings (8, 23, 25, 30, 44).
Data emerging from the oral microbiome project may suggest a shift in the paradigm for infection-induced periodontal diseases. Bacteria like Porphyromonas gingivalis, Prevotella intermedia, Aggregatibacter (Actinobacillus) actinomycetemcomitans, Tannerella forsythia, and Treponema denticola have previously been demonstrated to be major pathogens associated with periodontal diseases (49, 51, 52). However, recent developments, including novel, culture-independent techniques, have allowed the identification of as yet culturable and fastidious organisms in patients suffering from periodontitis (1, 13, 19, 43). Collectively, these studies have demonstrated that changes in periodontal status are associated with shifts in the composition of the bacterial community in the periodontal pocket. Filifactor alocis, a Gram-positive, asaccharolytic, obligate anaerobic rod, is one of the marker organisms that is now identified to be significant to the pathogenetic structure of biofilms associated with periodontal inflammation and should be considered an important organism for periodontal disease (12, 16, 50). Further, in comparison to the other traditional periodontal pathogens, F. alocis is present in the periodontal pocket in higher numbers and is least detected in healthy or periodontitis-resistant patients (31, 32, 63). This organism, first isolated in 1985 from the gingival sulcus in gingivitis and periodontitis patients, was classified as Fusobacterium alocis (7). However, based on phylogenetic analysis using 16S rRNA sequences, it was reclassified in 1999 into the genus Filifactor (24).
The etiology of periodontitis with both Gram-positive and Gram-negative bacteria suggests a complex heterogeneous microbial population where a coordinated microbial response is essential for growth and survival in the periodontal pocket. Although F. alocis possesses general virulence attributes common to Gram-positive bacteria and several proteases such as metal-dependent proteases, CaaX proteases, sialoglycoproteases, and calcium-dependent proteases (http://www.ncbi.nlm.nih.gov/genomeprj/46625), there is a gap in our understanding of its pathogenicity and ability to interact with other periodontal pathogens. In the present study we have evaluated the virulence potential of F. alocis and its ability to interact with the periodontal pathogen P. gingivalis. A proteomic analysis of F. alocis has identified several hypothetical proteins and those known to be important virulence factors in other bacteria.
MATERIALS AND METHODS
Bioinformatics analysis.
The DNA and amino acid sequences were retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov/genomeprj/46625) and aligned using Bioedit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The phylogenetic relationships between the oral pathogens were analyzed using MEGA software version 4.0 (59). The phylogenetic distance was calculated using the Kimura 2-parameter model, and clustering used the neighbor-joining method with bootstrap values based on 1,000 replicates (53). The amino acid sequences were analyzed using ClustalW version 2.0 (http://www.ebi.ac.uk/). Protein subcellular localization was predicted using the PSORT and iPSORT programs (39). Prediction of the conserved domains and other specific domains was carried out using the NCBI conserved domain database (37).
Bacterial strains and growth conditions.
F. alocis ATCC 35896 was purchased from the American Type Culture Collection (Manassas, VA). F. alocis clinical isolates (D-62D, D-62A, and F-176) were a gift from Floyd Dewhirst, the custodian of Moore's anaerobic microbial collection (The Forsyth Institute, Boston, MA). The identity of the clinical isolates was confirmed by 16S rRNA gene sequencing (D-62D, accession no. GU968904; D-62A, accession no. GU968903; F-176, accession no. GU968902). F. alocis strains were grown initially in Robertson's bullock heart medium followed by adaptation to brain heart infusion (BHI) broth supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), cysteine (0.1%), and arginine (100 μm). P. gingivalis strains were grown in BHI broth (Difco) supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%). Blood agar medium was prepared by the addition of sheep blood (5%) and agar (2%). Experiments with hydrogen peroxide were performed in BHI broth without cysteine. All cultures, unless otherwise stated, were incubated at 37°C. F. alocis and P. gingivalis strains were maintained in an anaerobic chamber (Coy Manufacturing) in 10% H2, 10% CO2, and 80% N2. Growth rates were determined spectrophotometrically (optical density at 600 nm [OD600]). Antibiotics were used at the following concentrations: clindamycin, 0.5 μg/ml; erythromycin, 300 μg/ml; carbenicillin, 100 μg/ml.
Growth analysis during oxidative stress conditions.
Actively growing cultures of F. alocis (ATCC 35896) and P. gingivalis W83 were incubated at 37°C under anaerobic conditions for 24 h. To test for adaptation to oxidative stress conditions, all the strains of F. alocis and P. gingivalis were grown in BHI broth (without cysteine) plus 0.25 mM H2O2. Controls were grown in the absence of H2O2.
Assay for estimation of protease activity.
Assay of nongingipain protease activity was carried out using an EnzChek protease assay kit (Molecular Probes) in accordance with the manufacturer's instructions. This method uses a fluorescence resonance energy transfer (FRET)-based method. Briefly, the protease substrate comprises a fluorophore and a quencher moiety separated by an amino acid sequence. Upon sequence cleavage by protease(s), the fluorophore separates from the quencher and is free to emit a detectable fluorescent signal (excitation and emission maxima of 502 and 528 nm, respectively). The magnitude of the resultant signal is proportional to the degree of substrate cleavage and can therefore be used to quantitate the enzyme activity present. The assay is performed in a simple mix-incubate-read format. The optical density of the assay was read at a wavelength of 600 nm.
The presence of Arg-X- and Lys-X-specific gingipain activities of cells grown to exponential phase (OD600 = 0.8) was determined using a microplate reader (Bio-Rad) by the method previously described by Olango et al. (41).
Sialidase assay.
Sialidase estimation was carried out using an Amplex Red neuraminidase (sialidase) assay kit (Molecular Probes) as previously described (3). The assay utilizes Amplex Red to detect H2O2 generated by galactose oxidase oxidation of desialylated galactose, the end result of neuraminidase action. The H2O2, in the presence of horseradish peroxidase (HRP), reacts with Amplex Red reagent to generate the red fluorescent oxidation product resorufin. The assay was read as the optical density at 492 nm.
Amino acid supplementation assay.
The amino acid utilization assay was performed according to the method of Uematsu et al. (61). Briefly, the amino acids arginine, lysine, valine, threonine, isoleucine, glycine, and cystine (Sigma-Aldrich) were used at 100 μM concentration in BHI medium supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%). The cultures were grown for 8 h, and the growth rates were determined spectrophotometrically (OD600). F. alocis grown without any amino acid supplementation was used as a control.
Static biofilm formation assays.
Static biofilm formation was assayed using the protocol of Hinsa and O'Toole (21) with slight modification. Briefly, overnight cultures of F. alocis and/or P. gingivalis ATCC 33277 were diluted with fresh BHI medium to obtain a concentration of 5 × 107 CFU/ml (confirmed by colony count). The cells were aliquoted into a 96-well microtiter plate (260 μl per well) and incubated anaerobically at 37°C for 24 h. The supernatant of the culture was aspirated, and then the well was washed twice with phosphate-buffered saline (PBS; 150 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, and 1.5 mM KH2PO4, pH 7.4). The biofilms were stained by incubation of each well with 100 μl of 0.1% crystal violet for 5 min. The plate was then washed twice with distilled water and destained with 95% ethanol (200 μl per well) for 5 min. The solubilized crystal violet was transferred to a new microtiter plate, and the OD540 was determined spectrophotometrically. Biofilm formation was qualitatively determined to be proportional to the absorbance of the crystal violet. Medium alone was used as a control to normalize the data. The poorly biofilm-forming P. gingivalis W83 strain was used as a negative control.
Epithelial cell culture.
HeLa cells were grown and maintained at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 IU/ml), streptomycin (100 IU/ml), and amphotericin B (2.5 mg/ml) (Invitrogen, Carlsbad, CA). Confluent stock cultures were trypsinized, adjusted to approximately 5 × 103 cells/ml, seeded (1 ml per well) into 12-well plates (Nunc, Rochester, NY), and further incubated for 48 h to reach semiconfluence (105 cells per well).
Adherence and standard antibiotic protection assay.
Invasion of epithelial cells was quantified using the standard antibiotic protection assay (65). Briefly, an isolated bacterial colony harvested from a solid agar plate was grown to exponential phase in BHI broth. The bacterial cells were then centrifuged, washed three times in PBS, and adjusted to 107 CFU/ml of bacteria in DMEM. The epithelial cell monolayer was washed three times with PBS, infected with bacteria at a multiplicity of infection (MOI) of 1:100 (105 epithelial cells), and then incubated at 37°C for 45 and 90 min under 5% CO2. Nonadherent bacteria were removed by washing with PBS, while cell surface-bound bacteria were killed with metronidazole (200 μg/ml, 60 min). F. alocis and P. gingivalis were sensitive to 100 μg/ml of metronidazole. After removal of the antibiotic, the internalized bacteria were released by osmotic lysis of the epithelial cells in sterile distilled water. Lysates were serially diluted, plated (in duplicate) on BHI agar, and incubated for 6 to 10 days. The number of bacterial cells recovered was expressed as a percentage of the original inoculum. The number of adherent bacteria was obtained by subtracting the number of intracellular bacteria from the total bacteria obtained in the absence of metronidazole (6). Coinfection was performed as described previously (52). F. alocis and P. gingivalis inocula were prepared by mixing equal volumes (1 × 107 cells/well) of bacterial suspension. The inocula were then incubated for 5 min in the anaerobic chamber prior to infection. The serially diluted lysate was plated on BHI blood agar and incubated for 6 to 10 days. The bacterial colonies were phenotypically identified.
FISH and confocal microscopy.
Fluorescence in situ hybridization (FISH) was performed by the method of Sunde et al. (58) with modification. The trypsinized culture of HeLa epithelial cells (5 × 103 cells/ml) was grown on coverslips in Leighton tubes (16 by 150 mm; Bellco, NJ) and also in 4-well BD Falcon (Bedford, MA) culture slides in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 IU/ml), streptomycin (100 IU/ml), and amphotericin B (2.5 mg/ml) (Invitrogen, Carlsbad, CA) at 37°C under 5% CO2. The cultures showing 80% confluence were used for the experiment. F. alocis D-62D and P. gingivalis W83 were used in this study after coculture at 20 min, 30 min, 45 min, and 1 h postinfection (p.i.).
The slides bearing the samples were fixed and processed for FISH by the method of Manz et al. (36). The probes used have been deposited in ProbeBase (34) (http://www.microbial-ecology.net/probebase/index.html). Species-specific probes were custom made by Integrated DNA Technologies (IDT) (San Diego, CA). F. alocis-specific probe 5′-TCT TTG TCC ACT ATC GTT TTG A-3′ was 5′ labeled with TYE563 (excitation coefficient of 208,100 liters/mol·cm). P. gingivalis-specific probe CAA TAC TCG TAT CGC CCG TTA TTC-3′ was 5′ labeled with TYE 665 (excitation coefficient of 256,800 liters/mol·cm). The specificities of the oligonucleotide probes were compared using Ribosomal Database Project II (9) and the OLIGO, version 4.0, program. The probes were used in the presence of 20 to 30% (vol/vol) formamide. The probe hybridization procedure described previously (49) was followed with modifications. The hybridization mixture was prepared by adding 1 volume of the probe working solution (50 ng/μl) to 9 volumes of the hybridization buffer, which was kept in the dark on ice. Ten microliters of the hybridization mix was added to the samples in each compartment of the BD Falcon culture slide, which was then incubated in a polyethylene tube in the horizontal position at 46°C for 90 min. The slides were then rinsed with prewarmed wash buffer (5 M NaCl, 1 M Tris-HCl, 0.5 M EDTA, 10% SDS made up in 50 ml distilled water) and later transferred to preheated wash buffer and incubated for 25 min at 48°C. DAPI (4′,6-diamidino-2-phenylindole) solution (1 μg/ml) was used for counterstaining. The air-dried sample was mounted using SlowFade Gold antifade reagent (Invitrogen, Carlsbad, CA). The culture slides were visualized using the inverted Axio Observer Z1 confocal microscope (Zeiss, Jena, Germany) equipped with a green diode-pumped solid-state (DPSS) laser, red HeNe laser, and Cohert Chameleon Vision II Ti sapphire laser under EC Plan NEO 20×, 40×, and 63× objectives. The images were analyzed using Zeiss LSM 710 software. The numbers of adherent and internalized bacteria, corresponding to the fluorescence intensity level, were determined in duplicate samples as described above.
Cell fractionation and SDS-PAGE analysis.
Bacterial cells were fractionated as described previously (18). Briefly, an overnight liquid culture of F. alocis was harvested by centrifugation (5,000 × g, 10 min, 4°C) and resuspended in ice-cold PBS. Cells were lysed in a French pressure cell at 20,000 lb/in2, and the lysate was cleared of unlysed cells by centrifugation (5,000 × g, 10 min, 4°C). To separate the membrane fractions from the cytosolic fractions, the lysate was further ultracentrifuged (100,000 × g, 1 h, 4°C). The supernatant containing cytosolic fractions was decanted from the pellet that contained the membrane fraction.
SDS-PAGE used a 4 to 12% Bis-Tris separating gel in MOPS (morpholinepropanesulfonic acid)-SDS running buffer according to the manufacturer's instructions (NuPAGE Novex gels; Invitrogen). Samples were prepared (65% sample, 25% 4× NuPAGE lithium dodecyl sulfate [LDS] sample buffer, 10% NuPAGE reducing agent), heated at 72°C for 10 min, and then electrophoresed at 200 V for 65 min in the XCell SureLock minicell system (Invitrogen). The protein bands were visualized with SimplyBlue safe stain (Invitrogen).
2D PAGE analysis.
Two-dimensional (2D) gel electrophoresis was carried out using 2D gel strips (7 cm; pI 3 to 10) in a Protean isoelectric focusing (IEF) cell (Bio-Rad) by the method of Poznanovic et al. (45). Briefly, the protein concentration of the sample was measured using a Bio-Rad spectrophotometer. The protein sample was diluted to a final concentration of 350 μg protein, and 20 μl of the sample was added to solubilization buffer consisting of 7 M urea, 2 M thiourea, 2% (wt/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 65 mM dithiothreitol (DTT), 0.002% bromophenol blue, and 1% (wt/vol) Zwittergent 3-10.The first-dimension immobilized pH gradient (IPG) strip was run by adding 100 μl of the diluted sample in the rehydration buffer (2 ml) (730 mg DTT, 70 mg iodoacetemide) for 8 h at a voltage gradient of 3 h at 300 V, 5 h at a linear gradient of 300 to 3,500 V, and 18 h at 3,500 V. After equilibration the IPG strips were loaded onto the gel and electrophoresed at 200 V and 0.3 A for 4 to 5 h and stained with Coomassie SimplyBlue safe stain (Invitrogen).
MS and data analysis.
An LCQ Deca XP Plus system with nanoelectrospray technology was used to analyze the extracted peptides from each gel piece. A four-part protocol was used for the mass spectrometry (MS) and tandem MS (MS/MS) analyses (62). This included one full MS analysis (from 450 to 1,750 m/z) followed by three MS/MS events, where the most intense ion from a given full MS scan was subjected to collision-induced dissociation, followed by the second and third most intense ions. The nanoflow buffer gradient was extended over 45 min in conjunction with the cycle repeating itself every 2 s using a 0 to 60% acetonitrile gradient from buffer B (95% acetonitrile with 0.1% formic acid) developed against buffer A (2% acetonitrile with 0.1% formic acid) at a flow rate of 250 to 300 μl/min. Twenty microliters of sample from the autosampler was moved using a Michrom nanotrap column. The spray voltage and current were set at 2.2 kV and 5.0 μA, with a capillary voltage of 25 V in positive ion mode. Data collection using the Xcalibur software (Thermo Electron) was screened with Bioworks 3.1. MASCOT software was used for each analysis to produce unfiltered data and output files. Statistical validation of peptide and protein findings was achieved using X TANDEM (www.thegmp.org) and SCAFFOLD 2 meta-analysis software. The presence of two different peptides at a probability of at least 95% was required for consideration as being positively identified. Confirmation of individual peptide matches was achieved using the NCBI F. alocis genome project database (http://www.ncbi.nlm.nih.gov/genomeprj/46625).
Electron microscopy.
Transmission electron microscopy (TEM) was performed using the FEI Tecnai G2 TEM by the method of Harris (20). Briefly, Formvar carbon-coated grids were prepared; the Formvar support was removed by placing the grids in an atmosphere of solvent vapor. Grids were then placed on a wire mesh in a glass petri dish, with carbon tetrachloride below the wire mesh. Five to 10 ml of the sample was placed under the carbon side of a 4- by 5-mm square of mica (approximately twice the size of an EM grid). The grid was washed in 0.5% acetic acid, then acetone. The carbon film was broken to free the specimen grid, after which the grid was placed in stain solution-neutral 1% aqueous phosphotungstic acid for 30 s. After being blotted dry, the grid was examined using the Tecnai TEM.
Ultrathin sections were made in accordance with the method of Massey (38). After a 2-h incubation with bacteria, HeLa cell monolayers were washed four times in PBS and detached from the plastic surface with trypsin. The cell slurry was then immediately centrifuged for 4 min at 15,000 × g. The cell pellet was washed twice with PBS, fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate, and postfixed in 1% OsO4 in 0.1 M sodium cacodylate for 1 h, and the ultrathin sections were contrasted with lead citrate and uranyl acetate before examination by the FEI Technai G2 transmission electron microscope.
Data and statistical analysis.
All experiments were performed in triplicate for each condition and repeated at least three times unless otherwise stated. The adherence and invasion assays were repeated four times (n = 4). Statistical comparison was performed using the Prism 5.0 software package (Graphpad Software Inc., La Jolla, CA). The Mann-Whitney-Wilcoxon test and Bonferroni posttest were performed for multiple comparisons.
RESULTS
Phylogenetic analysis using 16S rRNA from several major oral pathogens showed F. alocis to be clustered under the Gram-positive clad consisting of the clostridial spp., Fusobacterium nucleatum, and Enterococcus faecalis (see Fig. S1 in the supplemental material). In contrast to the Streptococcus genus, the Gram-negative oral pathogens such as Treponema spp., P. gingivalis, Bacteroides fragilis, Tannerella forsythia, and Actinomyces spp. were found to be closely related to F. alocis. It is noteworthy that the F. alocis clad included the late colonizers of the periodontal pocket. Genome-wide amino acid sequence analysis showed F. alocis to have more than 18% homology with Clostridium difficile (24) (data not shown).
Growth of F. alocis is stimulated by oxidative stress.
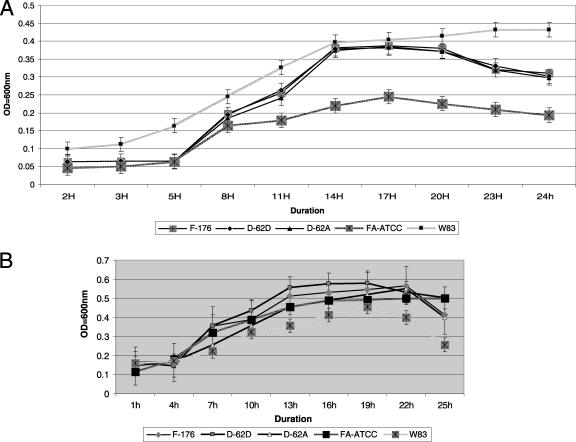
Growth of F. alocis on BHI blood agar showed nonhemolytic mucoid colonies after an incubation of 5 to 6 days. In broth culture the generation time was approximately 13 h for the F. alocis ATCC 35896 strain in contrast to 11 h for the clinical isolate D-62D (Fig. 1 A). Consistent with previous reports (3), a generation time of 3 h was observed for P. gingivalis. In the presence of H2O2, the generation time for F. alocis was approximately 6 to 7 h compared to 10 h for P. gingivalis (Fig. 1B). This indicates that F. alocis is more resistant to oxidative stress than P. gingivalis. Furthermore, its growth appears to be stimulated under oxidative stress conditions.
Fig. 1.
(A) Growth rates of Filifactor alocis strains. Cultures of F. alocis and P. gingivalis were incubated at 37°C under anaerobic conditions. Growth rates were determined by measuring OD600 over time. The data are means ± standard deviations (SD) of three independent experiments. The generation time (G) was calculated using the equation G = t/n, where t is the time interval and n is the number of generations. n = (log Nt − log N0)/log 2, where N0 is the number of bacteria at culture start time and Nt is the number of bacterial at the end of log phase. FA-ATCC, F. alocis ATCC 35896. (B) Growth of Filifactor alocis under oxidative stress. F. alocis and P. gingivalis W83 strains grown to early log phase were treated with 0.25 mM H2O2 and further incubated for 24 h. The results shown are representative of three independent replicates.
Sialidase activity is present in F. alocis strains.
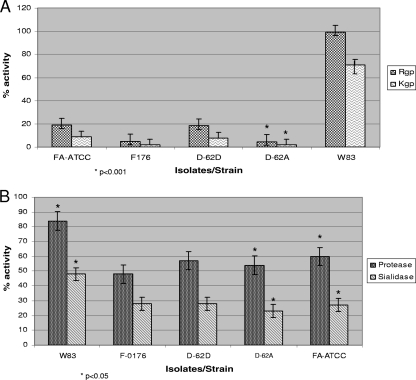
Sialidase activity in periodontal pathogens is important for their survival, persistence, and pathogenesis (4, 44). Sialidase activity was evaluated using whole-cell cultures of F. alocis and compared to that of P. gingivalis. As shown in (Fig. 2 B), sialidase activity was present in all the F. alocis strains. This activity, however, was reduced by approximately 40% compared to that of P. gingivalis W83.
Fig. 2.
Comparison of gingipain, nongingipain protease, and sialidase activities of F. alocis strains and P. gingivalis W83. F. alocis and P. gingivalis were grown to exponential phase (OD600 of 0.8). (A) Gingipain activities of F. alocis and P. gingivalis strains were tested against BAPNA (N-α-benzoyl-l-tyrosine p-nitroanilide; Rgp) and ALNA (acetyl-Lys-p-nitroanilide-HCl; Kgp) using whole-cell culture. The results shown are from three independent experiments. The clinical strains are statistically compared with the ATCC strain. (B) Nongingipain protease and sialidase activities of F. alocis strains. Nongingipain protease activity was estimated using a FRET-based assay method. Sialidase activity was tested using the Amplex Red neuraminidase assay kit. Percent activity was determined using the standard supplied with the kit. The results shown are from three independent experiments. Filifactor alocis strains are statistically compared with P. gingivalis.
Protease activity in F. alocis.
Several periodontal pathogens, including P. gingivalis, T. denticola, and F. alocis, were shown to have trypsin-like proteolytic activity (35). In comparison to P. gingivalis, there was 80 to 90% reduction in gingipain activity among the F. alocis strains (Fig. 2A). Among these strains D-62D and ATCC 33896 showed approximately 2-fold more Rgp and Kgp activity than strains F-176 and D-62A. Non-trypsin-like proteolytic activity in F. alocis was reduced by approximately 30 to 40% compared to that in P. gingivalis (Fig. 2B).
Amino acid utilization by F. alocis.
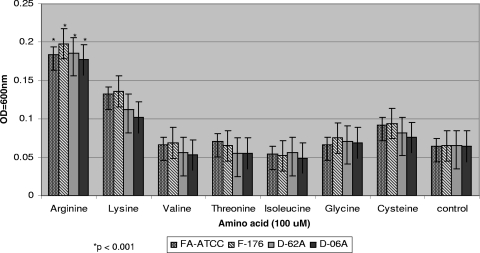
To satisfy the asaccharolytic requirement of F. alocis, the presence of amino acids is of importance for growth and survival (61). Several amino acids, including arginine and lysine, were evaluated for their ability to enhance the growth of F. alocis. As shown in the Fig. 3, among the amino acids supplemented, arginine, lysine, and cysteine (in order of decreasing ability) were able to enhance the growth of the F. alocis strains. In the absence of any supplemented amino acid, there was minimal growth in the same time period. Throughout the rest of our study, arginine at 100 μM concentration was used in the medium for growth of F. alocis.
Fig. 3.
Amino acid utilization by Filifactor alocis strains. Amino acids arginine, lysine, valine, threonine, isoleucine, glycine, and cystine were used at 100 μM concentration in BHI medium supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%). The cultures were grown for 8 h, and OD600 was determined. F. alocis grown without any amino acid supplementation was used as a control. The data are means ± SD of three independent experiments. Arginine supplementation was compared statistically with the other amino acids used.
F. alocis enhances biofilm formation in coculture.
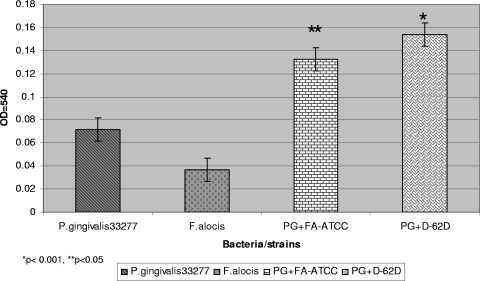
F. alocis can colocalize with other periodontal pathogens and can likely play an important role in biofilm formation (31, 32, 54). To assess its biofilm formation capacity, F. alocis was compared with P. gingivalis ATCC 33277, a biofilm-producing strain (28). Biofilm formation by coculturing F. alocis with P. gingivalis ATCC 33277 showed a biofilm formation index that was 2-fold higher than that for a P. gingivalis monoculture and 3-fold higher than that for an F. alocis monoculture. The clinical strains of F. alocis in coculture with P. gingivalis showed a 13 to 15% increase in biofilm-forming capacity in comparison with F. alocis ATCC 35896 cocultured with P. gingivalis ATCC 33277 (Fig. 4).
Fig. 4.
Biofilm formation by mono- and cocultures of Filifactor alocis and P. gingivalis ATCC 33277 (PG). Overnight cultures of F. alocis and/or P. gingivalis were diluted with fresh BHI medium to obtain a concentration of 5 × 107 CFU/ml. The cells were aliquoted to a 96-well microtiter plate (260 μl per well) and incubated anaerobically at 37°C for 24 h. The biofilms were stained with crystal violet. The solubilized crystal violet was transferred to a new microtiter plate, and the OD540 was determined spectrophotometrically. Medium and P. gingivalis W83 were used as controls. Coculture was statistically compared with P. gingivalis monoculture.
Adherence assay.
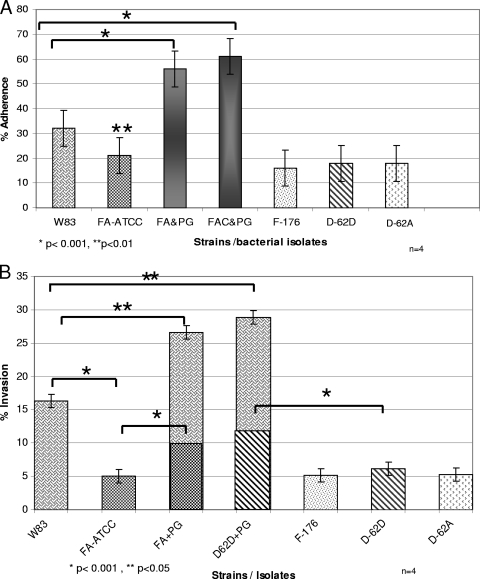
F. alocis and P. gingivalis in monoculture and coculture were evaluated for their capacity for adherence to epithelial cells. While all the monocultures of F. alocis strains showed less adherence (10 to 15%) than P. gingivalis, the coculture showed a 22% increase with the F. alocis ATCC 35896 strain (P < 0.001) and a 29% increase with F. alocis clinical strains (P < 0.001) (Fig. 5 A). There was no significant interstrain variation in the adherence capacity among the clinical strains of F. alocis; however, the ATCC 35896 strain showed a 3 to 4% increase in adherence capacity. In comparison with P. gingivalis W83 all strains of F. alocis showed a reduction in their ability to adhere to the HeLa cells in monoculture.
Fig. 5.
(A) Adherence by mono- and cocultures of Filifactor alocis strains and P. gingivalis W83 in epithelial cells. HeLa cells were grown and maintained at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum with antibiotics. Confluent stock cultures were trypsinized, adjusted to approximately 5 × 103 cells/ml, seeded (1 ml per well) into 12-well plates, and further incubated for 48 h to reach semiconfluence. Epithelial cells (105 cells) were infected with 107 bacteria (MOI = 100). Error bars represent mean ± SD of four independent experiments. Statistical significance was determined using analysis of variance (ANOVA) with the Bonferroni posttest. FA-ATCC, F. alocis ATCC 35896; PG, P. gingivalis; FAC, F. alocis clinical strain D-62D. (B) Invasion by mono- and cocultures of Filifactor alocis strains and P. gingivalis W83 into epithelial cells. Bacteria were grown to exponential phase in BHI broth, washed three times in PBS, and adjusted to 107 CFU/ml in DMEM. The epithelial cell monolayer was washed and infected with bacteria at an MOI of 1:100 (105 epithelial cells) and then incubated at 37°C for 45 min under 5% CO2. Error bars represent mean ± SD of four independent experiments. Statistical significance was determined using ANOVA with the Bonferroni posttest.
Invasion by P. gingivalis is increased during coculture with F. alocis.
F. alocis strains showed only 25% of the invasion capacity of P. gingivalis (P < 0.001). The F. alocis clinical strains showed an approximately 20% increase in invasion compared with F. alocis ATCC 35896 (P < 0.01). Invasion by P. gingivalis in coculture with F. alocis showed a 17 to 19% increase (P < 0.05) compared to the monoculture. Similarly, an increase of 50 to 53% in the invasion capacity of F. alocis in coculture with P. gingivalis was observed (Fig. 5B). At 90 min p.i. there was no significant variation in the capacity to invade epithelial cells infected with a monoculture of P. gingivalis or F. alocis compared to invasion capacity at 45 min p.i. (data not shown). Although not statistically significant, there was an increase of 5% in the invasion by P. gingivalis W83 and 2% increase in invasion by F. alocis during coculture at 90 min p.i.
FISH.
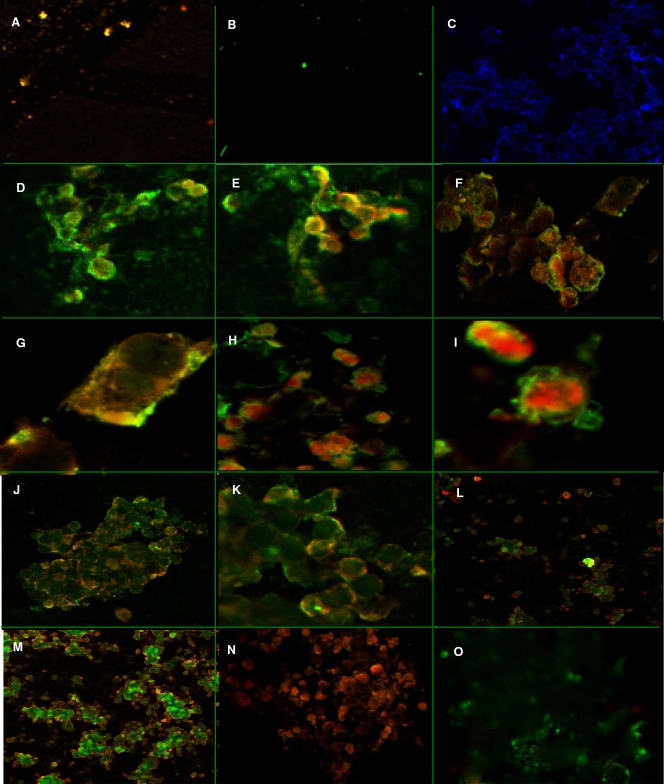
Fluorescence in situ hybridization (FISH) showed specific fluorescence of the infected epithelial cells. The P. gingivalis (Fig. 6A) and F. alocis bacterial cell controls (Fig. 6B) showed orange-red and green fluorescence, respectively. The uninfected epithelial cells showed no fluorescence in a DAPI mount (Fig. 6C). In epithelial cells infected with F. alocis there was green fluorescence around the margin of the cells at 20 min p.i. (Fig. 6D). At 30 min p.i., orange-red fluorescence was found to be more intense in the cytoplasm (Fig. 6E). At 45 min p.i., these epithelial cells, while showing orange-red fluorescence in the cytoplasm, also showed peripheral green fluorescence (Fig. 6F and G). In addition, F. alocis, depicted by green fluorescence, was also observed in the cytoplasm (Fig. 6H and I). At 60 to 90 min p.i., the epithelial cells showed an even distribution of orange-red and green fluorescence in the cytoplasm (Fig. 6J, K, L, and M). Monoculture-infected epithelial cells showed probe-specific fluorescence for P. gingivalis (Fig. 6N) and F. alocis (Fig. 6O).
Fig. 6.
Fluorescence in situ hybridization. FISH was performed using a trypsinized culture of HeLa cells (5 × 103 cells/ml) grown on coverslips. Probe-hybridized slides were air dried, mounted using the SlowFade Gold antifade reagent, and visualized using an inverted Axio Observer Z1 confocal microscope. The images were analyzed using Zeiss LSM 710 software. The images shown are representative of more than 12 viewing fields under a given magnification. F. alocis D-62D and P. gingivalis W83 were used in this study. (A) P. gingivalis W83 control cells showing specific red-orange fluorescence (645-nm absorbance/665-nm emission) (magnification, ×20). (B) F. alocis D-62D control cells showing specific green fluorescence (549-nm absorbance/563-nm emission) (magnification, ×20). (C) Negative control showing no fluorescence using DAPI (magnifi- cation, ×20). (D) Coinfection producing marginal green fluorescence around the epithelial cells, showing adherence of F. alocis D-62D seen after 20 min p.i. (magnification, ×40). (E) Coinfection producing a shift in the fluorescence, showing invasion by P. gingivalis W83 over F. alocis D-62D at 30 min p.i. (magnification, ×40). (F) Coinfected epithelial cells showing peripheral adherence of F. alocis D-62D and early invasion by P. gingivalis W83 (magnification, ×40) at 45 min p.i. (G) Coinfected individual epithelial cell showing early invasion by P. gingivalis W83 (magnification, ×63) at 45 min p.i. (H) Coinfected individual epithelial cell showing early invasion by P. gingivalis W83 (intense red-orange fluorescence in the cytoplasm) (magnification, ×40) at 45 min p.i. (I) Coinfected epithelial cell showing sparse adherence of F. alocis D-62D and cytoplasm showing the invading P. gingivalis W83 (magnification, ×40) at 45 min p.i. (J) Coinfection of F. alocis D-62D and P. gingivalis W83 showing generalized invasion of epithelial cells at 1 h p.i. (magnification, ×20). (K) Coinfection of F. alocis D-62D and P. gingivalis W83 showing general invasion of epithelial cells at 1 h p.i. (magnification, ×40). (L and M) Equal invasion was noticed after 1 h p.i. (magnification, ×20). (N) Invasion by P. gingivalis W83 monoculture in epithelial cells (magnification, ×20). (O) Invasion by F. alocis monoculture in epithelial cells (magnification, ×20).
Ultrastructural studies on invasion by F. alocis.
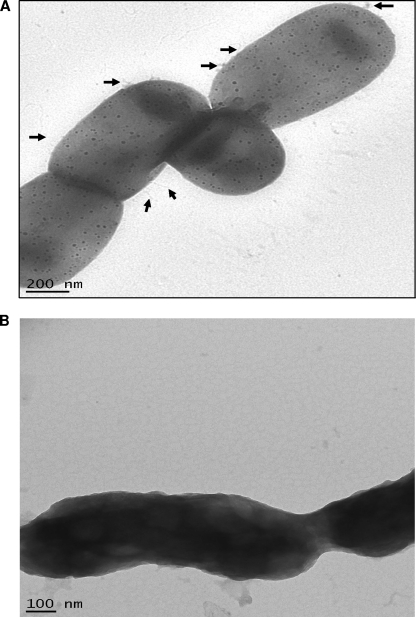
Transmission electron microscopic studies on the F. alocis strains revealed surface modifications between the ATCC 35896 strain and the clinical isolates. The external surfaces of the bacteria showed rudimentary markings and slender external projections resembling pili (Fig. 7A). However, the clinical isolates showed a thick slimy covering around the bacilli with no external projections and rudimentary marks (Fig. 7B).
Fig. 7.
Transmission electron microscopy was performed using the FEI Tecnai G2 TEM and Formvar carbon-coated grids. Conventional electron microscopy was performed by staining the sample with 1% phosphotungstic acid. (A). The external surface of Filifactor alocis ATCC 35896 shows rudimentary markings and slender projections resembling pili (arrows). (B) Filifactor alocis clinical strain D-62D showing thick slimy outer cover with no external projections or rudimentary marks.
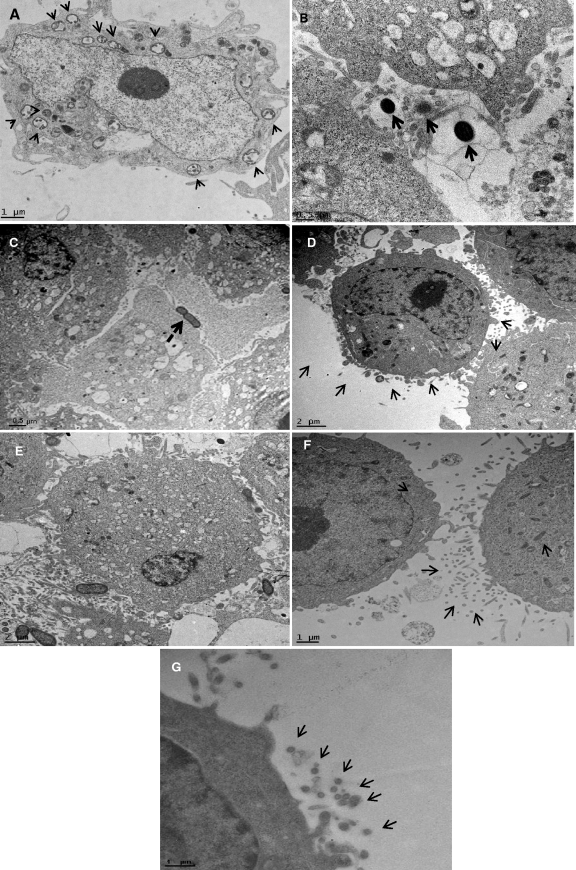
Cytoskeleton variations during mono- and coculture invasion by P. gingivalis W83 and F. alocis in epithelial cells were studied using transmission electron microscopy of ultrathin sections. Coculture showed a unique pattern of internalization, where both the pathogens were internalized together. A clear vacuolation of cytoplasm of the invaded epithelial cell showing vesicle-mediated internalization was noticed. The endocytic vesicles contained both P. gingivalis and F. alocis (Fig. 8A) at 45 min p.i. Such internalization was not observed in monocultures with P. gingivalis (Fig. 8B) or with F. alocis (Fig. 8C). Formation of such endocytic vesicles was observed once the bacteria came in contact with the epithelial cells (Fig. 8D). Epithelial cells infected by both strains showed initial invasion by P. gingivalis into the epithelial cells at 30 min p.i. (Fig. 8E). P. gingivalis monocultures were found to gain internalization by formation of pseudopodial filaments by the epithelial cells (Fig. 8F and G).
Fig. 8.
Ultrathin sections of the infected epithelial cell monolayer were washed with PBS, fixed with 2.5% glutaraldehyde, and stained with lead citrate and uranyl acetate. The grids were examined using by the FEI Tecnai G2 transmission electron microscope. Arrows show vesicles in the cytoplasm containing both Filifactor alocis D-62D and P. gingivalis W83. (A) Ultrathin sections of the coculture-infected epithelial cell monolayer at 45 min p.i. Arrows show endocytic vesicles in the cytoplasm containing both bacteria. (B) Monoculture of P. gingivalis W83 at 45 min p.i., showing invading organisms without an external envelope or covering. (C) Monoculture of Filifactor alocis D-62D at 45 min p.i., showing invading organisms without a covering. (D) Invasion by a coculture of F. alocis D-62D and P. gingivalis W83 at 30 min p.i., showing formation of the vesicular covering (arrows). The covering was noticed when the bacteria were in contact with the epithelial cells. Such receptor-mediated endocytosis was found only during coculture. (E) Coculture of P. gingivalis W83 and F. alocis D-62D at 30 min p.i., showing a higher number of invading P. gingivalis W83 cells. (F and G) P. gingivalis W83 monoculture at 30 min p.i., showing cytoskeleton variation through formation of microvilli, filamentous projections of the epithelial cells facilitating invasion by the bacteria.
SDS-PAGE analysis of F. alocis cell fractions.
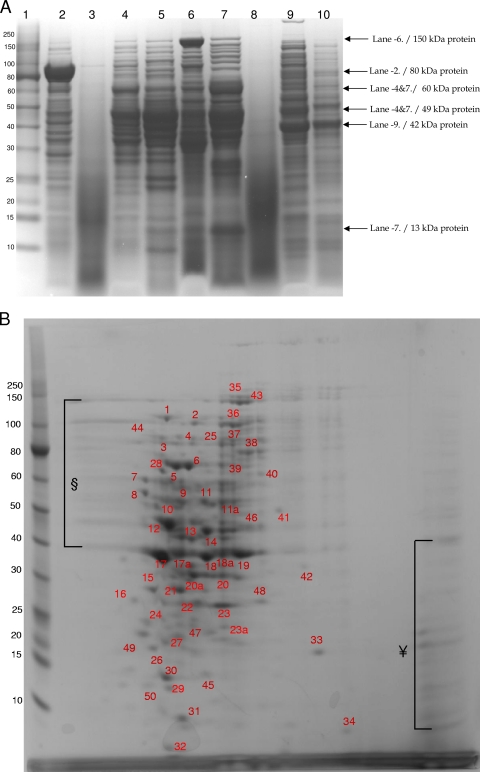
To identify some of the major proteins from F. alocis that may be important in its virulence, cell fractions of F. alocis ATCC 35896 and D-62D were subjected to SDS-PAGE analysis. The secretory fraction of F. alocis showed several low-molecular-weight proteins. Clinical isolate D-62D showed three protein bands in the range of 35 to 50 kDa that were missing in the ATCC 35896 strain (Fig. 9A, lanes 3 and 8). The membrane fraction of the F. alocis ATCC 35896 strain showed intense higher-molecular weight-bands above 150 kDa. While these were missing in the clinical strain, more-intense protein bands between 40 and 60 kDa were observed (Fig. 9A, lanes 6 and 10). The cytosolic fraction of F. alocis ATCC 35896 showed comparatively fewer protein bands than clinical strain D-62D. D-62D showed an intense 42-kDa protein band that was missing in the ATCC 35896 strain (Fig. 9A, lanes 5 and 9). The total cell lysate fractions of both F. alocis ATCC 35896 and clinical strain D-62D showed variation in the protein profile. Intense protein bands between 49 and 60 kDa were observed in both fractions. A 13-kDa protein was noted only in F. alocis D-62D. The total cell fraction showed more protein bands in the clinical strain than in ATCC 35896 (Fig. 9A, lanes 4 and 7). The particle-free pellet fraction of F. alocis 35896 showed more prominent protein bands between 35 and 80 kDa than the D-62D strain. An intense band of approximately 80 kDa was observed only in the ATCC 35896 strain (Fig. 9A, lane 2). Two major protein bands of 80 and 150 kDa in the particle-free pellet and membrane fractions, respectively, were found only in F. alocis ATCC 35896 (Fig. 9A).
Fig. 9.
(A) SDS-PAGE of cell fractions of F. alocis. Cell fractions of F. alocis strains were analyzed by SDS-PAGE and stained using Coomassie SimplyBlue safe stain. Approximately 25 μg of protein was loaded onto each lane. Lane 1, molecular mass (kDa) markers; lane 2, particle-free pellet (ATCC 35896); lane 3, particle-free supernatant (ATCC 35896); lane 4, total cell lysate (ATCC 35896); lane 5, cytosolic fraction (ATCC 35896); lane 6, membrane fraction (ATCC 35896); lane 7, total cell lysate (D-62D); lane 8, particle-free supernatant (D-62D); lane 9, cytosolic fraction (D-62D); lane 10, membrane fraction (D-62D). (B) 2D gel electrophoresis of total cell lysate of F. alocis (ATCC 35896). 2D gel electrophoresis was carried out using 7-cm gel strips in a Protean IEF cell. The protein sample was diluted to a final concentration of 350 μg protein, and 20 μl of the sample was added and was run in two dimensions. After equilibration, the IPG strips were loaded onto the gel, electrophoresed at 200 V and 0.3 A for 4 to 5 h, and stained with Coomassie SimplyBlue safe strain. §, poorly resolved spots in the range of 30 to 80 kDa; ¥, poorly resolved spots in the range of 10 to 40 kDa. Numbers correspond to the discrete protein spots (see Table S1 in the supplemental material).
2D gel electrophoresis of F. alocis total cell lysate.
The total cell lysate proteins of F. alocis 35896 were separated by 2D PAGE (Fig. 9B). The patterns of spots were reproducible both in technical and biological replicates. It is noteworthy that the presence of several groups of very intense discrete spots were seen in the 10- to 80-kDa range. Some less intense and poorly resolved spots were noted in two areas, one between 10 and 40 kDa at the basic end of the gel and the other between 40 and 150 kDa at the acidic end of the gel (Fig. 9B). A total of 55 different spots were excised from the gel and identified using mass spectrometry (see Table S1 in the sup- plemental material). The intense spots corresponded to the fol- lowing proteins: ATP-dependent protease La (HMPREF0389_ 00279), V-type sodium ATP synthetase (HMPREF0389_01297), thiazole biosynthesis protein (HMPREF0389_01137), two membrane proteins (HMPREF0389_00019 and HMPREF0389_01657), serine/threonine protein kinase (HMPREF0389_00441), exonuclease (HMPREF0389_00984), two hypothetical proteins (HMPREF0389_ 00152 and HMPREF0389_01239), a putative uncharacterized protein (HMPREF0389_01381), dihydroorotate dehydrogenase (HMPREF0389_00787), MAC/perforin domain-containing protein (HMPREF0389_01644), and calcium binding acid repeat protein (HMPREF0389_01532). Of the 55 protein spots analyzed, 12 proteins were predicted to be secretory due to the presence of N-terminal signal peptide sequences, 20 proteins were predicted to be involved in amino acid metabolism, and 6 were hypothetical proteins. Important regulatory proteins such as the serine/threonine kinase, protease La, calcium acid repeat protein, and neutrophil activating protein A were also identified in the total cell lysate.
DISCUSSION
This study is one of the first to evaluate the virulence potential of F. alocis. This bacterium, now believed to be associated with periodontal disease, ranks higher in its prevalence than other established periodontopathogens, including P. gingivalis (12, 16, 60). This implies that, similar to these pathogens, F. alocis has unique virulence properties that may enhance its ability to survive and persist in the periodontal pocket.
Phylogenetic analysis showed that F. alocis belongs to a separate Gram-positive clad (clostridial clad) that diverges from the Streptococcus clad. The close relatedness of F. alocis to Clostridium and Fusobacterium could suggest similar and common virulence attributes. While Clostridium spp. are known to produce an array of exotoxins that are involved in virulence (64), all the related bacteria in the clad appear to possess common important virulence factors such as Xaa protease, metal-dependent proteases, trypsin-like proteases, and sialoglycoproteases (3, 15, 29, 46).
The fastidious nature of F. alocis could have contributed to its low level of detection using culture-based methods and perhaps its previously reduced significance in periodontal diseases compared to other periodontal pathogens. In this study, the generation time for F. alocis was at least 4 times longer that that for P. gingivalis. In contrast, it is noteworthy that its growth was stimulated under oxidative stress and it was relatively more resistant to hydrogen peroxide-induced oxidative stress than P. gingivalis. While a mechanism for the observed stimulation is unclear, it is likely that this attribute may contribute to its high-frequency presence in the inflammatory microenvironment of the periodontal pocket (12, 16, 50). It is noted that F. alocis is one of the least detected in healthy or periodontitis-resistant individuals (31, 51). This may also suggest that F. alocis has a more developed system to eliminate and/or repair damage caused by reactive oxygen species (ROS). This is under further investigation.
Consistent with previous observations (61), specific amino acids stimulated the growth of F. alocis. In the periodontal pocket these amino acids are likely made available from the degradation of various protein substrates by bacteria and host-derived proteases (13, 14). Further, it is important to note that arginine is abundantly available in the niche of the periodontal pocket (61). It is likely that F. alocis could play a role in this process. Our study suggests that, even though F. alocis showed low gingipain-type activity, the nongingipain protease activity potential, comprising metalloproteases, ATP-dependent proteases, CaaX aminoproteases, glycoprotease, sialoglycoproteases, and serine proteases, was elevated. Among the amino acids utilized by F. alocis, arginine and lysine were highly degraded, followed by cysteine. Interrogation of the genome of F. alocis revealed the presence of several genes that indicated a well-developed mechanism for arginine metabolism that is distinctly different from those of other associated organisms (http://www.ncbi.nlm.nih.gov/genomeprj/30485). While fulfilling its metabolic requirements, this activity may lead to the maintenance of an environment conducive to the survival of several periodontal pathogens. The F. alocis arginine metabolic pathway predicts the enzymatic degradation of arginine by arginine deaminase and ornithine carbamoyltransferase, leading to the conversion of arginine to ornithine without forming ammonia (61). Seemingly, arginine degradation could favor a rise in the pH that will counteract acidic conditions generated from carbohydrate catabolism in a mixed bacterial oral flora (26).
The microenvironment of the periodontal pocket contains an abundance of sialylated glycoproteins such as fibronectin and immunoglobulins, found in saliva and gingival crevicular fluid (56). While a number of periodontal bacteria are known to exploit host sialylated glycoproteins as a nutrient source, sialidase activity can also play an important role in their pathogenicity (3, 4, 10, 17, 28). For example, sialidase activity can modify host glycoconjugates and expose potential binding receptors for bacterium-host interactions (22), sialidase treatment of immunoglobulins can make them more susceptible to proteolytic degradation, and cleavage of desialylated proteins can also provide the substrate for the proteases to generate the peptides needed to satisfy the asaccharolytic property of some periodontal pathogens. The released sialic acid can act as a ROS scavenger to reduce the oxidative stress in the inflammatory environment of the periodontal pocket (22). The presence of sialidase activity in F. alocis could suggest an important virulence attribute, as recently demonstrated in P. gingivalis (57) and other periodontal pathogens (5, 48, 57).
In addition to other biological factors, oral biofilms are primary initiating factors for periodontal disease. Biofilm formation involving F. alocis has been demonstrated both in periodontic and endodontic cases (54). While some interspecies interaction can inhibit biofilm formation (33), P. gingivalis ATCC 33277 cocultured with F. alocis showed a significant increase in biofilm formation. This enhanced biofilm-forming capacity may involve the ability of both these species to autoaggregate and/or express unique components (47). This may also indicate a symbiotic relationship between F. alocis and P. gingivalis. Thus, F. alocis and P. gingivalis, each with distinctly different growth rates, could form a mixed-species biofilm and coexist (55, 57). Hence, a coculture with P. gingivalis can proliferate and disseminate from such biofilms, which is a mechanism central for its virulence (11).
The abilities to adhere to and invade host cells are important attributes of a successful pathogen (40). In the genome of F. alocis there are genes that encode specific CaaX aminopeptidases that in other organisms are reported to be involved in masking the host ubiquitin system, thus facilitating invasion of host cells (42). In this study, F. alocis was shown to adhere to and invade epithelial cells. These abilities, which were enhanced in the presence of P. gingivalis, suggest the involvement of interacting proteins that could be modulated by bacterium-bacterium interaction (40) or contact with the host cell (35, 38). Similar enhancement of invasion has been observed during coculture of P. gingivalis with Fusobacterium nucleatum and Prevotella intermedia (51), Fusobacterium nucleatum with Streptococcus cristatus (14), and Fusobacterium nucleatum with Pseudomonas aeruginosa (42). Collectively, this suggests that there could be variations in host susceptibility due to the presence of specific coinhabitants (66).
The dynamics of invasion of the epithelial cells cocultured with F. alocis and P. gingivalis were examined by fluorescent in situ hybridization. In these studies, early rapid invasion by P. gingivalis compared that by F. alocis was observed. It is also likely that F. alocis and P. gingivalis utilize different invasion mechanisms. Previous reports suggest that, following adhesion to the integrins, P. gingivalis is captured by cellular pseudopodia, which enables invagination through an actin-mediated pathway (1). This invasive event has been reported to require host cellular dynamin, actin fibers, microtubules, and lipid rafts (2). Consistent with these reports, monoculture invasion of epithelial cells by P. gingivalis in our study involved microvilli forming filamentous projections, which is believed to mediate bacterial internalization. In contrast, microvilli forming filamentous projections were not observed during the invasion of F. alocis-infected epithelial cells. Even though a membrane ruffling mechanism of invasion is commonly noted among the Gram-positive bacteria (40), this was not observed in epithelial cells infected with F. alocis in monoculture or in coculture with P. gingivalis. Ultrastructural changes during invasion of epithelial cells by coculture showed vesicle-mediated internalization of both P. gingivalis and F. alocis. Such vesicle-mediated internalization is commonly seen in Gram-positive bacteria and may protect the pathogen after invasion, a crucial step in pathogenesis (40). The enhanced invasion by P. gingivalis in coculture with F. alocis could be attributed to such vesicle-mediated internalization. Taken together, our studies may suggest that, during the early stages of infection, the two bacteria could compete for invasion. As reported previously (reviewed in reference 60), the rapid-invasion ability of P. gingivalis, which is facilitated by its potent invasins, triggers an integrin-associated signaling cascade, resulting in its specific internalization. Because these vesicles were observed only during invasion by P. gingivalis in coculture with F. alocis, it is likely that the interaction of both strains may lead to bacterially derived protein(s) that may inhibit or slow vesiculation or escape from these vesicles. Alternatively, we cannot rule out the possibility that vesicle-mediated internalization could be a process that is rapidly resolved in the presence of F. alocis but is inhibited in the presence of P. gingivalis. P. gingivalis has properties that can modulate the trafficking in host cells (1). In the later stages of infection, it could be that another F. alocis-derived effector protein(s) induces vesicle-mediated internalization of both organisms. Vesicle-mediated endocytic internalization of Gram-positive bacteria is generally mediated by type II and III exotoxins (40). Several exotoxins have been identified in F. alocis; however, their role in invasion is unclear.
In the gingival sulcus, the ability of various bacteria to express specific surface molecules mediating interaction with other bacterial species and host cells is a critical virulence attribute (16). Our study on the proteome of F. alocis identified several hypothetical proteins and other proteins that, as observed in other bacteria, have important virulence properties. Several of these proteins were predicted to be involved in various secretory pathways, many of which, as shown in F. nucleatum, play a role in invasion (19). One protein of note that was identified was the putative neutrophil activating protein A. In Helicobacter pylori neutrophil-activating protein is a major virulence factor that plays a central role in the pathogenesis of mucosal inflammation by neutrophil attraction, activation, and Th1 cytokine response polarization (reviewed in reference 12). An emerging concept of periodontitis emphasizes the destructive role of the hyperactive/primed neutrophil, which leads to tissue damage and bone resorption (27; reviewed in reference 50). The results of the hyperactive/primed neutrophil could involve increased neutrophil adhesion, enzyme release, and, perhaps most importantly, an elevated oxidative burst that leads to the release of superoxide, hydroxyl radicals, and hydrogen peroxide. It is tempting to speculate that this neutrophil activating protein A in F. alocis may be an important virulence factor that also leads to survival and persistence of the bacterium given its relative resistance to oxidative stress and its stimulated growth under those conditions. Furthermore, there is evidence that the secretion of proinflammatory cytokines, including interleukin 1β (IL-1β), IL-6, and tumor necrosis factor alpha (TNF-α), from gingival epithelial cells and apoptosis of these cells can be induced by F. alocis (R. J. Lamont, personal communication). The role of neutrophil activating protein A or another putative virulence factor(s) in these processes is unclear. These important attributes are under further investigation in the laboratory.
Collectively, the data from this study may suggest a model for F. alocis pathogenesis which will form the basis for an exploration to further understand the relative significance of several of its virulence factors. F. alocis has unique properties that contribute to its relative resistance to oxidative stress. This oxidative stress, which is partially induced by the neutrophil activating protein A, will allow the organism to outcompete other, more-oxidation-sensitive bacteria. The sialoglycoproteases and proteases, including the dipeptidases, can facilitate its colonization. The sialoglycoproteases can modify host glycoconjugates and expose potential binding receptors for bacterium-host or bacterium-bacterium interactions. Further, cleavage of desialylated proteins can provide the substrate for the dipeptidases to generate the peptides needed to satisfy the bacterium's asaccharolytic property. In addition, the released sialic acid can act as a ROS scavenger to reduce the oxidative stress in the inflammatory environment of the periodontal pocket. The ability of F. alocis to form biofilms and invade host cells can facilitate its protection from the immune defense system, which will lead to its persistence in the periodontal pocket. Characterization of the genes involved in these processes could lead to the development of novel therapeutic approaches for the treatment of F. alocis-associated diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Loma Linda University and Public Health Grants DE-13664 and DE-019730 from NIDCR (to H.M.F.).
We thank Floyd E. Dewhirst for the gift of the F. alocis clinical isolates.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 8 August 2011.
REFERENCES
- 1. Amano A. 2007. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front. Biosci. 12:3965–3974 [DOI] [PubMed] [Google Scholar]
- 2. Amano A., Furuta N., Tsuda K. 2010. Host membrane trafficking for conveyance of intracellular oral pathogens. Periodontol. 2000 52:84–93 [DOI] [PubMed] [Google Scholar]
- 3. Aruni W., et al. 2011. Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis. Infect. Immun. 79:2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brigham C., et al. 2009. Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase. J. Bacteriol. 191:3629–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byers H. L., Homer K. A., Tarelli E., Beighton D. 1999. N-acetylneuraminic acid transport by Streptococcus oralis strain AR3. J. Med. Microbiol. 48:375–381 [DOI] [PubMed] [Google Scholar]
- 6. Castaneda-Roldan E. I., et al. 2004. Adherence of Brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cell. Microbiol. 6:435–445 [DOI] [PubMed] [Google Scholar]
- 7. Cato E. P., Moore L. V. H., Moore W. E. C. 1985. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int. J. Syst. Bacteriol. 35:475–477 [Google Scholar]
- 8. Chawla K., Mukhopadhayay C., Shetty P. V. 2010. First report of submandibular and parotid abscess due to Nocardia asteroides. Braz. J. Infect. Dis. 14:544–545 [DOI] [PubMed] [Google Scholar]
- 9. Cole J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Costello A. H., Cisar J. O., Kolenbrander P. E., Gabriel O. 1979. Neuraminidase-dependent hamagglutination of human erythrocytes by human strains of Actinomyces viscosus and Actinomyces naeslundii. Infect. Immun. 26:563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davey M. E. 2006. Techniques for the growth of Porphyromonas gingivalis biofilms. Periodontol. 2000 42:27–35 [DOI] [PubMed] [Google Scholar]
- 12. D'Elios M. M., Amedei A., Cappon A., Del P. G., de B. M. 2007. The neutrophil-activating protein of Helicobacter pylori (HP-NAP) as an immune modulating agent. FEMS Immunol. Med. Microbiol. 50:157–164 [DOI] [PubMed] [Google Scholar]
- 13. Dewhirst F. E., et al. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edwards A. M., Grossman T. J., Rudney J. D. 2006. Fusobacterium nucleatum transports non invasive Streptococcus cristatus into human epithelial cells. Infect. Immun. 74:654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Endo J., Otsuka M., Ohara E., Sato M., Nakamura R. 1989. Cleavage action of a trypsin-like protease from Bacteroides gingivalis 381 on reduced egg-white lysozyme. Arch. Oral Biol. 34:911–916 [DOI] [PubMed] [Google Scholar]
- 16. Feuille F., Ebersole J. L., Kesavalu L., Stepfen M. J., Holt S. C. 1996. Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum in a murine lesion model: potential synergistic effects on virulence. Infect. Immun. 64:2094–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fraser A. G., Brown R. 1981. Neuraminidase production by Bacteroidaceae. J. Med. Microbiol. 14:63–76 [DOI] [PubMed] [Google Scholar]
- 18. Fulkerson J. F., Jr., Mobley H. L. 2000. Membrane topology of the NixA nickel transporter of Helicobacter pylori: two nickel transport-specific motifs within transmembrane helices II and III. J. Bacteriol. 182:1722–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gross E. L., et al. 2010. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 48:4121–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harris J. R. 2007. Negative staining of thinly spread biological samples. Methods Mol. Biol. 369:107–142 [DOI] [PubMed] [Google Scholar]
- 21. Hinsa S. M., O'Toole G. A. 2006. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 152:1375–1383 [DOI] [PubMed] [Google Scholar]
- 22. Iijima R., Takahashi H., Namme R., Ikegami S., Yamazaki M. 2004. Novel biological function of sialic acid (N-acetylneuraminic acid) as a hydrogen peroxide scavenger. FEBS Lett. 561:163–166 [DOI] [PubMed] [Google Scholar]
- 23. Jakubovics N. S. 2010. Talk of the town: interspecies communication in oral biofilms. Mol. Oral Microbiol. 25:4–14 [DOI] [PubMed] [Google Scholar]
- 24. Jalava J., Eerola E. 1999. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int. J. Syst. Bacteriol. 49:1375–1379 [DOI] [PubMed] [Google Scholar]
- 25. Jenkinson H. F., Lamont R. J. 2005. Oral microbial communities in sickness and in health. Trends Microbiol. 13:589–595 [DOI] [PubMed] [Google Scholar]
- 26. Kanapka J. A., Kleinberg I. 1983. Catabolism of arginine by the mixed bacteria in human salivary sediment under conditions of low and high glucose concentration. Arch. Oral Biol. 28:1007–1015 [DOI] [PubMed] [Google Scholar]
- 27. Kantarci A., Van Dyke T. E. 2005. Resolution of inflammation in periodontitis. J. Periodontol. 76:2168–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kelm S., Schauer R., Crocker P. R. 1996. The Sialoadhesins—a family of sialic acid-dependent cellular recognition molecules within the immunoglobulin superfamily. Glycoconj. J. 13:913–926 [DOI] [PubMed] [Google Scholar]
- 29. Kim H. S., et al. 2010. Application of rpoB and zinc protease gene for use in molecular discrimination of Fusobacterium nucleatum subspecies. J. Clin. Microbiol. 48:545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kolenbrander P. E., et al. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar P. S., et al. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338–344 [DOI] [PubMed] [Google Scholar]
- 32. Kumar P. S., et al. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 44:3665–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuramitsu H. K., He X., Lux R., Anderson M. H., Shi W. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loy A., Maixner F., Wagner M., Horn M. 2007. probeBase—an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 35:D800–D804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maiden M. F., Tanner A., Macuch P. J. 1996. Rapid characterization of periodontal bacterial isolates by using fluorogenic substrate tests. J. Clin. Microbiol. 34:376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manz W., Amann R., Ludwig W., Wagner M., Schleifer K. H. 2011. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593–600 [Google Scholar]
- 37. Marchler-Bauer A., et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Massey B. W. 1953. Ultra-thin sectioning for electron microscopy. Stain Technol. 28:19–26 [DOI] [PubMed] [Google Scholar]
- 39. Nakai K., Kanehisa M. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95–110 [DOI] [PubMed] [Google Scholar]
- 40. Nitsche-Schmitz D. P., Rohde M., Chhatwal G. S. 2007. Invasion mechanisms of Gram-positive pathogenic cocci. Thromb. Haemost. 98:488–496 [PubMed] [Google Scholar]
- 41. Olango G. J., Roy F., Sheets S. M., Young M. K., Fletcher H. M. 2003. Gingipain RgpB is excreted as a proenzyme in the vimA-defective mutant Porphyromonas gingivalis FLL92. Infect. Immun. 71:3740–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pan Y., Teng D., Burke A. C., Haase E. M., Scannapieco F. A. 2009. Oral bacteria modulate invasion and induction of apoptosis in HEp-2 cells by Pseudomonas aeruginosa. Microb. Pathog. 46:73–79 [DOI] [PubMed] [Google Scholar]
- 43. Paster B. J., et al. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Periasamy S., Kolenbrander P. E. 2009. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J. Bacteriol. 191:6804–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Poznanovic S., Schwall G., Zengerling H., Cahill M. A. 2005. Isoelectric focusing in serial immobilized pH gradient gels to improve protein separation in proteomic analysis. Electrophoresis 26:3185–3190 [DOI] [PubMed] [Google Scholar]
- 46. Price C. T., Kwaik Y. A. 2010. Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front. Microbiol. 1:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rickard A. H., Gilbert P., High N. J., Kolenbrander P. E., Handley P. S. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94–100 [DOI] [PubMed] [Google Scholar]
- 48. Roy S., Douglas C. W., Stafford G. P. 2010. A novel sialic acid utilization and uptake system in the periodontal pathogen Tannerella forsythia. J. Bacteriol. 192:2285–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rudney J. D., Chen R., Sedgewick G. J. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69:2700–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ryder M. I. 2010. Comparison of neutrophil functions in aggressive and chronic periodontitis. Periodontol. 2000 53:124–137 [DOI] [PubMed] [Google Scholar]
- 51. Saito A., Inagaki S., Ishihara K. 2009. Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis. Microb. Pathog. 47:329–333 [DOI] [PubMed] [Google Scholar]
- 52. Saito A., et al. 2008. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 54:349–355 [DOI] [PubMed] [Google Scholar]
- 53. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 54. Schlafer S., et al. 2010. Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol. 10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Skillman L. C., Sutherland I. W., Jones M. V., Goulsbra A. 1998. Green fluorescent protein as a novel species-specific marker in enteric dual-species biofilms. Microbiology 144(Pt. 8):2095–2101 [DOI] [PubMed] [Google Scholar]
- 56. Stefanovic G., et al. 2006. Hypogalactosylation of salivary and gingival fluid immunoglobulin G in patients with advanced periodontitis. J. Periodontol. 77:1887–1893 [DOI] [PubMed] [Google Scholar]
- 57. Stinson M. W., Safulko K., Levine M. J. 1991. Adherence of Porphyromonas (Bacteroides) gingivalis to Streptococcus sanguis in vitro. Infect. Immun. 59:102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sunde P. T., et al. 2003. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology 149:1095–1102 [DOI] [PubMed] [Google Scholar]
- 59. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 60. Tribble G. D., Lamont R. J. 2010. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol. 2000 52:68–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Uematsu H., Sato N., Hossain M. Z., Ikeda T., Hoshino E. 2003. Degradation of arginine and other amino acids by butyrate-producing asaccharolytic anaerobic Gram-positive rods in periodontal pockets. Arch. Oral Biol. 48:423–429 [DOI] [PubMed] [Google Scholar]
- 62. Veith P. D., et al. 2009. Outer membrane proteome and antigens of Tannerella forsythia. J. Proteome Res. 8:4279–4292 [DOI] [PubMed] [Google Scholar]
- 63. Wade W. G. 2011. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J. Clin. Periodontol. 38(Suppl. 11):7–16 [DOI] [PubMed] [Google Scholar]
- 64. Wells C. L., Wilkins T. D. 1996. Clostridia: sporeforming anaerobic bacilli, p. 1012–1045 In Baron S. (ed.), Medical microbiology, 4th ed. University of Texas, Galveston, TX: [PubMed] [Google Scholar]
- 65. Yilmaz O., Young P. A., Lamont R. J., Kenny G. E. 2003. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology 149:2417–2426 [DOI] [PubMed] [Google Scholar]
- 66. Yin L., et al. 2010. Differential effects of periopathogens on host protease inhibitors SLPI, elafin, SCCA1, and SCCA2. J. Oral Microbiol. doi:10.3402/jom.v2i0.5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.