Abstract
The pattern of global gene expression in Salmonella enterica serovar Typhimurium bacteria harvested from the chicken intestinal lumen (cecum) was compared with that of a late-log-phase LB broth culture using a whole-genome microarray. Levels of transcription, translation, and cell division in vivo were lower than those in vitro. S. Typhimurium appeared to be using carbon sources, such as propionate, 1,2-propanediol, and ethanolamine, in addition to melibiose and ascorbate, the latter possibly transformed to d-xylulose. Amino acid starvation appeared to be a factor during colonization. Bacteria in the lumen were non- or weakly motile and nonchemotactic but showed upregulation of a number of fimbrial and Salmonella pathogenicity island 3 (SPI-3) and 5 genes, suggesting a close physical association with the host during colonization. S. Typhimurium bacteria harvested from the cecal mucosa showed an expression profile similar to that of bacteria from the intestinal lumen, except that levels of transcription, translation, and cell division were higher and glucose may also have been used as a carbon source.
INTRODUCTION
Salmonella enterica serovars Typhimurium and Enteritidis are the two S. enterica serovars most frequently associated with human food poisoning, with 1.4 million cases reported in the United States in 1999 (26) and an estimated 192,703 cases in the European Union in 2004 (4). Poultry and poultry products are generally considered to be major sources of human infection (3, 65). Healthy adult chickens generally show no clinical disease following oral infection with these serovars (6, 70). Infection of birds more than a few days old with S. Typhimurium or S. Enteritidis results in asymptomatic cecal colonization with persistent shedding of organisms, resulting in carcass contamination at slaughter and entry into the human food chain. The ecology of colonization of birds of this age is complex (21). In contrast, infection within a few hours of hatching, as can occur in hatcheries, when the chicken is immunologically immature and possesses a rudimentary gut flora, not only results in massive multiplication in the alimentary tract but can also result in severe systemic disease in the bird (6, 73).
Although intestinal colonization is central to entry into the human food chain, either through carcass contamination or by preceding systemic infection and subsequent egg contamination, the mechanism whereby S. enterica serovars colonize and interact with the host in the early stages of infection is still poorly understood. Screening of randomly generated mutant libraries of S. Typhimurium and more targeted studies have provided some insight into the bacterial genes required for colonization of chickens which are several weeks old and possess a gut flora. Type I and other fimbriae, including those encoded by the stb, csg, and sth operons (22, 31, 59), are thought to be involved in attachment of Salmonella and Escherichia coli bacteria to the mucosal layer or even to epithelial cells. Lipopolysaccharide is also thought to be involved, but it is unclear how (20, 59, 82). Additionally, global regulatory genes and a number of metabolic functions, including serine and citrate utilization, together with heat shock conditions, appear to contribute to the process in adult birds (59). Although some of the genes identified indicate that a close association with the gut mucosa is important in Salmonella colonization, the metabolic behavior of bacteria in the gut of newly hatched chickens is still poorly understood. Microbial behavior under these circumstances is very different from that in older birds. Viable numbers of Salmonella bacteria colonizing the cecum are much higher in younger than in older birds, and the interactions between the bacteria may more closely resemble those in stationary-phase broth cultures (100), where competition for nutrients under the prevailing redox conditions is at least known to be involved. Some studies also indicated the importance of proton-translocating proteins in colonization (44, 100; S. Muhammad, M. A. Jones, and P. Barrow, unpublished). Other factors, including some secreted proteins, contribute in different hosts, but it is again unclear how (52, 59, 82).
The numerical predominance of Salmonella bacteria in the ceca of young chicks following experimental infection allows effective analysis of the bacteria in the absence of other organisms, and gene transcription pattern analysis at the genome level is thus possible. A whole-genome array derived from S. Typhimurium was used to investigate gene expression of the virulent avian phage type 14 strain S. Typhimurium F98 (70, 82, 100), harvested directly from chick ceca and compared with expression patterns from bacteria grown in broth in vitro. This approach, at least with Campylobacter jejuni, has demonstrated successfully that expression profiles under these conditions do resemble those observed in older, fully colonized birds (92).
MATERIALS AND METHODS
Chick colonization and sample collection.
One hundred chickens from a brown-egg commercial laying line (Lohmann) were hatched in prefumigated incubators. Chickens were housed in fumigated cages and handled with sterile gloves to avoid contamination. One hundred chicks were infected orally within 12 h of hatching (to avoid the development of gut flora) by gavage with 0.1 ml of an S. Typhimurium F98 (70, 82, 100) culture grown for 16 h in LB broth at 37°C in a shaking incubator (150 rpm) and diluted to contain 107 CFU/ml. Only sterile water was provided, since the yolk sac is not fully resorbed for up to 3 to 4 days, providing sufficient food for the experimental period. At 16 h postinfection, the birds were killed individually and the cecal contents were removed immediately from the exposed ceca by syringe and mixed with Tri Reagent (Sigma). The cecal contents from seven of the birds were collected separately and stored on ice to be used for viable count estimations. The cecal contents from each group of birds, mixed with Tri Reagent, were pooled prior to extraction and purification. The purified RNA was further treated with DNase I and cleaned using RNeasy mini columns (Qiagen) and then concentrated further by RNA precipitation using 3 M sodium acetate. RNA was used only when the quality and concentration were optimal, as determined by spectrophotometer (Pharmacia). The experiment was repeated three times. Viable count estimations were made by plating decimal dilutions on MacConkey agar to allow the presence of any contaminating colonies among the predominant non-lactose-fermenting Salmonella bacteria to be detected. In the three experiments, the numbers of Salmonella bacteria were between 8.95 and 10.20 log10, and lactose fermenters or other colony types were not detected (<2 log10 per g).
Patterns of in vivo gene expression were compared with those of bacteria grown in vitro. For these controls, total RNA was extracted in the same way from three cultures of S. Typhimurium F98, in which 2 ml of an overnight LB broth culture was inoculated into 200 ml of prewarmed LB broth and incubated with shaking (150 rpm) for 3 h at 37°C. Cultures were pretreated with RNA Protect (Qiagen) before being centrifuged at 5,000 × g for 10 min at 20°C prior to RNA extraction.
Harvesting of Salmonella from the mucosal wall.
In addition to harvesting the cecal contents, material was taken from the cecal mucosa for analysis by microarray. Samples were extracted by emptying the ceca with gentle pressure and then opening the walls of the ceca lengthwise and shaking the cecal walls in RNA Protect (Qiagen) to release bacteria from the surface. RNA from the ceca and from the washings from the mucosal wall was isolated using standard cleanup procedures, and samples from the same experiments were pooled. RNA from both samples was amplified using a MessageAmp II-bacteria kit (Ambion) per the manufacturer's instructions. RNA quality and concentration were determined with a spectrophotometer (Pharmacia). Gene expression was compared with that of the luminal samples.
Microarray hybridization.
The S. Typhimurium array was printed as described previously (25). Total bacterial RNA was isolated from chicken ceca and from in vitro cultures grown in LB broth. The DNase-treated total in vivo- and in vitro-grown RNA was converted to fluorescently labeled cDNA using indirect labeling techniques (2, 25). Briefly, 15 μg of the total RNA samples from chick cecal contents was reverse transcribed (SuperScript II; Invitrogen) in the presence of 1 μl of deoxynucleoside triphosphates (dNTPs) (2.5 mM concentration each of dATP, dCTP, and dGTP and 1 mM concentration of dTTP [Amersham]), 1.5 μl of aminoallyl-dUTP (Sigma), and 30 μg of pd(N6) (Amersham) in a total volume of 12 μl. This mixture was incubated overnight at 42°C before the reaction was stopped, and the mixture was cleaned with 450 μl of water in triplicate using Microcon units (YM-30; Millipore).
Two cDNA probes were labeled with 100 mg of Cy3 (in vivo lumen sample) or Cy5 (in vitro or in vivo mucosal sample) (monofunctional dyes; Amersham). The Cy3- and Cy5-labeled probes were combined and cleaned using a QIAquick PCR purification kit (Qiagen). The probe was dried in a speed vacuum before it was resuspended in a total volume of 25 μl of hybridization buffer (3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 25 mM HEPES, yeast tRNA, 50× Denhardt's solution, 10% [wt/vol] SDS), heated for 2 min, and then cooled in the dark. The probe was applied directly to the array with a clean coverslip placed on top. The probe was hybridized for 16 h at 63°C in a humidified slide chamber (Telechem, Inc., CA). The slide was postprocessed as described previously (25). Slides were scanned using a commercial laser scanner (GenePix 4000A; Axon Instruments, MDS, Sunnyvale, CA).
Data analysis.
Fluorescence intensities of the signal and background were calculated for each spot using image analysis software (GenePix Pro 3.0; Axon Instruments). Three biological replicates each of both the in vitro-grown RNA and the in vivo-harvested RNA were compared. The data were analyzed using the Limma package (71). The data were first normalized within arrays using the Loess method (72) and then normalized between arrays in order to scale the log ratios to have the same median absolute deviation (MAD) across arrays (95). A linear model was then fitted for each spot across the series of arrays. The resulting P values were adjusted according to the false-discovery-rate method of Benjamini and Hochberg (8). Functional annotations were linked to the genes from the NCBI file NC_003197.ptt (http://www.ncbi.nlm.nih.gov/nuccore/16763390).
RT-PCR.
The data were validated by quantitative reverse transcriptase PCR (qRT-PCR) of 15 genes which were differently regulated in the lumen samples to confirm gene expression ratios (87). Primers (Table 1) and fluoroprobes were designed using Primer Express software (PE Applied Biosystems) and purchased from Sigma-Genosys Europe Ltd. (Cambridge, United Kingdom). One-step qRT-PCR was performed in triplicate by using a mix of 2 ng/μl DNase-treated total RNA, gene-specific primers (50 nM) and probes (100 nM), and reverse transcriptase qPCR master mix (RT-QPRT-032X; Eurogenetic, EGT Group, Belgium). The concentrations of primers and template in each reaction mixture were determined by construction of a standard curve, starting with 200 ng total RNA and 500 nM primer and using 10-fold dilutions from 10−1 to 10−5. Three total RNA samples were analyzed in triplicate in PCRs, and three replicate values were used to generate the standard curves. Amplification and detection of specific primers were performed using the ABI Prism 7700 sequence detection system (PE Applied Biosystems, Warrington, United Kingdom). The cycle parameters were as follows: an initial cycle of 48°C for 30 min and 95°C for 10 min and then 40 cycles of 95°C for 15s and 60°C for 1 min. The results were expressed in terms of threshold cycle value, the cycle at which the change in the reporter dye passes a significant threshold value above background. The fold changes in gene expression calculated from the qRT-PCR data were converted to log2 values and plotted against the changes calculated from the array data, which had also been log2 converted.
Table 1.
Oligonucleotide primers used for mutant production
| Gene | Oligonucleotide sequences of primersa | Enzyme | Resistance cassette |
|---|---|---|---|
| argA | TCACTCGAGGCAAAGAGGTGTGCCGTG | XhoI | Km |
| GCCGCTGGGCCGCTGGGGTACCARGACGGCGTGG ATT | KpnI | ||
| CTCGCCTCGTGCCATGGTACCCCAGCGGC | KpnI | ||
| CGCAGATCTTAACCCTAAATCCGCCATCA | BglII | ||
| potG | TCACTCGAGACGAAAGTGAAGAGCGGA | XhoI | Km |
| GATAAAAAGCTGGGTACCAGGATGCACCTTGAA | KpnI | ||
| CGACCACCGAGGCATGGTACCCAGCTTTTTATC | KpnI | ||
| CGGAGATCTCCGTCGGCACCACACAGCTC | BglII | ||
| csgA | TCACTCGAGGGATCAAAACTATTGTCCGT | XhoI | Km |
| AATGCTCAGGTACCGCCGTTATGATTACC | KpnI | ||
| ATAACGGCGGTACCTGAGCATTTATCAGT | KpnI | ||
| CGCAGATCTTAGCGCAGACGCTAAATTAA | BglII | ||
| metF | CGTCTCGAGGACATGAAGAAAATTCAACT | XhoI | Km |
| TTATTCCAGGTACCGCTCTTTGATGCCTT | KpnI | ||
| CAAAGAGCGGTACCTGGAATAACGGTATC | KpnI | ||
| TCCAGATCTTGGCAAATGGCATAACTCAT | BglII | ||
| ttrB | TCACTCGACCGCTGATTCTCTGGAGGA | XhoI | Km |
| CTTGTACCGGGTACCCCGGCACAC AGG | KpnI | ||
| GTCCCTGGATGCCTGGGTACCGGCCATAGG | KpnI | ||
| CGCAGATCTTGGCAATGTGGACGGGAG | BglII | ||
| ttrS | TCACTCGACCCCGGCTTGTTGTTGATC | XhoI | Km |
| ACTGGGCCGGGTACCCGTCCACCAGTC | KpnI | ||
| CCGCCTGAGCCGCATGGTACCCGCCCCAGT | KpnI | ||
| GCGAGATCTTCATCCAGTAGATGAAT | BglII | ||
| pduA | TCACTCGACCCATGCGAGGTCTTTATG | XhoI | Km |
| CGCGGCGATGTTACCCGGTCAAAG | KpnI | ||
| TGCATCGGTGGCCGCGGTAACATCGCCGCG | KpnI | ||
| CGCAGATCTCCACCAGCTGACTGCTGC | BglII | ||
| eutR | TCACTCGACGAGAGCCTCCCCATCAAT | XhoI | Km |
| GTGGCCAGCGTTACCTGCACAAAGCCC | KpnI | ||
| CTAGCGCTGGAGGTAGTTACCGCTGGCGAG | KpnI | ||
| CGGAGATCTGTCGGAGGGCCGGGCGTC | BglII | ||
| btuB | TCACTCGACAAGCCTGCGGCATCCTCC | XhoI | Km |
| CTCCGCTATGGTACCTTCCGATGCTAT | KpnI | ||
| GCGCTTTGTAGGAGGGGTACCATAGCGGAG | KpnI | ||
| CGGAGATCTCGGTGGGACGAGGTTCAG | BglII | ||
| cobS | GATCTAGAACGAATCTGCTGTTTGCGCT | XhoI | Km |
| CAGCAGGGTACCTAGCGGAATACCACACCAG | KpnI | ||
| CCGCTAGGTACCCTGCTGACCGGTGGTTTTCA | KpnI | ||
| AGTCTAGAACAGAGCCAGCAGAAAGATC | BglII | ||
| CAGCAGGGATCCTAGCGGAATACCACACCAGG | BamH1 | Spc | |
| CCGCTAGGATCCCTGCTGACCGGTGGTTTTCA | BamH1 | ||
| cbiA | CATCTAGAAAGGCATCACGCATTTATTC | XhoI | Km |
| CGTTATGGTACCAATGGCATTTTTGAGGAGCT | KpnI | ||
| GCCATTGGTACCATACGGTGATGTTAAAACAT | KpnI | ||
| TGTCTAGACAGCCAGTGCTGCACCATTT | BglII | ||
| TAACATCACCGGATCCGCCGCCAG | BamH1 | Spc | |
| AGCAATCATGGCATGGGATCCGGTGATGTT | BamH1 |
Underlining and boldface indicate enzyme sites.
Creation of mutants.
Insertion mutants using kanamycin or streptomycin/spectinomycin resistance cassettes were prepared as single mutants using standard procedures detailed elsewhere (82, 83, 100). Briefly, oligonucleotide primers were used to amplify upstream and downstream fragments, which were then joined together by an additional overlap extension PCR using the same two fragments as a template. This allowed the introduction of a KpnI site in the middle of the combined fragment and an XhoI and BglII (or, in the case of the cobS and cbiA mutants, XbaI) site at each end. This construct was incorporated into the suicide vector pDM4 (54), and the Kmr GenBlock insertion was introduced into the KpnI site. Spectinomycin and streptomycin (Spc-Str) resistance insertions were made in the same way. The cassette was in pHP45ΩSpc (H. Krisch, Département de Biologie Moleculaire, Université de Genèva, Switzerland). A single-base-pair change generated a BamHI site in the middle of the fragment that enabled an Spc-Str resistance cassette to be inserted after base 406 of the open reading frame (ORF), and XbaI sites were incorporated into each end of the fragment for cloning into pDM4. Oligonucleotide primers are shown in Table 1. These pDM4 derivatives were maintained in E. coli strain SM10λpir (83) and were introduced into the recipient Salmonella strains by conjugation. Transconjugants were isolated on selective medium supplemented with either streptomycin or kanamycin (25 μg/ml), and their sensitivities to chloramphenicol were then tested to identify those that resulted from a recombinational double-crossover event that had not incorporated any pDM4 DNA. The mutation was transduced into a fresh culture using P22 HT int (5). Transductants were checked by PCR using primers from the 3′ end of the cassette and the 5′ end of the structural gene, which generated a single DNA fragment in each of the mutants but not in the parent strain.
Double mutants were prepared with the creation of the additional mutation in a single mutant background using the alternative resistance cassette.
Assessment of colonization ability.
Colonization was checked in specific-pathogen-free (SPF) day-old Light Sussex chickens obtained from the Poultry Production Unit, Institute for Animal Health. Birds were maintained in cages at 33°C with water and received no food prior to oral inoculation.
Colonization ability was assessed in two ways. First (100), groups of 10 chickens were inoculated orally within 24 h of hatching with 0.1 ml of an undiluted broth culture of the strain (mutant or parent of nalidixic acid-resistant [Nalr] S. Typhimurium F98) to be tested. They were then given access to a vegetable protein-based diet (SDS, Manea, Cambridgeshire, United Kingdom). Twenty-four hours later, 3 birds were killed and the numbers of bacteria of the inoculated strain in the ceca were enumerated. The remaining 7 birds were inoculated orally with 0.1 ml of a 1:1,000 dilution of a broth culture of an Spcr mutant of the parent F98 strain. Three days later, all birds were killed and the numbers of bacteria of both strains in the cecal contents were counted on brilliant green containing either sodium nalidixate (20 μg/ml) and novobiocin (1 μg/ml) or spectinomycin (50 μg/ml) (Sigma).
Second, at 1 day of age, groups of 20 chickens were inoculated orally with 0.1 ml of an overnight LB broth culture of cecal contents obtained from healthy, adult SPF chickens to prevent the development of systemic disease. They were then given access to feed, as described above. Twenty-four hours later, the chickens were infected orally with 108 CFU of either a spontaneous Nalr mutant of S. Typhimurium F98 or a Nalr mutant with a single or double insertion mutation in selected genes in 0.1 ml of LB broth. At 1, 2, and 3 weeks after inoculation, cloacal swabs were taken from each bird and plated in a standard manner (6) on brilliant green agar containing sodium nalidixate (20 μg/ml) and novobiocin (1 μg/ml) to obtain a semiquantitative enumeration of the bacteria excreted.
Virulence assays.
Selected mutants of S. Typhimurium F98 were tested for their virulence for newly hatched Rhode Island Red chickens. The mutations were transferred by P22 transduction (5) to strain 4/74, which is virulent for mice (SL1344) (83), for assessment of virulence in BALB/c mice. Virulence was assessed by oral inoculation of groups of 20 newly hatched chickens with 0.1 ml or of 10 BALB/c mice with 50 μl of a broth culture diluted to contain 106 CFU in this volume. Morbidity and mortality were recorded over a 3-week period. Signs in chickens included anorexia and a disinclination to drink, standing with head and wings lowered, and caked feces around the vent. Mice became unsteady and had a “starry” coat. These signs are generally predictive of severe disease and death, and animals with signs of disease were killed humanely. Animals showing signs typical of salmonellosis were killed humanely, and their livers were cultured on MacConkey agar. Differences in mortality were analyzed by a χ2 test.
Microscopy of cecal contents.
Eight newly hatched chickens were inoculated orally with 0.1 ml of a 1/1,000 dilution of an overnight LB broth culture of S. Typhimurium within 8 h of hatching. Eighteen hours later, all birds were killed and cecal contents were harvested into universal bottles and stored at 4°C. They were diluted 1:100 in phosphate-buffered saline (PBS) and observed within 1 to 2 h by phase microscopy. The number of bacterial cells that showed evidence of division, expressed as a proportion of the total, was counted for each sample. Bacteria which were attached or had a visible septum were regarded as in the process of division. Motility and general cell shape were also observed.
Microarray data accession numbers.
Raw data have been deposited in GEO (http://www.ncbi.nlm.nih.gov/pubmed/11752295), platform GPL6439 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL6439), and series GSE10337 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10337).
RESULTS
Transcription profile from within the cecal contents.
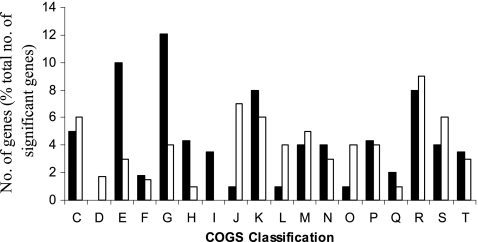
RNA extracted from S. Typhimurium bacteria from the luminal contents of the ceca of day-old chicks was compared to that from the in vitro cultures. The genes were grouped by clusters of orthologous groups of proteins (COGs) classification and are shown in Fig. 1. This overarching classification indicated major changes resulting from adaptation to the cecal environment. Overall, 17% of the 4,457 S. Typhimurium coding sequences (CDS) present on the array showed changes in expression during infection. Of these, 282 CDS were upregulated more than 2-fold, including genes associated with amino acid, carbohydrate, coenzyme, and lipid transport. A total of 464 CDS were downregulated more than 2-fold, including genes associated with cell cycle regulation, translation, and DNA replication. Total RNA was extracted from five noninfected birds to determine if the cecal contents alone produced a cross-reaction with the array; no cross-reaction was detected (data not shown).
Fig. 1.
Comparison of S. Typhimurium genes expressed in the lumens of newly hatched chicks with those expressed in in vitro-grown bacteria, classified according to COGs. Black bars, cecal contents; white bars, in vitro. The classified genes were found to be significantly different, with a >2-fold change in expression and a P value of less than 0.05. COGs classification abbreviations: C, energy production and conversion; D, cell cycle control, mitosis, and meiosis; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms.
Genes which showed statistically significant differential expression between in vivo and in vitro conditions (2-fold change, P < 0.05) were considered to be of interest. The genes with increased and decreased levels of expression which fulfilled this criterion are listed in Tables 2 and 3, respectively.
Table 2.
S. Typhimurium genes of interest which were upregulated during colonization of the cecal lumen, compared to gene expression in broth culturesa
| COGs class | Locus tag | Gene | Function or product | Change in expression level (fold) | P value |
|---|---|---|---|---|---|
| Not in COGs | STM1144 | csgA | Major curlin subunit precursor | 3.9 | 0.002 |
| STM1601 | ugtL | Putative exported protein | 2.9 | 0.02 | |
| STM1602 | sifB | Secreted effector | 3.4 | 0.01 | |
| STM1384 | ttrC | Tetrathionate reductase complex subunit C | 2.3 | 0.03 | |
| STM0550 | fimY | Putative regulatory protein | 3.1 | 0.01 | |
| STM1143 | csgB | Minor curlin subunit precursor | 4.2 | 0.04 | |
| STM3758 | fidL | Putative inner protein | 2.2 | 0.04 | |
| Amino acid transport and metabolism | STM0878 | potG | Putrescine transporter | 5.6 | 0.03 |
| STM2992 | argA | N-α-acetylglutamate synthase | 4.6 | 0.03 | |
| STM4296 | adi | Catabolic arginine decarboxylase | 4.4 | 0.047 | |
| STM1094 | pipD | Pathogenicity island-encoded protein D | 4.1 | 0.05 | |
| STM4105 | metF | 5,10-Methylenetetrahydrofolate reductase | 3.6 | 0.03 | |
| STM3965 | metE | 5-Methyltetrahydropteriryltriglutamate-homocystein S-methyltransferase | 2.2 | 0.04 | |
| STM3086 | speA | Arginine decarboxylase | 3.6 | 0.02 | |
| STM0887 | artJ | Arginine transport system component | 2.4 | 0.02 | |
| STM2055 | pduU | Polyhedral body protein | 3.7 | 0.05 | |
| STM2056 | pduV | Propanediol utilization protein | 2.8 | 0.01 | |
| STM2469 | eutP | Putative ethanolamine utilization protein | 3.5 | 0.0006 | |
| STM2468 | eutQ | Putative ethanolamine utilization protein | 3.0 | 0.007 | |
| STM0877 | potF | Putrescine transporter | 2.5 | 0.05 | |
| STM0878 | potG | Putrescine transporter | 5.6 | 0.03 | |
| Carbohydrate transport and metabolism | STM1928 | otsA | Trehalose-6-phosphate synthase | 4.3 | 0.03 |
| STM4298 | melA | α-Galactosidase | 3.2 | 0.04 | |
| STM3674 | lyxK | l-Xylulose kinase | 2.0 | 0.047 | |
| STM3675 | sgbH | Putative 3-hexulose-6-phosphate isomerase | 3.1 | 0.02 | |
| STM0018 | Putative exochitinase | 3.0 | 0.014 | ||
| STM1560 | Putative α-amylase | 2.9 | 0.008 | ||
| STM3254 | Putative fructose-1-phosphate kinase | 2.7 | 0.009 | ||
| STM3671 | Putative transporter | 3.0 | 0.02 | ||
| Energy production and conversion | STM0369 | prpC | Putative citrate synthase | 6.0 | 0.004 |
| STM1383 | ttrA | Tetrathionate reductase complex subunit A | 4.6 | 0.03 | |
| STM2057 | pduW | Propionate kinase | 3.4 | 0.02 | |
| General function | STM0370 | prpD | 2-Methylcitrate dehydratase | 7.1 | 0.001 |
| Inorganic ion transport | STM2862 | sitB | Putative ATP-binding protein | 2.8 | 0.04 |
| STM0206 | butF | Putative periplasmic cobalamin-binding protein | 2.6 | 0.01 | |
| STM2863 | sitC | Putative permease | 2.3 | 0.05 | |
| Lipid | STM0371 | prpE | Putative acetyl-CoA synthetase | 3.5 | 0.003 |
| Motility | STM0339 | stbB | Putative fimbrial chaperone | 2.8 | 0.01 |
| STM0195 | stfA | Putative fimbrial subunit | 3.4 | 0.02 | |
| STM4593 | sthB | Putative fimbrial usher protein | 2.6 | 0.05 | |
| STM0198 | stfE | Putative minor fimbrial subunit | 3.1 | 0.02 | |
| STM0199 | stfF | Putative minor fimbrial subunit | 3.7 | 0.03 | |
| STM0200 | stfG | Putative minor fimbrial subunit | 3.5 | 0.02 | |
| Replication | STM2150 | stcC | Putative outer membrane protein | 2.3 | 0.04 |
| STM0395 | sbcC | ATP-dependent dsDNA exonuclease | 2.9 | 0.01 | |
| STM1992 | dcm | DNA cytosine methylase | 3.4 | 0.01 | |
| STM2996 | recC | Exonuclease V subunit | 2.3 | 0.02 | |
| Secondary metabolites biosynthesis, transport and catabolism | STM2046 | pduK | Polyhedral body protein | 4.1 | 0.03 |
| STM2054 | pduT | Polyhedral body protein | 2.6 | 0.04 | |
| STM2047 | pduL | Propanediol utilization protein | 3.5 | 0.03 | |
| STM2465 | eutM | Putative detox protein | 2.4 | 0.02 | |
| STM2464 | eutN | Putative detox protein | 2.4 | 0.02 | |
| Transcription | STM3964 | metR | metE/metH regulator | 2.2 | 0.03 |
| STM3756 | rmbA | Putative cytoplsmic protein | 3.1 | 0.04 | |
| STM0552 | fimW | Putative fimbrial protein | 3.0 | 0.02 | |
| Translation | STM1909 | argS | Arginine tRNA synthetase | 2.0 | 0.04 |
| Function unknown | STM1088 | pipB | Secreted effector protein | 5.5 | 0.01 |
| STM3764 | mgtC | Mg2+ transport protein | 2.5 | 0.05 | |
| STM0884 | Putative inner membrane protein | 4.8 | 0.04 |
Genes selected as genes of interest showed a >2-fold increase in expression levels and a P value of <0.05.
Table 3.
S. Typhimurium genes of interest which were downregulated during colonization of the cecal lumen, compared with expression in broth culturesa
| COGs class | Locus tag | Gene | Function or product | Change in expression level (fold) | P value |
|---|---|---|---|---|---|
| Not in COGs | STM2770 | fljA | Phase 1 flagellin repressor | 2.5 | 0.03 |
| STM2304 | pmrD | Polymyxin resistance protein B | 6.02 | 0.005 | |
| Amino acid transport and metabolism | STM | dsdA | d-Serine deaminase | 6.65 | 0.0036 |
| STM3244 | tdcB | Threonine dehydratase | 4.9 | 0.01 | |
| STM3240 | tdcG | l-Serine deaminase | 2.6 | 0.03 | |
| Carbohydrate transport and metabolism | STM2433 | crr | Glucose-specific IIA component | 10.78 | 0.026 |
| STM4231 | lamB | Maltoporin precursor | 4.4 | 0.04 | |
| STM2190 | mglB | Galactose transport protein | 5.6 | 0.001 | |
| STM0684 | nagB | Glucosamine-6-phosphate deaminase | 2.5 | 0.05 | |
| STM0685 | nagE | N-Acetylglucosamine-specific enzyme IIABC | 2.1 | 0.04 | |
| STM2431 | ptsH | Phosphohistidinoprotein-hexose phosphotransferase | 9.00 | 0.014 | |
| Cell cycle | STM3569 | ftsX | Putative cell division protein | 3.3 | 0.008 |
| STN3570 | ftsE | Putative cell division ATPase | 3.3 | 0.03 | |
| STM0960 | ftsK | Cell division protein | 3.5 | 0.02 | |
| Energy production and conversion | STM2320 | nuoJ | NADH dehydrogenase I chain J | 2.3 | 0.03 |
| STM2321 | nuoI | NADH dehydrogenase I chain I | 2.4 | 0.03 | |
| STM2255 | napC | Periplasmic nitrate reductase | 3.0 | 0.02 | |
| STM0440 | cyoD | Cytochrome o ubiquinol oxidase subunit IV | 3.4 | 0.04 | |
| STM4340 | frdD | Fumarate reductase membrane anchor polypeptide | 3.6 | 0.05 | |
| STM2325 | nuoE | NADH dehydrogenase I chain E | 4.1 | 0.03 | |
| STM2324 | nuoF | NADH dehydrogenase I chain F | 3.5 | 0.05 | |
| STM0441 | cyoC | Cytochrome o ubiquinol oxidase subunit III | 6.96 | 0.013 | |
| STM0740 | cydA | Cytochrome d terminal oxidase polypeptide subunit I | 6.33 | 0.0062 | |
| General function | STM4361 | hfq | Host factor I | 10.08 | 0.0005 |
| STM1751 | hns | DNA-binding protein HLP-II | 8.73 | 0.0033 | |
| Cell motility | STM1959 | fliC | Flagellin | 14.89 | 0.00055 |
| STM1171 | flgN | Putative FlgK/FlgL export chaperone | 7.79 | 0.007 | |
| STM1920 | cheW | Chemotaxis docking protein | 7.26 | 0.0027 | |
| STM1183 | flgK | Flagellar hook-associated protein 1 | 2.3 | 0.02 | |
| STM4533 | tsr | Methyl-accepting chemotaxis protein | 3.9 | 0.03 | |
| STM1915 | cheZ | Chemotactic response protein | 4.0 | 0.02 | |
| STM2771 | fljB | Phase 2 flagellin | 4.0 | 0.01 | |
| STM1921 | cheA | Chemotaxis sensory histidine protein kinase | 4.0 | 0.02 | |
| STM1174 | flgB | Flagellar basal body rod protein | 4.2 | 0.05 | |
| STM3577 | tcp | Methyl-accepting transmembrane citrate/phenol | 5.2 | 0.002 | |
| chemoreceptor | |||||
| Replication, recombination, and repair | STM1339 | himA | Integration host factor alpha subunit | 3.9 | 0.013 |
| STM3185 | yqiE | ADP-ribose pyrophosphatase | 3.6 | 0.007 | |
| STM0484 | dnaX | DNA polymerase III tau/gamma subunits | 3.1 | 0.01 | |
| STM4170 | hupA | DNA-binding protein HU-alpha | 6.36 | 0.00079 | |
| Signal transduction mechanisms | STM1916 | cheY | Chemotaxis regulator | 6.62 | 0.01 |
| Transcription | STM0900 | Putative helicase | 12.5 | 0.002 | |
| STM2875 | hilD | Invasion protein regulatory protein | 10.3 | 0.008 | |
| STM1172 | flgM | Anti-FliA factor | 7.71 | 0.011 | |
| STM2867 | hilC | Invasion regulatory protein | 5.89 | 0.014 | |
| STM3245 | tdcA | Transcriptional activator | 3.0 | 0.005 | |
| STM1956 | fliA | Sigma 28 | 3.0 | 0.025 | |
| Translation | STM3728 | rpmB | 50S ribosomal subunit protein L28 | 5.92 | 0.02 |
| STM3440 | rplC | 50S ribosomal subunit protein L3 | 2.1 | 0.04 | |
| STM3437 | rplB | 50S ribosomal subunit protein L2 | 2.22 | 0.02 | |
| STM3425 | rplF | 50S ribosomal subunit protein L6 | 2.6 | 0.02 | |
| STM3414 | rplQ | 50S ribosomal subunit protein L17 | 3.3 | 0.01 | |
| STM3438 | rplW | 50S ribosomal subunit protein L23 | 2.9 | 0.01 | |
| STM3430 | rplN | 50S ribosomal subunit protein L14 | 3.2 | 0.008 | |
| STM3422 | rplP | 50S ribosomal subunit protein L16 | 3.3 | 0.01 | |
| STM4393 | rpsR | 30S ribosomal subunit protein S18 | 2.2 | 0.03 | |
| STM3441 | rpsJ | 30S ribosomal subunit protein S10 | 2.5 | 0.02 | |
| STM0981 | rpsA | 30S ribosomal subunit protein S1 | 3.2 | 0.03 | |
| STM3447 | rpsG | 30S ribosomal subunit protein S7 | 3.6 | 0.03 | |
| STM3448 | rpsL | 30S ribosomal subunit protein S12 | 3.7 | 0.045 | |
| STM3436 | rpsS | 30S ribosomal subunit protein S19 | 3.9 | 0.012 | |
| STM3419 | rpmJ | 50S ribosomal subunit protein X | 3.6 | 0.02 | |
| STM1335 | rpmI | 50S ribosomal subunit protein L35 | 3.9 | 0.01 | |
| Translocation | STM3321 | yhbH | Putative sigma N modulation factor | 16.11 | 0.0049 |
| Function unknown | STM2390 | yfcZ | Putative cytoplasmic protein | 9.52 | 0.0051 |
| STM3995 | yihD | Putative cytoplasmic protein | 6.99 | 0.00041 | |
| STM2697 | Phage tail-like protein | 6.87 | 0.003 | ||
| STM4088 | yiiU | Putative cytoplasmic protein | 6.26 | 0.0017 |
Genes selected as genes of interest showed a >2-fold change in expression levels and a P value of <0.05.
Compared with in vitro-grown luminal bacteria, significant changes were observed in genes associated with the following factors. (i) Relating to cell division, 12 genes associated with recombination, gene regulation, transcription, and chromosome replication, including hupA, himA, ygiE, and dnaX, were downregulated in vivo, compared to in vitro-grown bacteria. In addition, seven genes involved in cell division (including ftsEKX) were downregulated. There was a significant reduction in expression of 32 genes associated with translation, including rplB to rplW, rpsAGJSP, and rpmBIJ, following analysis of gene expression within the lumen of the cecum. Genes associated with DNA repair (including dcm, encoding DNA cytosine methylase, recC, and sbcC) were upregulated.
(ii) Regarding energy sources, the prpBCDE locus, but not prpR, its regulator, was significantly upregulated in the lumen. A number of genes in the pdu operon were upregulated, particularly the latter part, pduK-pduV. However, there was no associated upregulation of the cob or cbi genes. The btuF gene was found to be expressed, indicating utilization of an external source of cobalamin. Increased expression of genes in the eut (ethanolamine degradation) operon (eutPQTDMN) was detected. The low redox environment of the lumen is indicated by the significant upregulation of ttrABC, although phs and asr gene products were not significantly upregulated. Other genes associated with respiration with oxygen as the terminal electron acceptor, including cydA, cyoCD, nuoEFIJ, frd, and napC, were downregulated.
(iii) Regarding carbohydrates, a number of different loci involved in the utilization of carbohydrates showed different levels of up- and downregulation. Expression of melA was significantly upregulated in the lumen, although the changes in expression of melB and melR were not statistically significant. Four of the 11 genes (yiaM, yiaN, lyxK, sgbH) required for the catabolism of l-ascorbate to d-xylulose were upregulated. The gene encoding trehalose phosphate synthase, otsA, was also upregulated in the lumen, as were some unidentified genes, including STM0018, STM1560, and STM3254, classified as having a role in carbohydrate utilization. Interestingly, there was significant downregulation in glucose utilization genes, including crr and ptsH and genes involved in N-acetylglucosamine utilization, such as nagBE. The genes lamB and mglB were also downregulated.
(iv) With respect to amino acid utilization, there was a significant level of upregulation of expression of metE, metF, and metR, and upregulation of adiA, speA, argA, and argS indicated that arginine was being utilized by bacteria in the lumen. Interestingly, there was a significant upregulation in the expression of the potFGHI operon (putrescine transport) within the lumen. tdcAB, the transcriptional activator, and tdcB, involved in threonine utilization, were downregulated. In addition, dsdA and tdcG, involved in serine utilization, were also downregulated.
(v) For the bacterial surface, the majority of genes involved in flagellum production were downregulated in the lumen, including flgM, flgN, flgK, flgB, fliC, fljB, and fliA. There was also significant downregulation of chemotaxis genes cheAWZ, tcp, and tsr. Several fimbrial genes were significantly upregulated, including stfAEFG, stbB, stjB, stcC, sthB, csgA, and csgB. Parts of the fim operon, fimY and fimW, were also upregulated.
(vi) With respect to virulence factors, a small number of genes from Salmonella pathogenicity island 1 (SPI-1) were significantly upregulated, including sitBC, sipD, and spaS. The sit genes encode a part of an ABC transport system for the uptake of iron into the periplasmic space, indicating a potential function in colonization (94, 101). Two genes from SPI-1, hilC and hilD, were significantly downregulated in expression. Both of these genes are involved in the transfer of environmental signals to the central virulence gene regulator, HilA (23). This result is surprising, given the predicted effect of downregulating HilA. No significant change was observed for hilA expression, and a number of genes in SPI-1 were upregulated. This strongly suggests that in the lumen of the gut, a number of different factors are acting on the regulation of HilA. Little change in expression was detected within SPI-2 and SPI-4. Within SPI-3, mgtC, rmbA, and fidL and the colonization-associated genes shdA and misL, and in SPI-5, pipB, were found to be upregulated in the lumen. While a role for mgtC has been described for growth in low-magnesium environments (58), the requirement for pipB, rmbA, and fidL expression may represent redundant gene expression from the same island. There also seems no obvious reason for pipB expression to be required, as it is involved in intracellular kinesin binding (37).
Validation by quantitative RT-PCR.
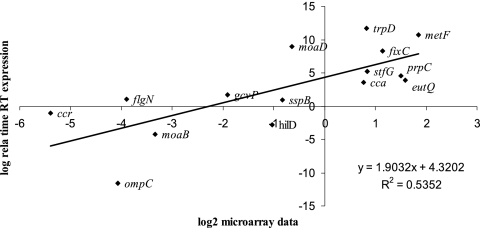
To validate the microarray results, RT-PCR was carried out on 15 selected genes showing different levels of expression within the lumen. The data for the 15 genes (Fig. 2) gave an r2 value of 0.53, which was a good fit (P = 0.0019). The slope (2.27) indicated higher values by RT-PCR than by microarray.
Fig. 2.
Correlation between microarray and real-time RT-PCR expression values. Log2-transformed expression values for 15 genes from total bacterial RNA extracted from day-old-chick cecal contents in triplicate. The best-fit linear regression line is shown together with the r2 value and the calculated slope equation.
Transcription profile from the mucosal wall.
Patterns of gene expression in RNA extracted from S. Typhimurium bacteria from the washings of the cecal mucosa were compared to the data arising from the RNA harvested from the cecal lumen of day-old chicks. The genes were grouped by COGs classification and are shown in Fig. 3. A total of 33 genes were significantly (change of 2-fold, P < 0.005) upregulated at the mucosa, and 16 genes were significantly downregulated (Tables 4 and) 5.
Fig. 3.
Comparison of S. Typhimurium genes expressed at the mucosal wall with those expressed in the lumens of newly hatched chicks, classified according to COGs. Black bars, lumen; gray bars, mucosal wall. The classified genes were found to be significantly different, with a >2-fold change in expression and a P value of less than 0.05. COGs classification abbreviations: C, energy production and conversion; D, cell cycle control, mitosis, and meiosis; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms.
Table 4.
S. Typhimurium genes upregulated at the mucosal walla
| COGs class | Locus tag | Gene | Function or product | Change in expression level (fold) | P value |
|---|---|---|---|---|---|
| Amino acid transport and metabolism | STM3592 | yhiP | Putative peptide transport protein | 2.08 | 0.049 |
| Carbohydrate transport and metabolism | STM0310 | gmhA | Phosphoheptose isomerase | 2.06 | 0.037 |
| STM1558 | Putative glycosyl hydrolase | 2.13 | 0.017 | ||
| STM2431 | ptsH | Phosphohistidinoprotein-hexose phosphotransferase | 2.29 | 0.017 | |
| STM2433 | crr | Glucose-specific IIA component | 2.4 | 0.036 | |
| Cell cycle control, mitosis, and meiosis | STM0132 | ftsA | ATP-binding cell division protein | 2.37 | 0.019 |
| STM3371 | yhdE | Putative inhibitor of septum formation | 2.26 | 0.0198 | |
| STM3374 | mreB | Rod shape-determining protein | 2.13 | 0.014 | |
| Cell motility | STM1177 | flgE | Flagellar hook protein | 2.64 | 0.03 |
| Cell wall/membrane biogenesis | STM0124 | murF | d-Alanine-d-alanine ligase | 2.09 | 0.044 |
| Energy production and conversion | STM4439 | cybC | Cytochrome b562 | 2.06 | 0.03 |
| Function unknown | STM0119 | yabB | Putative cytoplasmic protein | 2.28 | 0.042 |
| STM1088 | pipB | Secreted effector protein | 2.35 | 0.016 | |
| STM3347 | yhcB | Putative periplasmic protein | 2.42 | 0.037 | |
| General function prediction only | STM1581 | yddE | Putative phenazine biosynthetic protein | 2.02 | 0.045 |
| STM2580 | era | GTPase | 2.09 | 0.028 | |
| STM2969 | ygdH | Putative nucleotide binding | 3.18 | 0.012 | |
| Inorganic ion transport and metabolism | STM4324 | cutA | Putative periplasmic divalent cation tolerance protein | 2.3 | 0.02 |
| Not in COGs | STM0471 | ylaC | Putative inner membrane protein | 2.06 | 0.003 |
| STM1059 | ycbW | Putative cytoplasmic protein | 2.71 | 0.03 | |
| STM1092 | Putative cytoplasmic protein | 2.32 | 0.024 | ||
| STM1601 | ugtL | Putative membrane protein | 2.1 | 0.04 | |
| STM2983 | orfX | Putative lipoprotein | 2.09 | 0.01 | |
| Nucleotide transport and metabolism | STM1163 | pyrC | Dihydroorotase | 2.09 | 0.013 |
| Posttranslational modification, protein turnover, and chaperones | STM1682 | tpx | Thiol peroxidase | 2.79 | 0.038 |
| STM3342 | sspA | Stringent starvation protein A | 2.23 | 0.041 | |
| Transcription | STM1704 | Putative regulatory protein | 2.73 | 0.022 | |
| STM3320 | rpoN | Sigma 54 | 2.09 | 0.022 | |
| STM3515 | yciT | Transcriptional activator | 2 | 0.026 | |
| STM4318 | Putative acetyltransferase | 2.198 | 0.019 | ||
| Translation | STM3418 | rpsM | 30S ribosomal subunit protein S13 | 2.73 | 0.026 |
Genes selected as genes of interest showed a >2-fold increase and a P value of <0.05.
Table 5.
S. Typhimurium genes downregulated at the mucosal walla
| COGs class | Locus tag | Gene name | Function or product | Change in level of expression (fold) | P value |
|---|---|---|---|---|---|
| Amino acid transport and metabolism | STM0067 | carB | Carbamoyl-phosphate synthase large subunit; putative ABC transporter | 2.9 | 0.035 |
| STM1255 | Periplasmic binding protein | 2.49 | 0.044 | ||
| STM2055 | pduU | Polyhedral body protein | 2.097 | 0.031 | |
| STM1257 | Putative ABC transporter protein | 2.03 | 0.006 | ||
| STM3594 | prlC | Oligopeptidase A | 2.09 | 0.0496 | |
| Carbohydrate transport and metabolism | STM3784 | Putative phosphotransferase system mannitol/fructose-specific IIA domain | 2.32 | 0.0199 | |
| Cell wall/membrane biogenesis | STM2120 | asmA | Suppressor of OmpF assembly mutants | 2.12 | 0.049 |
| Energy production and conversion | STM2057 | pduW | Propionate kinase | 2.1 | 0.006 |
| PSLT027 | ccdA | Antidote | 2.14 | 0.045 | |
| STM0813 | ybhP | Putative cytoplasmic protein | 2.09 | 0.025 | |
| STM2007 | Tetratricopeptide repeat protein | 2.33 | 0.034 | ||
| Not in COGs | STM0903 | Putative chaperone | 2.79 | 0.003 | |
| STM2041 | pduD | Propanediol dehydratase medium subunit | 2.58 | 0.04 | |
| STM3688 | Putative cytoplasmic protein | 2.16 | 0.0499 | ||
| Replication, recombination, and repair | STM4168 | nfi | Endonuclease V | 2.22 | 0.011 |
| Transcription | STM3773 | Putative transcriptional regulator | 2.07 | 0.019 | |
| Translation | STM2665 | yfiA | Ribosome stabilization factor | 2.75 | 0.01 |
Genes selected as genes of interest showed a >2-fold change in expression levels and a P value of <0.05.
Potentially significant changes in the mucosa, compared with luminal bacteria, were observed in genes associated with the following factors. (i) Relating to carbohydrate transport and metabolism, genes associated with glucose utilization were significantly upregulated at the mucosa, including gmhA, ptsH, and crr. A phosphotransferase suppressor of ompF was downregulated at the mucosal wall.
(ii) Regarding amino acid transport and metabolism, only one gene, yhiP, encoding a putative peptide transport protein, was significantly upregulated at the mucosa. However, three genes were significantly downregulated, including two encoding ABC transporter proteins and carB.
(iii) With respect to energy production and conversion, the cytochrome b562 gene, cybC, was significantly upregulated at the mucosal wall. Four genes, pduDUW and ybhP, plus a gene coding for a tetratricopeptide repeat protein were significantly downregulated.
(iv) With respect to cell division and transcription, three genes associated with cell division were upregulated, namely, ftsA, yhdE, and mreB. Four genes associated with transcription were also upregulated (including rpoN and yciT).
(v) Regarding translation and posttranslational modification, two genes, tpx and sspA, were significantly upregulated at the mucosal wall, as was rpsM, which encodes a 30S ribosomal subunit protein. However, only one gene, the ribosome stabilization factor gene yfiA, was downregulated significantly.
(vi) Interestingly, pipB was upregulated at the mucosal wall. ugtL, which provides resistance to antimicrobial peptides, was also upregulated.
It is appreciated that the number of genes showing changes in expression at the mucosa is very small compared with that of the bacteria in the lumen and that the overall patterns of expression in the two populations were almost identical.
Microscopy of bacteria from the mucosa.
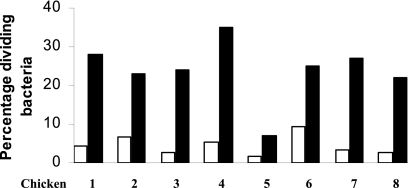
Phase-contrast microscopy of the cecal lumen and cecal mucosa from the above-described samples was used to estimate the numbers of dividing bacteria in these two sites in the cecum. Samples taken from eight chickens indicated that the percentage of dividing bacteria was higher at locations close to the mucosal wall than within the lumen (Fig. 4). There was no difference in bacterial cell size. No bacterial cells from the lumen showed evidence of motility (directional movement) in 10 microscopic fields, supporting the results of the gene expression studies above.
Fig. 4.
Percentage of bacteria showing evidence of cell division out of the total number of bacteria observed by phase microscopy from the cecal lumens (white bars) or cecal mucosae (black bars) of 8 chickens infected orally with S. Typhimurium when less than 24 h old and killed 24 h later.
Colonization of chickens.
Assessing the contribution to intestinal colonization of genes which were upregulated in the intestine was difficult in newly hatched chickens, since even serovars such as Salmonella enterica serovar Choleraesuis, which is unable to colonize the alimentary tract of adult birds, are nevertheless able to multiply in the guts of newly hatched chickens. We therefore decided to use a competition assay in which selected mutants are assessed for their ability to exclude a superinfecting parent strain inoculated 24 h later (100). This method is preferred to an assay in which both strains are inoculated simultaneously, because it allows an assessment of whether the mutant is utilizing the same nutrients under stationary-phase redox conditions as the parent strain which will compete with it. Our experience is that mutants which are sometimes noninhibitory in our assay are nevertheless frequently able to grow to equally high numbers as the parent strain when inoculated simultaneously (P. Barrow, M. Lovell, and M. A. Jones, unpublished results). Mutants were selected because the genes were relatively highly upregulated (metF, csgA, argA, and potG) or because they were linked metabolically (ttrS, ttrB, pduA, and eut). At the time of challenge, all mutants tested (metF, csgA, ttrS, ttrB, pduA, eut, argA, and potG) colonized the gut well, judging from the counts in the ceca of three birds killed at the time of challenge (Table 6). When the birds were killed 3 days after challenge, most mutants were still colonizing well, with the mean cecal count ranging from 7.03 to 7.71 log g−1. Only the argA, pduA, ttrS, and potG mutants were found in low numbers. Despite this, all the mutants tested were able to exclude the parental challenge strains, with 6 of the 7 birds killed having challenge counts of <2 log, whereas the mean count of the challenge strain in birds which had not been previously inoculated with another strain was 5.11 (range, 4 to 7.20).
Table 6.
Viable counts of test (Nalr) and challenge (Spc) mutants of S. Typhimurium F98 in the ceca of newly hatched chickens in a competition assaya
| Mutation | Viable count ofb: |
||
|---|---|---|---|
| Test strain |
Challenge strain postmortem | ||
| At time of challenge | Postmortem | ||
| metF | 8.32, 8.42, 8.59 | 7.17 (6.60–7.82) | <2 (<2–<2) |
| csgA | 8.40, 7.64, 8.04 | 7.79 (7.18–8.18) | <2 (<2–<2) |
| ttrS | 7.73, 6.30, 7.08 | 6.95 (6.00–7.79) | <2 (<2–2.85) |
| ttrB | 7.93, 8.43, 9.11 | 7.74 (7.00–8.00) | <2 (<2–<2) |
| pdu | 8.08, 8.04, 8.00 | 6.60 (6.00–7.65) | <2 (<2–5.2) |
| eut | 8.2, 8.88, 9.00 | 7.59 (7.18–7.68) | <2 (<2–<2) |
| argA | 7.11, 7.88, 7.28 | 6.00 (6.00–6.90) | <2 (<2–2.78) |
| potG | 7.90, 7.83 | 6.95 (6.70–7.28) | <2 (<2–<2) |
| Parent | 8.80, 8.18, 8.28 | 7.00 (6.60–8.57) | <2 (<2–<2) |
| None | <2, <2, <2 | <2 (<2–<2) | 4.3 (4.00–7.82) |
Ten chickens were inoculated with the test strain. Three chickens were killed to enumerate this strain 24 h later at the time of challenge. All chickens were killed 3 days later to enumerate both strains in the ceca.
Viable counts at time of challenge are presented for all three chickens. Viable counts postmortem of all other chickens are the means and ranges.
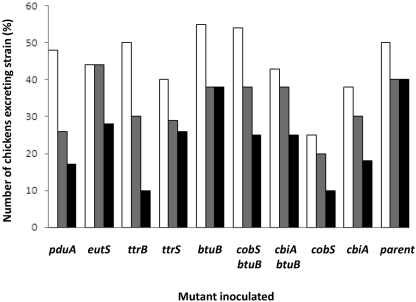
Because pdu, ttr, and btu genes were all upregulated in the intestine, and because these genes are all related to the anaerobic catabolism of 1,2-propanediol and ethanolamine, mutants with inactivated ttr, pdu, eut, or btu genes, and also cob and cbi operons, were tested for their ability to colonize the guts of 1-day-old chickens which had received gut flora preparations. The patterns of fecal excretion are shown in Fig. 5. The greatest reductions in fecal excretion from that of the parent strain were seen with the pduA, ttrB, and cbiA genes and the cobS btuB double mutant. Statistical significance was assessed using the χ2 test. Statistically significant reductions in colonization were observed only with the ttrB mutant (P < 0.01). Additional reductions which were less significant were observed with pduA (P = 0.03) and cobS (P = 0.1) mutants.
Fig. 5.
Numbers of chickens, expressed as a percentage, which were excreting the inoculated mutant of a nalidixic acid-resistant derivative of S. Typhimurium F98 at 1 week (white bars), 2 weeks (gray bars), and 3 weeks (black bars) after oral inoculation of chickens possessing gut floras.
None of these mutations produced any significant attenuation in the virulence of S. Typhimurium for mice or newly hatched chickens. Signs of severe systemic disease were observed in 8 to 10 of the 10 inoculated mice and in 15 to 20 of the 20 inoculated chickens, regardless of whether the strain was the virulent parent or a mutant strain (P = 0.25). Pure, heavy growth of Salmonella was obtained by culturing the livers of animals which were killed humanely.
DISCUSSION
The results here demonstrate that extensive transcriptional changes occur following infection of day-old chicks with S. Typhimurium, with many genes being downregulated in expression, indicating decreased metabolic activity from that of the broth culture. Those genes which were upregulated reflect a degree of adaptation to the luminal environment.
To study gene expression in Salmonella during colonization of chickens, the most appropriate model is generally regarded to be animals that are 2 to 6 weeks old and that have established gut floras which would be more dominant numerically than the colonizing pathogen. The constraints imposed by studying gene expression by microarray meant that experiments had to be performed in newly hatched chickens to avoid false-positive signals from the presence of numerically dominant flora components, such as E. coli. This model reflects the situation that occurs during infection in newly hatched chickens which does take place within hatcheries. Despite the shortcomings of this approach, the patterns of expression were closer to our preconceptions than we imagined. Similarly, patterns of global gene transcription in Campylobacter jejuni in a similar model were found to resemble those in older birds with gut floras (92), and other similar models (e.g., a streptomycin-treated mouse) have been used with E. coli with success (15, 44, 45).
The requirement for a large number of chickens to generate sufficient RNA also meant that bacteria present in the ceca of different birds would also likely have been present at different stages of the growth cycle, depending on whether the ceca were full, had just emptied, or were freshly filled (P. Barrow, unpublished). This potential variation had implications for measuring the expression of genes associated with logarithmic versus stationary-phase growth, but it did not appear to have a profound effect, judging from the patterns of expression observed.
Although we measured luminal gene expression, we were aware that the ceca contained heterogeneous environments, as indicated by the differing rates of cell division in the lumen and close to the mucosa. This was supported by differences in expression levels in genes associated with cell division, transcription, and translation. What was perhaps more surprising was that so few other genes were affected. The metabolic and virulence profiles from the bacteria harvested from the lumen fairly well reflected those from the mucosa, where most of the cell multiplication was taking place. This was reassuring. The small number of changes in expression at the mucosa from that of the luminal contents suggested that the two populations were very similar, but it offers some insight into the lifestyle of the bacteria close to the mucosa. The involvement of genes (crr and gmhA) in the uptake and metabolism of glucose, galactose, or mannose suggests that sialic acid from host cell membranes would likely act as a potential carbon source for bacteria close to the mucosa.
The data presented were also validated by the similar changes in expression observed in selected genes tested by RT-PCR, as has been found by other authors (2, 92).
The reduced level of cell division within the lumen, indicated by microscopy, together with the reduced expression of genes associated with cell division, transcription, and translation, suggests a greatly reduced rate of metabolism and growth at this site. There is a direct dependence of transcription and translation rates and gene doses on bacterial growth rates (49, 53), in addition to the dependence on total RNA quantity and ribosomal proteins (35, 47). The relationship between cell growth rates and expression of genes associated with cell division is less clear, since a key gene associated with the formation of the Z ring, ftsZ, is expressed independently of growth rate (88). The ftsEKX genes interact and form part of the divisome. Although functionality of several fts genes is not required for cell growth, as indicated by continued filamentous growth in fts mutants, ftsE mutants do show reduced growth rates, which can be suppressed at high osmolarities (18). Despite the apparent low rate of cell division and the shortage of nutrients in the lumen, there was evidence that propionate, 1,2-propanediol, and ethanolamine acted as important carbon sources. This was less apparent at the mucosal wall, where the pduDUW genes were significantly downregulated and the main source of nutrients was unclear, although there was some evidence for use of glucose in this niche but not in the lumen (see below). Interestingly, expression of SspA, stringent starvation protein A, was upregulated at the mucosal wall. This protein in E. coli was found to be induced during stationary phase and starvation for carbon, amino acids, nitrogen, and phosphate (34). It is thought to act as a global regulator. Its expression suggests that the bacteria were experiencing conditions of starvation, and though most bacterial multiplication is occurring close to the mucosa, this itself is not an ideal or static environment and it indicates the complexity of the environmental niches in the gut.
Within the lumen, degradation of 1,2-propanediol appeared to be occurring, although this generally requires endogenous adenosyl cobalamin (coenzyme B12) biosynthesis. The pdu genes are contiguous and coregulated with cobalamin biosynthetic genes (cob and cbi) (49, 66). However, in the current experiments, there was no significant upregulation of the cob or cbi operons within the lumen. Some vitamin B12 is thought to be present in egg yolk (17) and would be present in the gut of newly hatched chickens, as the yolk sac is not fully resorbed for 3 to 4 days. This could be scavenged by BtuF (85), and BtuF in Salmonella is a periplasmic binding protein with a high affinity for vitamin B12 which was expressed within the lumen.
Genes involved in the catabolism of ethanolamine, which is derived in part from host cells and membranes, are encompassed in the eut operon (67, 75). The eutT gene encodes an adenosyltransferase, which is used to activate EutR, and in turn triggers transcription of the operon (68). Two of the genes, eutM and eutN, partially encode a metabolosome with the products of eutSLK, which encode the shell proteins of the metabolosome (74). The role of this structure was proposed to be to concentrate low levels of ethanolamine catabolic enzymes (10). The eutD gene encodes a phosphotransacetylase, which acts as a safety valve to minimize flux variations in a system which converts ethanolamine into acetyl coenzyme A (acetyl-CoA). The roles of eutP and eutQ remain unclear, though they were significantly upregulated in the cecal lumen.
Tetrathionate is one of the electron acceptors of choice for the utilization of ethanolamine and 1,2-propanediol (64) in the absence of oxygen. Other genes associated with respiration, including cydA, cyoCD, nuoEFIL, frd, and napC, were downregulated, suggesting that an anaerobic environment is present in the cecal lumen. This is in contrast to the findings of Jones et al. (44) showing that cytochrome bd oxidase was required for colonization of the streptomycin-treated mouse intestine by E. coli. These models are not strictly comparable, since the streptomycin-treated mouse will retain some gut flora, whereas there was virtually none in this series of experiments. In addition, we have found a degree of host specificity related to the likely route of respiration during intracellular Salmonella infection in chickens and mice (83) and the redox conditions implied therein. Tetrathionate is reduced to thiosulfide and further to H2S with the products of ttr, phs, and asr genes. It is likely that the tetrathionate results, in part, from material from the yolk sac, which is rich in sulfur. The role of sulfur-based electron acceptors in respiration in the gut has been shown recently by Winter et al. (90), who demonstrated that in mice with acute intestinal infection, reactive oxygen is released, which generates thiosulfate to be used as an electron acceptor. The model used here involved birds in which, at the time of harvesting, no inflammation was visible. It seems likely that during a more established infection when inflammation and gut damage will also occur, similar events are likely to take place.
Expression of ackA, encoding acetate kinase, which balances acetate and acetyl coenzyme A production, and an alternative phosphate donor acetyl phosphate, was upregulated 2-fold. A significant role of substrate-level phosphorylation in chickens is further supported by the poor colonization ability of ackA and pta mutants (P. Barrow and M. A. Lovell, unpublished findings).
The results from in vivo studies with mutations affecting the complex interactions between propanediol and ethanolamine as carbon sources, tetrathionate as the electron acceptor, and cobalamin as a cofactor were ambiguous, probably indicating the degree of redundancy in these nutrients as carbon sources. Thus, although the pduA mutant, like the other mutants, was fully inhibitory in the competition assay, it colonized the gut less well in these birds and also colonized the birds with the floras less well, albeit with a reduction of marginal significance. The eutS mutant colonized the gut well, again indicating the degree of redundancy in carbon source availability in this complex niche. Thus, although genes may be upregulated, indicating metabolic activity, their mutation will divert metabolic activity to other catabolic pathways. Both the ttrB and ttrS mutants colonized less well in this assay, with only the ttrB being significantly reduced. The picture is confused by the fact that the double mutants with a btuB mutation colonized well, whereas the single cobS and cbiA mutants colonized less well, although not significantly so. The interaction between propanediol utilization with tetrathionate and with cobalamin is highly complex, and much of the nature of these interactions in vivo remains to be determined.
The breakdown of propionate occurs via the 2-methylcitrate cycle using the prpBCDE locus (33), encoding the propionate-degrading enzymes and carrying prpR, a transcriptional regulator (38) which was previously thought to act as a sensor for 2-methylcitrate, an intermediate of the breakdown pathway (60, 61, 81). Although cobB expression was also thought to be required (79), there was no significant difference between expression in vivo and in vitro. The absence of cobB expression may be compensated for by expression of pduW, which encodes propionyl coenzyme A, a precursor of 2-methylcitrate, and which was upregulated 4-fold. The prpE mutant showed no reduction in colonization ability from that of the parent strain (tested in a different assay; results not presented). However, given the other energy sources available to Salmonella within the lumen, this was not unexpected.
d-Glucose is taken up and concomitantly phosphorylated either by the glucose-specific enzyme II (EII) transporter or by the phosphoenol-pyruvate-dependent transporter (97). The phosphoryl group is transferred to glucose through enzyme I (encoded by ptsI) and the phosphohistidine carrier protein (encoded by ptsH) to sugar-specific EII, which consists of two subunits, crr and ptsG. At the mucosal wall, glucose may be a more important carbon source, with upregulation of ptsH and crr, though expression of ptsI and ptsG was not significant. However, in the cecal lumen, we think that a number of other carbohydrates may also have been utilized, most significantly melibiose and l-ascorbate, suggesting, with the downregulation of crr and ptsH, that glucose was not an available source. The breakdown of melibiose utilizes two genes, melA (α-galactosidase) and melB (transporter), and their expression is stimulated by MelR (77). Expression of melA was significantly upregulated in the lumen, although expression of melB and melR was not statistically significant. With the high levels of expression of melA, it suggests either that this compound may already have been present in the cell or that the product of the melA gene was being used to break down a second carbohydrate source.
Four of the 11 genes required for the catabolism of l-ascorbate to d-xylulose, which enters the pentose phosphate pathway, were upregulated. Generation of internal trehalose also appears to occur in the lumen, with the upregulation of otsA. This would fit with a model where the bacteria in the lumen are growing slowly or are under stress, as the trehalose operon is induced under these conditions in an RpoS-dependent manner (76, 84). Trehalose has been demonstrated to play a role in cell protection against stressful environmental conditions, such as osmotic stress and heat shock, and was proposed to have a role in survival but not virulence (39).
Several other sources of carbohydrates were not utilized in the lumen, including maltose and galactose, as indicated by downregulation of lamB (30) and mglB, respectively. Again, this is in contrast to the findings of Jones et al. (45), which showed that maltose was important for E. coli colonization of the mouse intestine. These authors also found, in contrast to our previous findings, that glycogen was a significant carbon source (57). These results indicate the different responses in terms of gene expression and metabolism to colonizing different hosts, as recognized by Chang et al. (15) and as is found in those genes responsible for respiration during systemic Salmonella infection in chickens or mice (83).
Bacteria from the ceca demonstrated a requirement for methionine with significant levels of expression of metE, metF, with which metH forms the folate branch of the methionine pathway, and metR. The MetR protein acts as an activator for the transcription of metE, metA, metF, and metH (93). Homocysteine functions as a coregulator for MetR-mediated regulation and has a positive effect on the expression of metE, which encodes a transmethylase, and metF, which encodes 5,10-methylenetetrahydrofolate reductase but has a negative effect on metA and metH. The methylation of homocysteine, the final reaction prior to formation of methionine, is carried out via the vitamin B12-independent enzyme MetE (94). Genes involved in the utilization of other amino acids, including threonine (tdcB) and serine (dsdA and tdcG), were downregulated in the cecal lumen. The downregulation of tdcA, the transcriptional activator of the tdc operon, suggests that there is little requirement for threonine or serine within the lumen. There was also a significant downregulation of genes involved in the biosynthesis of glycine and one-carbon units (gcvH and gcvP), suggesting that these amino acids were not essential for growth and survival in the lumen.
The environment within the chick cecum was thought to be very weakly acidic, at pH 6.5 to 7 depending on diet, in addition to being anaerobic. Several mechanisms of survival of Salmonella under acidic conditions have been well documented (27, 29), although it is fairly certain that this pH would not induce a strong acid tolerance response. Three acid-resistant (AR) mechanisms have been identified in Escherichia coli, including AR1, which involves RpoS and cyclic AMP (cAMP) enabling cells to resist a pH as low as 2.5 (28). The AR3 system involves an arginine decarboxylase and has recently been identified in Salmonella and expressed under anaerobic conditions (48). S. Typhimurium DT104 was found to induce an arginine-dependent AR response involving transcriptional activation of adiA and adiC genes by adiY (48). In the present study, expression of adiA and speA was detected at high levels in vivo, suggesting that Salmonella was degrading arginine to agamatine. Upregulation of the transcriptional regulator adiY and of speB, which converts agmatine to putrescine, was not detectable in the ceca. However, Salmonella was actively generating arginine, as indicated by the upregulation of argA and argS, and scavenging arginine, as indicated by expression of artJ, which encodes a binding protein for arginine. Interestingly, Salmonella expressed significant levels of genes from the potFGHI operon, which encodes an ATPase-binding, putrescine-specific uptake system. Polyamines have been found to increase survival in extremely acidic and other inimical environments (96). Whether these data indicate low pH at the microenvironmental level or resistance to another factor inimical to metabolism in a gross environment where the pH is close to neutral remains to be determined. Mutation of argA did not alter colonization ability or survival in day-old chicks, although a role for speA in the colonization of 2-week-old chickens was suggested by Morgan et al. (59).
Mutation of potG did not alter colonization of day-old chicks. Similarly, Morgan et al. (59) found that mutation of potH did not reduce colonization ability. This suggests that the potFGHI operon was not functioning to transport putrescine into the cell but may have been playing an alternative role.
As the evidence suggested an environment where oxygen concentrations were very low, a 3-fold increase in dcm, associated with DNA repair, was unexpected. Heithoff et al. (36) reported previously that dam mutants were virulent in mice, but the role of cytosine methylation (dcm) was unclear. It is important in the regulation of biological processes in plants and animals, but the role of dam in the methylation of adenine is more important. However, the results here suggest that in chicks, dcm may contribute to the survival of Salmonella within that environmental niche. Given the probably low oxygen content of the cecal lumen, suggested by the downregulation of cydA, cyoCD, nuoEFIJ, and frd, the expression of recC was unexpected. The protein encoded by this gene functions to repair damage to DNA caused by host-synthesized compounds. Mutations in recA and recBC were found to be highly sensitive to oxidative compounds synthesized by macrophages and avirulent in mice (11). Similarly, the expression of sbcC was unexpected. The protein encoded by this gene acts to restore recombination and to resist DNA damage. It suggests that radical oxygen molecules, which could be damaging to the chromosome, may exist within the lumen. Interestingly, expression of tpx was detected at the mucosal wall, suggesting a gradient of oxygen across the cecum itself. Bacteria protect themselves from reactive oxygen species with a range of antioxidant defense enzymes, including thiol peroxidase. It was found that tpx acts as a lipid peroxidase to inhibit bacterial membrane oxidation and acts as a principle antioxidant for E. coli during anaerobic growth (14). It is possible that tpx may be functioning in a similar way here. Again, the recent work by Winter et al. (90) is relevant here, since it indicates that the release of reactive oxygen species into the gut results from inflammation. Although there was no indication of any gross inflammatory response here, the induction of proinflammatory cytokines by invading bacteria is a rapid event (46) and begins to be apparent by 16 to 24 h postinfection of newly hatched chickens (91). This process would undoubtedly have started in the gut of the chickens examined here.
Bacteria in the lumen displayed poor motility compared to in vitro-grown bacteria, as demonstrated by phase-contrast microscopy. The lack of motility is further supported by the downregulation of a number of genes involved in flagellar structure and function in the cecal lumen. The majority of the genes involved in the process were downregulated in the lumen, including two regulatory genes, flgM and flgN, which act to regulate gene expression. flgM acts as anti-sigma factor 28, which binds sigma factor 28 until the completion of the hook-basal body unit (2). flgN has two roles (1): it acts as a sensor for late gene expression in flagellar assembly by promoting expression of flgM translation, and it is associated with hook-associated proteins to inhibit its translation on flagellar completion. The first hook-filament junction protein, encoded by flgK, was downregulated, as was flgB, which forms part of the rod protein (95). Interestingly, fliC and fljB, which encode flagellin, were downregulated, as was fljA, which acts as a negative regulator for fliC expression (95). This suggests that no flagellin was produced in the chick lumen and, with the lack of expression of chemotaxis genes (cheAWZ, tcp, tsr), suggests that there is no major chemoattractant in the lumen which Salmonella bacteria would move toward. The downregulation in expression of tcp and tsr (41) suggests that neither citrate (tcp) nor serine (tsr) is present in the lumen. Stecher et al. (74) have shown that motility increases closer to the mucosa in the inflamed mouse intestine, although flagellation was less important in the noninflamed gut. We did not look at motility at the mucosa, but there would certainly not have been any gross inflammation during the short period of the experiments here.
Up to 13 different fimbrial operons have been suggested to be elaborated by Salmonella (40, 56). Some fimbrial genes are only expressed in particular environments (24). Within the chick lumen, several fimbrial genes were expressed, including stfAEFG, stbB, stjB, stcC, and sthB, suggesting that they may have a role in colonization or survival outside the host. The stf operon was found not to be essential for colonization by Clayton et al. (16). Morgan et al. (59) suggested that stbC and sthB contributed to colonization of older chickens. Genes required for biosynthesis of thin, curled fimbriae (csgB and csgA) were upregulated in the lumen as in macrophages (24). These are thought to have a role in adhesion, becoming associated with extracellular matrix, and are known to have a role in pathogenesis in E. coli (31). They appeared to play little role in our in vivo model.
The fim operon, encoding type 1 fimbriae, was downregulated in the lumen due to the upregulation in the expression of two regulatory genes, fimY and fimW. The role of the fimY gene in S. Typhimurium remains unclear, though it is essential for fimbrial production and acts as a coactivator with fimZ (79). fimW acts as a negative regulator and interacts with fimZ-mediated activation of fimA expression (78).
Salmonella pathogenicity islands (SPI) contain genes which confer virulence-associated functions upon the host bacterium, often mediated by secreted proteins. In Salmonella, many pathogenicity islands and other gene clusters have been well characterized, and expression of a number of genes from the 5 major islands has been detected. A small number of genes from SPI-1 were upregulated, including sitBC, which encodes an iron uptake system (101). The sitABCD operon is induced under iron-deficient conditions and is thought to play a role in iron acquisition in mice (42, 98). Interestingly, hilC and hilD are downregulated in the lumen. These genes encode transcriptional activators, which can bind to hilA and induce expression of three operons within SPI-1, namely, inv-spa, prg-org, and sic-sip (23). The high levels of repression of hilD suggest that expression of SPI-1 is inhibited, though expression of sipD and spaS was detected. hilD also plays a role in mediating the activities of SPI-1 and SPI-2 (12). The role of SPI-1 genes, and secreted proteins in general, in colonization in day-old chicks has not been widely investigated and is of considerable interest. Most SPI-1 genes were found not to be required for colonization of the cecal lumen of older birds by Morgan et al. (59), although they were required for colonization of the intestinal mucosa of calves. Recently, Jones et al. (43) found that SPI-1 did not play an essential role in systemic infection in 1-day-old birds. A subset of SPI-2 genes, including ttrAC, ssaBCDMSU, and sseC, were upregulated significantly in the lumen. Interestingly, gene expression was detected throughout SPI-2, though most of the changes were not significant. Regulation of expression of SPI-2 genes is thought to involve OmpR-EnvZ and PhoP-PhoQ (9), but none of the genes encoding these proteins showed significant alterations in their levels of expression. Again, Morgan et al. (59) found very few of these genes to be required for colonization of older birds. Interestingly, Wigley et al. (89) found in day-old chickens that SPI-1 contributed to and SPI-2 was essential for the virulence of Salmonella enterica serovar Pullorum in newly hatched chicks, where gut colonization does represent an early phase of the infection process in this infection model (73).
In SPI-3, mgtC was upregulated, along with rmbA and fidL. The mgtC gene forms a part of the mgtBC operon, which is positively regulated by magnesium, although the exact role of mgtC has not yet been clearly defined. This gene does not have a role in magnesium uptake, though it may have a role in long-term survival in macrophage cell lines (58), suggesting that mgtC may have a similar role here. Statistically nonsignificant increases in expression were also observed with mgtA and mgtB. No genes were expressed from SPI-4, and no role in colonization of chickens was observed by Morgan et al. (59). However, in SPI-5, pipB, whose role is unclear, was found to be upregulated in the lumen and at the mucosal wall. These authors also found that pipB contributed to colonization of older chickens (59). Although its role is unclear, it has a link with SPI-2 since this SPI is required for its secretion (50), though here those genes were not significantly upregulated.
It is worth noting that expression of ugtL was upregulated at the mucosal wall. This gene is required for resistance to the antimicrobial peptides maginin 2 and polymyxin B (69). Within the lumen, magnesium limitation appears to be occurring, but it is interesting that a defense peptide is being expressed by Salmonella, suggesting that a rapid response by the host to infection is occurring.
The exact role for secreted proteins in colonization is unclear. Older work (82) supported the hypothesis that cecal colonization, of chickens at least, by Salmonella was largely a physiological characteristic, since there seemed little ecological advantage in adhesion to the mucosa in an organ where the rate of flow of chyme was very low. However, the identification of some SPI genes in colonization (59, 82) suggested that colonization, as a virulence trait, might not be as straightforward as originally thought. More recent information indicated that a T cell-mediated response, rather than secretory antibodies, was central to immune clearance of S. Typhimurium from the chicken gut (7), suggesting that a close association with the mucosa may indeed be involved. The upregulation of fimbrial genes (59) supports this assertion.
The microscopic observations indicated a higher rate of cell division in the mucosa than in the lumen of the cecum. This was supported by the increased expression at the mucosa of ftsA, a gene involved in cell division which acts to anchor the protofilaments of bacterial tubulin, encoded by ftsZ, to the membrane (62). However, the presence of ftsA alone is not sufficient for the Z ring to form, and zipA (32) and other downstream genes required were not expressed at significant levels at the mucosal wall. On the other hand, further support for active cell division occurring close to the mucosa comes from expression of mreB. The protein encoded by mreB has been found responsible for the rodlike shape of Salmonella bacteria (19). Recent work (86) suggests that the MreB protein directs the incorporation of new peptidoglycan into the wall, though the presence of ftsZ is required to direct the insertion. In addition, yhdE, which inhibits the formation of the septum, was expressed (12), suggesting that the cells were elongating after cell division at the point of sample collection. The MreB protein also contributes, it is thought, to chromosomal segregation (51).
These preliminary data suggest that S. Typhimurium bacteria in the cecal lumens of newly hatched chickens show downregulation of genes associated with transcription, translation, and cell division, all required for growth, whereas there was some expression of genes associated with cell division in bacteria harvested closer to the mucosa. It seems likely that concentrations of oxygen or of other electron acceptors and a variety of nutrients would be present closer to the mucosa than to the lumen, so these findings are not surprising. They are supported by earlier results with E. coli colonization of the mouse intestine, where there was evidence for most microbial growth taking place close to the mucosa (63), and our microscopic findings support this. The data suggest that several energy and carbohydrate sources are utilized which are different from those used in late-log-phase nutrient broth cultures, including propionate, ethanolamine, and 1,2-propanediol. Organisms in the lumen were poorly motile and showed a downregulation of genes associated with chemotaxis, though no genes associated with motility were identified or expressed at the mucosa. From our colonization studies, it was clear that several genes associated with propanediol catabolism under anaerobic conditions were involved in colonization, although the picture is obviously complex. However, in the light of previous findings where very few genes made great changes when tested as single mutations (16, 100), coupled with the degree of metabolic redundancy in enteric bacteria, this perhaps should not be surprising. This indicates that the colonization phenotype is a multifactorial characteristic with the main site of metabolic and other physiological activity close to the mucosa.
ACKNOWLEDGMENTS
We thank Alice Middleton, Marianne Goodchild, and Sandrine Touchard for assistance in a number of ways, Eirwen Morgan for fruitful discussions, and Jay Hinton, Arthur Thompson, and Saccha Luccinini at the Institute of Food Research (IFR) for assistance in establishing the microarray.
The work was funded by the European Union (RTD projects FP5-CT-2000-01126 SALARRAY and FP6-CT-2003 50523 SUPASALVAC).
Footnotes
Published ahead of print on 18 July 2011.
REFERENCES
- 1. Aldridge P., Hughes K. T. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160–165 [DOI] [PubMed] [Google Scholar]
- 2. Aldridge P. D., et al. 2006. The flagellar-specific transcription factor, sigma28, is the type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 20:2315–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anonymous. 1998. PHLS evidence to House of Commons Select Committee on Agriculture; enquiry into food safety. Fourth Report for the session 1997-98. Her Majesty’s Stationery Office, London, England: http://www.publications.parliament.uk/pa/cm199798/cmselect/cmagric/331iv /ag0402.htm [Google Scholar]
- 4. Anonymous. 2006. Trends and sources of zoonoses, Zoonotic agents and antimicrobial resistance in the European Union in 2004. European Food Safety Authority, Parma, Italy: http://www.efsa.europa.eu/en/efsajournal/doc/310ar.pdf [Google Scholar]
- 5. Barrow P. A., Hassan J. O., Berchieri A., Jr 1990. Reduction in faecal excretion by chickens of Salmonella typhimurium by immunization with avirulent mutants of S. typhimurium. Epidemiol. Infect. 104:413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrow P. A., Lovell M. A. 1991. Experimental infection of egg-laying hens with Salmonella enteritidis. Avian Pathol. 20:339–352 [DOI] [PubMed] [Google Scholar]
- 7. Beal R. K., Powers C., Davison T. F., Barrow P. A., Smith A. L. 2006. Clearance of enteric Salmonella enterica serovar Typhimurium in chickens is independent of B-cell function. Infect. Immun. 74:1442–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- 9. Bijlsma J. J., Groisman E. A. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57:85–96 [DOI] [PubMed] [Google Scholar]
- 10. Brinsmade S. R., Paldon T., Escalante-Semerena J. C. 2005. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 187:8039–8046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buchmeier N. A., Lipps C. J., So M. Y., Heffron F. 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7:933–936 [DOI] [PubMed] [Google Scholar]
- 12. Bustamante V. H., et al. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. U. S. A. 105:14591–14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reference deleted.
- 14. Cha M. K., Kim W. C., Lim C. J., Kim K., Kim L. H. 2004. Escherichia coli periplasmic thiol peroxidase acts as lipid hydroperoxide peroxidase and the principal antioxidative function during anaerobic growth. J. Biol. Chem. 279:8769–8778 [DOI] [PubMed] [Google Scholar]
- 15. Chang D. E., et al. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U. S. A. 101:7427–7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clayton D. J., et al. 2008. Analysis of the role of 13 major fimbrial subunits in colonisation of the chicken intestine by Salmonella enteric serovar Enteritidis reveals a role for a novel locus. BMC Microbiol. 8:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coates M. E., Gregory M. E., Porter J. W. G., Williams A. P. 1963. Vitamin B and its analogues in the gut contents of germ-free and conventional chicks. Proc. Nutr. Soc. 22:27–35 [Google Scholar]
- 18. Corbin B. D., Wang Y., Beuria T. K., Margolin W. 2007. Interaction between cell division proteins FtsE and FtsZ. J. Bacteriol. 189:3026–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Costa C. S., Anton D. N. 1993. Round-cell mutants of Salmonella typhimurium produced by transposition mutagenesis: lethality of rodA and mre mutations. Mol. Gen. Genet. 236:387–394 [DOI] [PubMed] [Google Scholar]
- 20. Craven S. E. 1994. Altered colonizing ability for the ceca of broiler chicks by lipopolysaccharide-deficient mutants of Salmonella typhimurium. Avian Dis. 38:401–408 [PubMed] [Google Scholar]
- 21. Dunkley K. D., et al. 2009. Food-borne Salmonella ecology in the avian gastro-intestinal tract. Anaerobe 15:26–35 [DOI] [PubMed] [Google Scholar]
- 22. Edelman S., Leskela S., Ron E., Apajalahti J., Korhonen T. K. 2003. In vitro adhesion of an avian pathogenic Escherichia coli O78 strain to surfaces of the chicken intestinal tract and to ileal mucus. Vet. Microbiol. 91:41–56 [DOI] [PubMed] [Google Scholar]
- 23. Ellermeier J. R., Slauch J. M. 2007. Adaptation to the host environment: regulation of the SPI-1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24–29 [DOI] [PubMed] [Google Scholar]
- 24. Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J. C. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 25. Fardini Y., et al. 2007. The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica serovar Enteritidis. Infect. Immun. 75:358–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Foley S. L., et al. 2006. Comparison of subtyping methods for differentiating Salmonella enterica serovar Typhimurium isolates obtained from food animal sources. J. Clin. Microbiol. 44:3569–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Foster J. W. 1999. When protons attack: microbial strategies of acid adaptation. Curr. Opin. Microbiol. 2:170–174 [DOI] [PubMed] [Google Scholar]
- 28. Foster J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898–907 [DOI] [PubMed] [Google Scholar]
- 29. Foster J. W., Hall H. K. 1990. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172:771–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gibbs K. A., et al. 2004. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol. Microbiol. 53:1771–1783 [DOI] [PubMed] [Google Scholar]
- 31. Gophna U., et al. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hale C. A., de Boer P. A. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hammelman T. A., et al. 1996. Identification of a new prp locus required for propionate catabolism in Salmonella typhimurium LT2. FEMS Microbiol. Lett. 137:233–239 [DOI] [PubMed] [Google Scholar]
- 34. Hansen A. M., et al. 2005. SspA is required for acid resistance in stationary phase by down-regulation of H-NS in Escherichia coli. Mol. Microbiol. 56:719–734 [DOI] [PubMed] [Google Scholar]
- 35. Haugen S. P., Ross W., Gourse R. L. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6:507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heithoff D. M., Sinsheimer R. L., Low D. A., Mahan M. J. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967–970 [DOI] [PubMed] [Google Scholar]
- 37. Henry T., et al. 2006. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc. Natl. Acad. Sci. U. S. A. 103:13497–13502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horswill A. R., Escalante-Semerna J. C. 1997. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179:928–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Howells A. M., et al. 2002. Role of trehalose biosynthesis in environmental survival and virulence of Salmonella enterica serovar Typhimurium. Res. Microbiol. 153:281–287 [DOI] [PubMed] [Google Scholar]
- 40. Humphries A. D., et al. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357–1376 [DOI] [PubMed] [Google Scholar]
- 41. Iwama T., et al. 2000. Mutational analysis of ligand recognition by Tcp, the citrate chemoreceptor of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1437–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Janakiraman A., Slauch J. M. 2000. The putative iron transport systems SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146–1155 [DOI] [PubMed] [Google Scholar]
- 43. Jones M. A., Hulme S. D., Barrow P. A., Wigley P. 2007. The Salmonella pathogenicity island 1 and Salmonella pathogenicity island 2 type III secretion systems play a major role in pathogenesis of systemic disease and gastrointestinal tract colonization of Salmonella enterica serovar Typhimurium in the chicken. Avian Pathol. 36:199–203 [DOI] [PubMed] [Google Scholar]
- 44. Jones S. A., et al. 2007. Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75:4891–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jones S. A., et al. 2008. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect. Immun. 76:2531–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaiser P., et al. 2000. Differential cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarum. Microbiology 146:3217–3226 [DOI] [PubMed] [Google Scholar]
- 47. Keener J., Nomura M. 1996. Regulation of ribosome synthesis, p. 1417–1431 In Neidhardt F. C., et al. (ed.) Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 48. Kieboom J., Abee T. 2006. Arginine-dependent acid resistance in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:5650–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klumpp S., Zhang Z., Hwa T. 2009. Growth-rate dependent global effects on gene expression in bacteria. Cell 139:1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Knodler L. A., et al. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43:1089–1103 [DOI] [PubMed] [Google Scholar]
- 51. Kruse T., Moller-Jensen J., Lobner-Olesen A., Gerdes K. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Latasa C., et al. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322–1339 [DOI] [PubMed] [Google Scholar]
- 53. Lewis N. E., et al. 2010. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol. 6:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lucas R. L., Lee C. A. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reference deleted.
- 56. McClelland M., et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 57. McMeechan A., et al. 2005. Glycogen production by different Salmonella enterica serotypes: contribution of functional glgC to virulence, intestinal colonization and environmental survival. Microbiology 151:3969–3977 [DOI] [PubMed] [Google Scholar]
- 58. Moncrief M. B. C., Maguire M. E. 1998. Magnesium and the role of mgtC in growth of Salmonella typhimurium. Infect. Immun. 66:3802–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morgan E., et al. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994–1010 [DOI] [PubMed] [Google Scholar]
- 60. Palacios S., Escalante-Semerena J. C. 2000. prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:905–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Palacios S., Starai V. J., Escalante-Semerena J. C. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J. Bacteriol. 185:2802–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pichoff S., Lutkenhaus J. 2007. Identification of a region of FtsA required for interaction with FtsZ. Mol. Microbiol. 64:1129–1138 [DOI] [PubMed] [Google Scholar]
- 63. Poulsen L. K., Licht T. R., Rang C., Krogfelt K. A., Molin S. 1995. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J. Bacteriol. 177:5840–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Price-Carter M., Tingey J., Bobik T. A., Roth J. R. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rodrigue D. C., Tauxe R. V., Rowe B. 1990. International increase in Salmonella enteritidis: a new pandemic? Epidemiol. Infect. 105:21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rondon M. R., Kazmierczak R., Escalante-Semerena J. C. 1995. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium LT2. J. Bacteriol. 177:5434–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Roof D. M., Roth J. R. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174:6634–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sheppard D. E., Penrod J. T., Bobik T., Kofoid E., Roth J. R. 2004. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J. Bacteriol. 186:7635–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shi Y., Cromie M. J., Hsu F. F., Turk J., Groisman E. A. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53:229–241 [DOI] [PubMed] [Google Scholar]
- 70. Smith H. W., Tucker J. F. 1980. The virulence of salmonella strains for chickens: their excretion by infected chickens. J. Hyg. (Lond.) 84:479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:article 3 [DOI] [PubMed] [Google Scholar]
- 72. Smyth G. K., Michaud J., Scott H. 2005. The use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075 [DOI] [PubMed] [Google Scholar]
- 73. Snoeyenbos G. H. 1991. Pullorum disease, p. 73–87 In Calnek B. W. (ed.), Diseases of poultry, 9th ed. Iowa State University Press, Ames, IA [Google Scholar]
- 74. Stecher B., Barthel M., Schlumberger M. C., Kremer M., Hardt W.-D. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell. Microbiol. 10:1166–1180 [DOI] [PubMed] [Google Scholar]
- 75. Stojiljkovic I., Baumler A. J., Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutH gene cluster. J. Bacteriol. 177:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Strom A. R., Kaasen I. 1993. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol. Microbiol. 8:205–210 [DOI] [PubMed] [Google Scholar]
- 77. Tamai E., Shimamoto T., Tsuda M., Mizushima T., Tsuchiya T. 1998. Conversion of temperature-sensitive to -resistant gene expression due to mutations in the promoter region of the melibiose operon in Escherichia coli. J. Biol. Chem. 273:16860–16864 [DOI] [PubMed] [Google Scholar]
- 78. Tinker J. K., Hancox L. S., Clegg S. 2001. FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tinker J. K., Clegg S. 2000. Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 68:3305–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Reference deleted.
- 81. Tsang A. W., Horswill A. R., Escalante-Semerena J. C. 1998. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J. Bacteriol. 180:6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Turner A. K., Lovell M. A., Hulme S. D., Zhang-Barber L., Barrow P. A. 1998. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66:2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Turner A. K., et al. 2003. Contribution of proton-translocating proteins to the virulence of Salmonella enterica serovars Typhimurium, Gallinarum, and Dublin in chickens and mice. Infect. Immun. 71:3392–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tweeddale H., Notley-McRobb L., Ferenci T. 1998. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 180:5109–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Van Bibber M., Bradbeer C., Clark N., Roth J. R. 1999. A new class of cobalamin transport mutants (btuF) provides genetic evidence for a periplasmic binding protein in Salmonella typhimurium. J. Bacteriol. 181:5539–5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Varma A., dePedro M., Young K. D. 2007. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 189:5692–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Watson M. 2005. ProGenExpress: visualization of quantitative data on prokaryotic genomes. BMC Bioinformatics 6:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Weart R. B., Levin P. A. 2003. Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 185:2826–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wigley P., Jones M. A., Barrow P. A. 2002. Salmonella enterica serovar Pullorum requires the Salmonella pathogenicity island 2 type III secretion system for virulence and carriage in chickens. Avian Pathol. 31:501–506 [DOI] [PubMed] [Google Scholar]
- 90. Winter S. E., et al. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Withanage G. S. K., et al. 2004. Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar Typhimurium. Infect. Immun. 72:2152–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Woodall C. A., et al. 2005. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73:5278–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wu W.-F., Urbanowski M. L., Stauffer G. V. 1992. Role of the MetR regulatory system in vitamin B12-mediated repression of the Salmonella typhimurium metE gene. J. Bacteriol. 174:4833–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wu W.-F., Urbanowski M. L., Stauffer G. V. 1995. Characterization of a second MetR-binding site in the metE metR regulatory region of Salmonella typhimurium. J. Bacteriol. 177:1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yamamoto S., Kutsukake K. 2006. FljA-mediated posttranscriptional control of phase 1 flagellin expression in flagellar phase variation of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:958–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang Y. H., et al. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yohannes E., Thurber A. E., Wilks J. C., Tate D. P., Slonczewski J. L. 2005. Polyamine stress at high pH in Escherichia coli K-12. BMC Microbiol. 5:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zaharik M. L., et al. 2004. The Salmonella enterica serovar Typhimurium divalent cation transport systems MntH and SitABCD are essential for virulence in an Nramp1G169 murine typhoid model. Infect. Immun. 72:5522–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Reference deleted.
- 100. Zhang-Barber L., et al. 1997. Influence of genes encoding proton-translocating enzymes on suppression of Salmonella typhimurium growth and colonization. J. Bacteriol. 179:7186–7190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhou D., Hardt W.-D., Galan J. E. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67:1974–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]