Abstract
The multisubunit SWI/SNF and RSC complexes utilize energy derived from ATP hydrolysis to mobilize nucleosomes and render the DNA accessible for various nuclear processes. Here we test the idea that remodeling involves intermediates with mobile DNA bulges or loops within the nucleosome by cross-linking the H2A N- or C-terminal tails together to generate protein “loops” that constrict separation of the DNA from the histone surface. Analyses indicate that this intranucleosomal cross-linking causes little or no change in remodeling-dependent exposure of DNA sequences within the nucleosome to restriction enzymes. However, cross-linking inhibits nucleosome mobilization and blocks complete movement of nucleosomes to extreme end positions on the DNA fragments. These results are consistent with evidence that nucleosome remodeling involves intermediates with DNA loops on the nucleosome surface but indicate that such loops do not freely diffuse about the surface of the histone octamer. We propose a threading model for movement of DNA loops around the perimeter of the nucleosome core.
INTRODUCTION
Assembly of the eukaryotic genome into nucleosomes and higher-order chromatin structures greatly reduces the accessibility of DNA and restricts various nuclear events, including DNA repair, recombination, replication, and transcription. Eukaryotic cells have developed several ways to disrupt or modulate chromatin structures to facilitate the binding of trans-acting factors to DNA, allowing such processes to occur. For example, the chromatin structure is directly or indirectly altered by posttranslational modifications such as phosphorylation, acetylation, methylation, and ubiquitylation, which primarily occur on the core histone tail domains. Although the precise functions of each of these highly conserved modifications have yet to be resolved, it is believed that the combination of distinct covalent modifications can be recognized by downstream protein factors which in turn regulate chromatin structure and DNA accessibility (37, 48, 56). A second critical process involves multiple-subunit enzymes that use energy derived from ATP hydrolysis to remodel chromatin structure and disrupt DNA-histone interactions, thereby stimulating DNA-dependent processes (5, 23, 47).
ATP-dependent remodeling complexes have been extensively studied and shown to have the ability to alter and rearrange nucleosomes in a manner that increases the accessibility of nucleosomal DNA (9, 16, 35). These complexes share a homologous ATPase domain that belongs to the SF2 superfamily of DNA-stimulated helicases and are generally divided into four different families: SWI/SNF, ISWI, INO80, and CHD (9, 13, 16). Typically, the isolated ATPase subunit can catalyze nucleosome remodeling independent of other remaining subunits, with ATPase activity stimulated by double-stranded DNA and/or nucleosomes (16, 34). These chromatin-remodeling enzymes can often recognize histone posttranslational modifications through auxiliary subunits, and they can regulate chromatin structures by assembly, disassembly, and translocation of nucleosomes in an ATP-dependent manner (9, 16).
The Saccharomyces cerevisiae SWI/SNF complex was the first remodeling enzyme to be identified, and it is required for expression of many inducible genes, such as HO, SUC2, and INO1 (10, 36, 53). Although SWI/SNF is not essential for yeast growth, a genome-wide analysis demonstrated that ∼5% of yeast genes are regulated by SWI/SNF, with functions that contribute to both gene activation and repression (19). Moreover, this complex plays a critical role in gene expression in late mitosis (24). The yeast RSC (remodels the structure of chromatin) complex is related to SWI/SNF but is more abundant and essential for cell growth (7). RSC has functions in stress response, gene activation in transcription, and chromosome segregation (11, 22). Both SWI/SNF and RSC have also been shown to function during recombinational repair of DNA double-strand breaks (8).
SWI/SNF and RSC have been the targets of extensive biochemical characterization. SWI/SNF was shown to destabilize approximately 40 bp of histone-DNA interactions at either edge of a nucleosome, based on electron energy loss microscopy and atomic force microscopy studies (4, 41). Furthermore, a photochemical mapping study demonstrates that SWI/SNF can mobilize a mononucleosome by as much as 50 bp off the ends of DNA fragments (21). ATP-dependent nucleosome sliding along DNA substrates occurs in cis and leads to the exposure of cognate DNA for trans-acting factors (17, 18, 51). However, crucial aspects of the mechanism of nucleosome remodeling remain undefined.
One proposed mechanism for how the nucleosome might be translocated along the DNA, referred to as twist diffusion, involves the ATP-dependent twisting of the DNA helix on the histone surface like a corkscrew, with as little as ±1 bp of DNA traveling through the core region (25, 50). This model is supported by crystal structures of a nucleosome core particle in which the DNA on one side is observed to contain a single-base-pair “twist defect” compared to the DNA at the other side of the core (12, 33) and has the energetic advantage that the gain or loss of a base pair internally within the nucleosome can occur without disruption of histone-DNA interactions. However, studies using DNA substrates that inhibit DNA rotation on the nucleosome surface indicate that nucleosome sliding catalyzed by human SWI/SNF (hSWI/SNF) likely does not occur solely via a twist-diffusion mechanism (1, 3, 49).
Recent studies have led to a proposal that remodeling enzymes use a DNA translocase mechanism to induce nucleosome sliding along DNA by creating transient DNA bulges or loops on the histone octamer surface (40, 57, 58). This bulge propagation model suggests that linker DNA can be drawn into the core to form an internal DNA loop. Indeed, several studies indicate that remodeling enzymes harbor DNA translocation activity (20, 39, 52), suggesting that a unifying feature of remodelers is catalyzing DNA movement relative to the remodeling complex. Alternatively, a related third model proposes that a loop may be formed within the nucleosome by a dissociation and recapture mechanism, involving linker DNA. In either of the last two models, the internal DNA loop is envisioned to propagate through the nucleosome core like a wave, resulting in the movement of the histone octamer along the DNA segment (26). This model is strengthened by recent single-molecule and biochemical studies demonstrating that both SWI/SNF and RSC are able to translocate on DNA and nucleosomal templates to produce loops in an ATP-dependent fashion and that nucleosome remodeling by RSC seems to produce a remodeled intermediate that contains internal DNA loops and more than 147 bp of DNA (29, 43, 57).
In this work we have tested whether SWI/SNF- and RSC-dependent nucleosome remodeling involves DNA bulge propagation by employing nucleosome substrates in which either the two N-terminal or C-terminal tails of H2A are cross-linked together in an intranucleosomal fashion. Cross-linking generates constraining loops around the superhelical gyres that should inhibit the propagation of DNA bulges or loops on the histone surface. Our results indicate that loops formed during SWI/SNF and RSC nucleosome remodeling likely are not freely propagated around the exterior of the nucleosome; rather, we propose such loops are translocated around the histone octamer via a threading mechanism.
MATERIALS AND METHODS
DNA templates.
A 215-bp EcoRI-DdeI DNA fragment and a 244-bp EcoRI-NsiI DNA fragment containing the Xenopus borealis somatic 5S RNA sequence were generated from plasmid pXP-10; the fragments included sequences −78 to +137 and −78 to +166 according to the 5S transcription start site, respectively. These fragments harbor a site for the restriction enzyme EcoRV at position +35, which is protected from digestion by the histone octamer after nucleosome assembly (Fig. 1A). A 343-bp DNA fragment containing the 601 nucleosome-positioning sequence was obtained from plasmid CP1024 by EcoRI and HindIII restriction enzyme digestion (54); this fragment contains a unique HhaI digestion site close to the predicted nucleosome dyad (55) (Fig. 1B). These three fragments were 5′ end radiolabeled with [γ-32P]ATP and polynucleotide kinase (PNK) (New England BioLabs) by standard techniques. For large-scale preparations, the 601 DNA fragments were amplified by standard PCR (31). In certain instances, 5′ ends of DNA fragments within reconstituted nucleosomes were radiolabeled with [γ-32P]ATP and PNK, followed by a second glycerol gradient purification (see below).
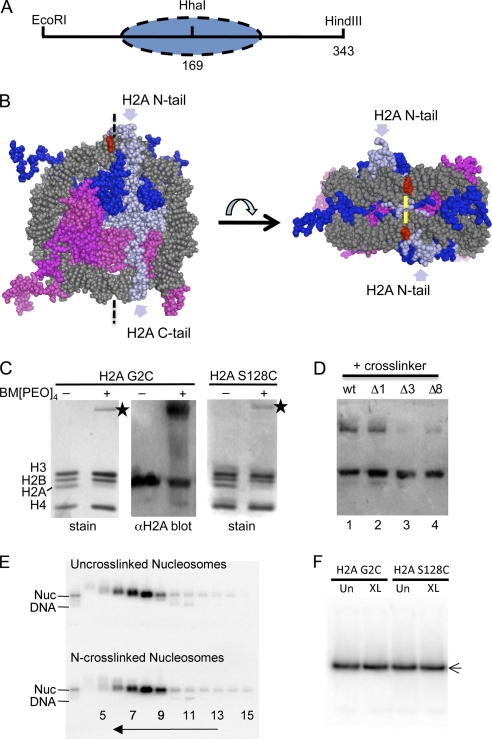
Fig. 1.
Intranucleosomal cross-linking. (A) The 343-bp 601 DNA fragment used for nucleosome reconstitution. The HhaI site is located near the nucleosome dyad at the center of the DNA fragment. The oval indicates the predicted location of the nucleosome. (B) Location of the N- and C-terminal tails of H2A within a nucleosome core model (PDB accession code 1kx5) (12). DNA is gray, H2A light blue, H2B dark blue, H3 light magenta, and H4 magenta. The model includes all of the N-terminal tail of H2A and all but the final two residues of the C-terminal tail (12). Bases contacted by the H2A N-terminal tail domain as determined by cross-linking studies are red (27). The predicted location of the bifunctional cross-linker bridging the two H2A N-terminal tail domains is indicated by yellow bars. (C) Intranucleosomal cross-linking occurs with high efficiency. Nucleosomes reconstituted with the 343-bp 601 DNA fragment and either H2A G2C or H2A S128C were cross-linked with BM[PEO]4, and the samples were analyzed by SDS-PAGE and Coomassie staining or Western blotting with anti-H2A antisera. Lanes 1 and 2 in each panel contain un-cross-linked and cross-linked nucleosomes, respectively. Stars indicate the cross-linked H2A-H2A band. (D) Nucleosomes containing H2A G2C (wild type [wt]) or H2A G2CΔ1 (Δ1), H2A G2CΔ3 (Δ3), or H2A G2CΔ8 (Δ8), in which 1, 3, or 8 amino acid residues, respectively, were deleted from the H2A N-terminal tail domains, were treated with BM[PEO]4, and the efficiency of cross-linking was analyzed by SDS-PAGE and Western blotting with anti-H2A antisera. (E) Cross-linked and un-cross-linked nucleosomes (Nuc) containing H2A G2C exhibit similar sedimentation profiles. Radiolabeled nucleosomes separated on glycerol gradient fractions were analyzed by 0.7% nucleoprotein gel electrophoresis and phosphorimaging. The direction of sedimentation and fraction numbers are indicated. Fraction 8 was chosen for further analysis in this preparation. (F) Nondenaturing polyacrylamide gel analysis of un-cross-linked (Un) and cross-linked (XL) nucleosomes. Gradient-purified cross-linked and un-cross-linked nucleosomes were run on the gel as indicated.
Preparation of histone proteins.
The coding sequences for the mutant H2As containing a cysteine substitution at either residue 2 (H2A G2C) or 128 (H2A S128C) were obtained by QuikChange site-directed mutagenesis (Stratagene). Xenopus H2A, H2B, and H2A mutants were expressed in bacterial cells and purified as preformed dimers as described previously (28). H2A deletion mutants were based on H2A G2C but with residue 3 (H2A G2CΔ1), residues 3 to 5 (H2A G2CΔ3), or residues 3 to 10 (H2A G2CΔ8) deleted.
Nucleosome reconstitution.
Nucleosomes were reconstituted with DNA fragments and either wild-type H2A or mutants in complex with H2B and H3/H4 in a large-scale format to allow analysis of protein-protein cross-linking in the same samples used for remodeling, as described previously (31). Briefly, 1 μg radiolabeled DNA fragments (1,000 kcpm), 500 μg of cold 601 343-bp DNA fragments, ∼200 μg H2A/H2B dimer, and 170 μg H3/H4 tetramer were combined in a 2-ml reaction mixture containing Tris-EDTA (TE)-2 M NaCl and 10 mM dithiothreitol (DTT). Alternatively, nucleosomes were reconstituted with linearized CP1024 plasmid DNA in an identical 2-ml reaction mixture. Nucleosomes were reconstituted by standard salt dialysis at 4°C with 10 mM β-mercaptoethanol in all dialysis buffers (31).
H2A-H2A intranucleosome cross-linking.
The cysteine residues within the N- or C-terminal tails of H2A mutants were cross-linked after nucleosome formation by reacting with BM[PEO]4 (Pierce). Cross-linking reactions were carried out by titrating in BM[PEO]4 to a final nucleosome/cross-linker ratio of 1:2 and incubating at 37°C for 30 min as described previously (31). Reactions were terminated by addition of DTT to a 10 mM final concentration, and the cross-linking was quantified by running a portion of the sample on 15% SDS-PAGE gel and by Western blotting (31). Cross-linked and un-cross-linked nucleosomes were loaded onto 10-ml 5 to 30% glycerol gradients (in 10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 10 mM DTT) and sedimented at 34,000 rpm for 15 to 18 h at 4°C with an SW41 type rotor. Fractions containing the purified nucleosomes were identified by running 10 μl on 0.7% native nucleoprotein gels (0.5× Tris-borate-EDTA [TBE]) and stored at 4°C.
RSC and SWI/SNF remodeling reactions.
Purified RSC from Saccharomyces cerevisiae was a kind gift from Blaine Bartholomew (Southern Illinois University). The yeast SWI/SNF complex was prepared as described previously (46). Both yeast RSC and SWI/SNF were aliquoted and stored at −80°C. RSC remodeling was carried out in a 20-μl reaction mixture with approximately 0.5 nM nucleosomes, with the amounts of RSC noted in the figure legends in RSC remodeling buffer (20 mM Na-HEPES [pH 7.8], 3 mM MgCl2, 0.08% NP-40, 1.7% glycerol, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM DTT), and with 0.8 mM ATP at 30°C. SWI/SNF remodeling reactions were carried out in the same manner but with SWI/SNF remodeling buffer (10 mM Tris, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 100 ng/μl bovine serum albumin), with the amounts of enzyme noted in the figure legends, and with 1 mM ATP at 30°C. Restriction enzyme digestions were performed by adding 10 U restriction enzymes to the above reaction mixtures either during or after the remodeling as indicated in the figure legends. In the latter case, remodeling reactions were stopped and remodelers were removed from nucleosomes by adding 300 ng sonicated calf thymus (CT) DNA as a nonspecific competitor. For analysis of DNA cleavage, reactions were stopped by adding 4 μl 6× SDS DNA loading buffer containing 2 mM EDTA. The results were analyzed on 5% polyacrylamide gels containing 0.04% SDS run at 120 V for 2.5 h at room temperature.
Nucleosome sliding assays.
Remodeling reactions were set up as described above. Remodelers were removed by addition of 300 ng of nonspecific competitor DNA and glycerol to a 5% final concentration and kept on ice for 5 min. Then the nucleosome translational positions were determined by separation on a nondenaturing 4% polyacrylamide gel (acrylamide/bisacrylamide ratio, 35:1) run at 100 V for 3 h at 25°C, including a prerun of 100 V for 1.5 h at the same temperature.
RESULTS
Facile cross-linking of the N- or C-terminal tails of histone H2A within a single nucleosome.
To test if bulge propagation/looping contributes to the mechanism of ATP-dependent-remodeling-induced nucleosome mobilization, we generated nucleosomes containing proteinaceous loops constraining the superhelical wraps of DNA on the outside of the nucleosome. This was accomplished by cross-linking cysteine residues placed within either the N- or C-terminal tail domains of H2A in reconstituted nucleosomes by reaction with the bifunctional reagent BM[PEO]4. The loops are expected to create steric hindrance to the formation and/or propagation of a DNA bulge loop on the nucleosome surface.
We generated H2A mutants H2A G2C and H2A S128C, in which either the 2nd or 128th (penultimate) residue in the protein is replaced by cysteine, near the end of either tail domain. Note that the H2A N- or C-terminal tails exit the nucleosome in opposite directions, approximately parallel to the nucleosome dyad, above and below one or two superhelical DNA gyres, respectively (Fig. 1B) (12, 33). Prior studies indicate that the ends of the H2A N-terminal tails wrap around the upper/lower DNA superhelices and cross-link to DNA bases about 25 Å apart (28) (Fig. 1B, red). Considering that Cα-Cα distances of cysteine residues cross-linked by BM[PEO]4 a are predicted to be ∼24 Å, the N-terminal tail domains of H2A G2C are thus likely to be cross-linked by this reagent in the native structure. A similar analysis indicates that the H2A C-terminal tail domains should be close enough to allow cross-linking in nucleosomes containing H2A S128C (12, 27). Indeed, we find that nucleosomes containing H2A G2C or H2A S128C are efficiently cross-linked in an intranucleosomal fashion. Nucleosomes containing these proteins were reconstituted in a large-scale format (see Materials and Methods) on a 343-bp 601 DNA fragment (Fig. 1A) to allow determination of the extent of cross-linking by SDS-PAGE. After removal of the reducing agent, the cross-linker was titrated into solutions of nucleosomes as described previously (31) and cross-linking was assessed by SDS-PAGE. We found that at optimal cross-linker/nucleosome ratios either H2A G2C or H2A S128C was cross-linked at an efficiency of ∼80% (Fig. 1C) (30). Since we noted that the cross-linked H2A-H2A protein species exhibited a somewhat reduced Coomassie staining intensity compared to monomeric histone proteins, the efficiency of cross-linking was confirmed by Western blotting with anti-H2A antibodies (Fig. 1C) (31).
To determine the extent to which cross-linking the N-terminal tail domains of H2A together constrains the DNA to the surface of the core histone octamer, we prepared nucleosomes in which these domains were shortened by 1, 3, or 8 amino acid residues. Interestingly, shortening the N-terminal tail domain by 1 residue resulted in a reduction in total cross-linking, while decreasing the length of the tails by 3 residues virtually eliminated cross-linking (Fig. 1D). Thus, assuming a maximum of about 3 Å is contributed per residue to the length of the tail domain (less if the tail adopts a secondary structure such as an α-helix), this implies that a loop formed by cross-linking the native tails is about 6 Å longer than that for H2A G2CΔ1, which represents the minimum length that allows cross-linking, or near to it. Therefore, the total size of the loop is nominally 10 by 2 by 3 Å (the H2A tail has 12 residues, cysteine at residue 2); adding 24 Å for the cross-linker gives a total of ∼84 Å. Assuming that the loop circumscribes at least 1/2 a circle around the DNA superhelices (33), the radius of the loop is about 27 Å. Given that tails 1 residue shorter can still be cross-linked and that the base of the H2A tail is near the histone surface upon which the DNA is wrapped, this implies that the loop formed by the native cross-linked H2A tail domains has about 3 Å of vertical “play,” less than 1/4 the diameter of one DNA helix. Thus, these results imply the there is very little clearance between the cross-linked loop and the two DNA superhelical gyres.
The cross-linked and un-cross-linked nucleosomes were then radiolabeled and isolated on 5 to 30% glycerol gradients. We found that intranucleosome cross-linked species represented the vast majority of the cross-linked products, since both cross-linked and un-cross-linked nucleosomes exhibited nearly identical sedimentation profiles through the gradients and internucleosome cross-linked species would be expected to sediment as dinucleosomes (Fig. 1E). In addition, analysis of gradient-isolated nucleosomes on native 4% acrylamide gels showed that the N- and C-terminal cross-linked nucleosomes exhibit the same migration through the gel as un-cross-linked nucleosomes, indicating that all species likely have identical histone compositions and overall conformations (Fig. 1F). These results indicate that cross-linking occurred in an intranucleosomal fashion and had little or no effect on the native nucleosome conformation and stability.
H2A-H2A intranucleosomal cross-linking has little effect on SWI/SNF or RSC nucleosome remodeling, as determined by restriction enzyme accessibility assays.
We first determined whether intranucleosomal cross-linking of the H2A N- or C-terminal tail domains affected the ability of SWI/SNF to increase the accessibility of a restriction enzyme to a cognate site buried within a nucleosome (32, 38). Reconstitution of the 343-bp DNA fragment containing the 601 nucleosome positioning element results in a nucleosome with an HhaI restriction enzyme site located close to the nucleosome dyad and about 100 bp of linker DNA on either side of the nucleosome core region (Fig. 1B). Un-cross-linked control nucleosomes incubated in remodeling buffer were resistant to HhaI cleavage, as expected, confirming that the 601 nucleosomes were efficiently reconstituted and located at the expected position on the 601 DNA fragment (Fig. 2A, lane 1). In the presence of SWI/SNF or ATP alone, little or no cleavage of the nucleosome DNA is observed (a small amount of cleavage is likely due to residual ATP that copurified with the SWI/SNF (Fig. 2A, lanes 2 and 3). However, when both SWI/SNF and ATP were added to the reaction mixture, HhaI digested nearly all of the nucleosomes in a 60-min incubation (Fig. 2A, lane 4), indicating that the ATP-dependent remodeling activity of SWI/SNF altered nucleosome structure and/or position to increase accessibility of the HhaI site.
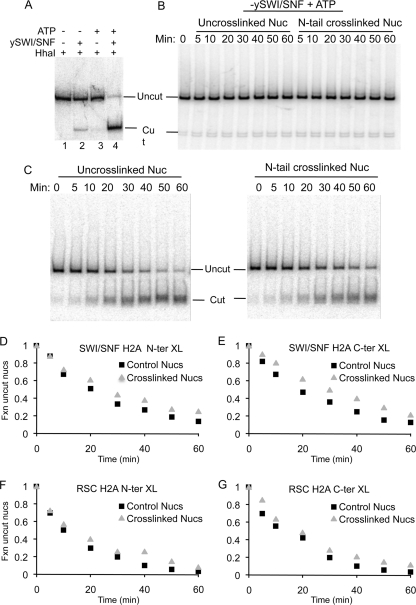
Fig. 2.
Intranucleosome cross-linking has little effect on SWI/SNF- and RSC-dependent exposure of restriction enzyme sites within the 601 nucleosome. (A) ATP- and SWI/SNF-dependent exposure of the HhaI site within the 343-bp 601 nucleosome. Radiolabeled, purified nucleosomes (0.5 nM) were incubated in remodeling buffer with 10 U HhaI (lane 1), with HhaI and 1 nM (1 nM) yeast SWI/SNF (ySWI/SNF) in the absence of ATP (lane 2), with 1 mM ATP without ySWI/SNF (lane 3), or with both ATP and ySWI/SNF (lane 4), as indicated above the gel. Reaction mixtures were incubated at 30°C for 60 min. Reactions were then stopped by addition of SDS loading buffer, and samples were analyzed on a 5% SDS-polyacrylamide gel. (B) Time course assay of nucleosome digestion by HhaI in the absence of SWI/SNF. Cross-linked and un-cross-linked nucleosomes containing H2A G2C were incubated with the restriction enzyme in remodeling buffer, reactions were stopped at the indicated times, and digestion was analyzed as in panel A. (C) Cross-linked and un-cross-linked nucleosomes were incubated as in panel B except reaction mixtures included 1 nM SWI/SNF. (D) The intensity of the undigested nucleosome band at each time point in panel C was quantified by phosphorimaging, and the fraction (Fxn) of uncut nucleosomes was plotted versus digestion time. The squares and triangles represent un-cross-linked and cross-linked nucleosomes, respectively. (E) As in panel D except that nucleosomes containing H2A S128C and cross-linked via the C terminus of H2A were examined. (F and G) As in panels D and E, respectively, except that nucleosomes were incubated with 0.1 nM RSC.
The effect of cross-linking on the ability of HhaI to digest nucleosome DNA was then investigated by a restriction enzyme accessibility (REA) time course experiment. In the absence of SWI/SNF, nucleosomes containing N-terminally cross-linked H2A were as resistant to HhaI digestion as control nucleosomes over the entire time course of the reaction, further indicating that the cross-linking did not alter nucleosome positioning or stability (Fig. 2B). Time course experiments carried out in the presence of SWI/SNF and ATP (Fig. 2C and D) showed that approximately 5 to 10% of the DNA in un-cross-linked and cross-linked nucleosomes was rapidly digested by HhaI in 5 min and the percentage of cut nucleosome DNA increased further with digestion time, with 80 to 90% of nucleosome DNA digested in 60 min. These results indicate that intranucleosome cross-linking of the N-terminal tail domains of H2A has little effect on the SWI/SNF remodeling-induced increase in HhaI site accessibility.
We next examined nucleosomes cross-linked via the H2A C-terminal tail domains, which creates a loop around the DNA superhelix that crosses the dyad axis of the nucleosome. Nucleosomes containing H2A S128C were reacted with BM[PEO]4 to cross-link the H2A C-terminal tail domains (Fig. 1E). Then substrates were remodeled with SWI/SNF, REA assays were performed as described above (Fig. 2C), and the extent of digestion was plotted versus time (Fig. 2E). Again, little or no effect of cross-linking within H2A S128C-containing nucleosomes on REA was detected, similar to results with the nucleosomes with cross-linked H2A N-terminal tail domains.
The RSC ATP-dependent remodeling complex is related to the SWI/SNF complex and mobilizes nucleosomes in a similar manner (9). In addition, recent work shows that RSC can generate a remodeling intermediate with DNA loops on the nucleosome surface (43). We therefore determined the effect of intranucleosomal cross-linking on the remodeling activities of RSC by REA assays. Control and cross-linked nucleosomes were remodeled by RSC in the presence of ATP and digested with HhaI, and cleavage was quantified by gel electrophoresis and phosphorimaging. Similar to results with SWI/SNF, nucleosomes containing cross-linked N- or C-terminal H2A tails were remodeled by RSC at rates nearly identical to those for un-cross-linked controls (Fig. 2F and G).
In the previous experiments, we noticed a small effect of intranucleosome cross-linking on the extent of REA at the HhaI site, primarily at the longest times of digestion. However, the cross-linked and un-cross-linked nucleosomes were digested in separate reactions, so small differences in remodeler or restriction enzyme activities may have contributed to these differences. To ensure that cross-linked and un-cross-linked nucleosomes were exposed to identical remodeling and restriction enzyme activities and to test another restriction enzyme and DNA template, distinguishable 5S nucleosome substrates were combined in the same reaction pot. Nucleosomes reconstituted with H2A G2C on a 215-bp fragment were cross-linked as described above, combined with an equal amount of un-cross-linked nucleosomes reconstituted with a 244-bp 5S DNA fragment (Fig. 3A), and then incubated in the same remodeling reaction mixture. In the absence of SWI/SNF, both nucleosomes were resistant to digestion with EcoRV (Fig. 3C). However, in the presence of SWI/SNF and ATP, the cross-linked and un-cross-linked nucleosomes were remodeled at virtually the same rate, but with the same small difference (∼6 to 8%) in the extent of digestion of un-cross-linked versus cross-linked nucleosomes detected in previous experiments (Fig. 3B and C).
Fig. 3.
Intranucleosome cross-linking has little effect on SWI/SNF-dependent exposure of restriction enzyme sites within the 5S nucleosomes. (A) Nucleosomes were reconstituted on 244-bp and 215-bp 5S DNA with H2A G2C and wild-type H3/H4, and the N-terminal tails within the 215-bp nucleosomes were cross-linked as described in the text. (B) The same amounts of cross-linked and un-cross-linked nucleosomes were mixed together and subjected to EcoRV digestion in the absence of SWI/SNF or presence of 1 nM SWI/SNF for the indicated times. (C) Plot of the fraction of uncut 5S nucleosomal DNAs versus digestion time.
H2A-H2A cross-linking inhibits remodeling-induced nucleosome mobilization.
Both SWI/SNF remodeling and RSC remodeling result in movement of nucleosomes from central positions to the ends of DNA fragments, in a bidirectional manner (9). Mobilization of nucleosomes can be monitored by nondenaturing polyacrylamide gel electrophoresis, where the end-positioned nucleosomes show more rapid mobility than center-positioned nucleosomes. To determine whether cross-linking alters the ability of SWI/SNF to mobilize nucleosomes, the un-cross-linked 244-bp and cross-linked 215-bp 5S nucleosomes were remodeled with limiting amounts of enzyme and translational positions were analyzed on native polyacrylamide gels. Nucleosomes on 5S DNA fragments adopt a distribution of distinct translational positions near the center and upstream ends of these fragments, as observed on the native gel (Fig. 4A, lanes 1 and 2), consistent with previous chemical and nuclease mapping studies (3, 28). When incubated with remodeling buffer and ATP (no SWI/SNF) or SWI/SNF (no ATP) alone, the nucleosomes exhibit unaltered distributions of translational positions in the gel (Fig. 4A, lanes 1 to 8). However, when nucleosomes were incubated with both ATP and SWI/SNF, the nucleosome translational positions were significantly changed. Almost all of the un-cross-linked 244-bp nucleosomes shifted to a rapidly migrating species corresponding to translational positions at the very ends of the DNA fragment as a result of SWI/SNF remodeling (Fig. 4A, compare lanes 7 and 9). In contrast, we found that about one-half of the 215-bp cross-linked nucleosomes were converted to a rapidly migrating species, while about one-half of the nucleosomes maintained more-central positions on the 215-bp DNA fragment (Fig. 4A, compare lanes 8 and 10). To confirm that the inhibition of nucleosome mobilization is due to the cross-linking of the H2A-H2A N-terminal tail domains rather than differences in fragment length, un-cross-linked nucleosomes reconstituted on the 215-bp 5S DNA fragment were assayed for nucleosome positions before and after remodeling. Importantly, nearly 100% of the 215-bp 5S nucleosomes were mobilized to the DNA ends in the presence of SWI/SNF and ATP, in contrast to the cross-linked nucleosomes (lanes 11 and 12). These results indicate that the intranucleosomal cross-linking of the H2A N-terminal tail domains inhibits SWI/SNF-induced mobilization to end positions on the 5S DNA fragment.
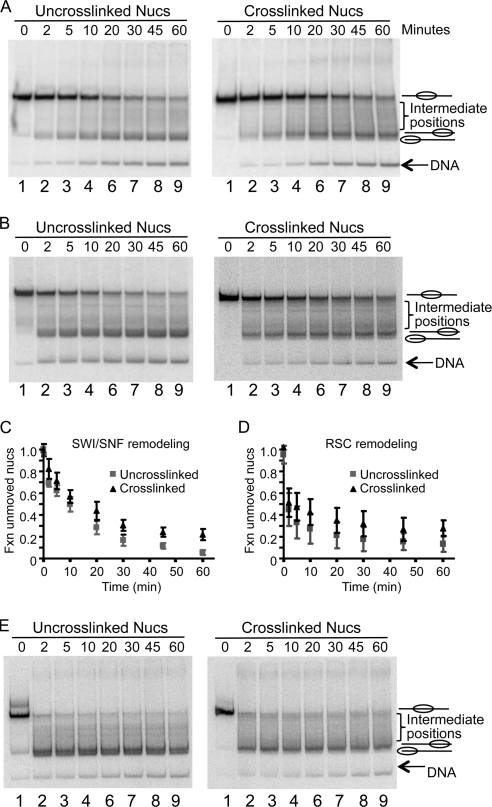
Fig. 4.
Intranucleosome cross-linking inhibits nucleosome mobilization by SWI/SNF. Nucleosomes cross-linked via the N-terminal tail domains of H2A and un-cross-linked controls were remodeled by incubation with 1 nM SWI/SNF, and translational positions were analyzed on 4% polyacrylamide translational gels as described in Materials and Methods. (A and B) Remodeling of un-cross-linked and cross-linked nucleosomes assembled on 244-bp and 215-bp 5S DNA fragments, respectively (A) or on the 343-bp 601 DNA fragment (B). Lanes 1, 3, 5, 7, and 9 show un-cross-linked 244-bp control nucleosomes, while lanes 2, 4, 6, 8, and 10 contain cross-linked nucleosomes on the 215-bp fragment. Samples were incubated either in TE (lanes 1 and 2), in remodeling buffer alone (lanes 3 and 4), or in remodeling buffer and ATP alone (lanes 5 and 6), SWI/SNF alone (lanes 7 and 8), or both SWI/SNF and ATP (lanes 9 and 10). Nucleosome positions represented by each of the bands are shown beside the gel. Lanes 11 and 12 in panel A show un-cross-linked nucleosomes on the 215-bp 5S template before and after remodeling, respectively. (C) Exonuclease III mapping of nucleosome positions before and after RSC remodeling. Remodeling reactions with nucleosomes containing H2A G2C and the 343-bp 601 DNA fragment were stopped by addition of 300 ng CT DNA and then subjected to Exo III digestion for 5 min, and the products were analyzed by sequencing gel electrophoresis and autoradiography. Lanes 1 and 2, unremodeled control nucleosomes incubated with or without 0.25 unit of Exo III, respectively; lanes 3 and 4, remodeled control nucleosomes incubated with 0.25 or 0.5 unit of Exo III; lanes 5 and 6, unremodeled cross-linked nucleosomes incubated with or without 0.25 unit of Exo III; lanes 7 and 8, remodeled, cross-linked nucleosomes incubated with 0.25 or 0.5 unit of Exo III, respectively.
We also examined nucleosome mobilization on the 343-bp 601 DNA fragment (Fig. 1B). Notably, nucleosomes containing N-terminally cross-linked H2As had translational positioning indistinguishable from those of un-cross-linked nucleosomes (Fig. 4B, lanes 1 and 2). Similar to what was found for the 5S nucleosomes, incubation with remodeling buffer or buffer containing SWI/SNF or ATP alone did not result in significant changes in the migration of the nucleosomes in the gel (Fig. 4B, lanes 1 to 8). However, incubation of un-cross-linked nucleosomes in the presence of both SWI/SNF and ATP resulted in efficient conversion to species with substantially faster electrophoretic mobility on the gel, consistent with a change of nucleosome positions from the center to the ends of the DNA templates (Fig. 4B, lane 9). Interestingly, two main remodeled products with closely related translational positions near the ends of the DNA fragment were observed on the gel. Previous work suggests that the lower band corresponds to a position abutting or shifted off the end of the DNA fragment (21), while the slower-migrating band corresponds to a related, more interior end position. We confirmed this assignment by restriction enzyme (data not shown) and exonuclease III (Exo III) mapping. Prior to remodeling a single Exo III-resistant band is detected, consistent with a nucleosome at the expected location at the center of the fragment (Fig. 4C, lane 1). After remodeling, Exo III barely trims the end of the DNA (Fig. 4C, compare lanes 2 and 3), while more-extensive digestion reveals a second digestion pause, about 20 bp further in from the end of the fragment (lane 4).
In contrast, when cross-linked 601 nucleosomes were remodeled, about 35% of nucleosomes retained their original centrally located translational positions, without any apparent movement, while ∼10% of the remodeled nucleosomes migrated at intermediate positions between the center and end-positioned nucleosomes (Fig. 4B, lane 10). Only about one-half of the cross-linked nucleosomes were converted to rapidly migrating species corresponding to positions near the end of the DNA fragment. Notably, only a small percentage (5%) of the nucleosomes were converted to the most rapidly migrating species, while 50% maintained the more interior position at the end of the DNA fragment (Fig. 4B, lane 10). Exo III mapping shows that, while cross-linking does not alter the location of the unremodeled nucleosome (Fig. 4C, lanes 1 and 5), the digestion pause after remodeling at the extreme end of the DNA fragment is considerably weakened (Fig. 4C, compare lanes 3 and 7). More-extensive Exo III digestion reveals that the majority of the nucleosomes are located at a more interior position, with much weaker protection at the extreme end of the fragment (Fig. 4C, lanes 4 and 8). These results demonstrate that the nucleosome mobilization activity of SWI/SNF is restricted by the H2A-H2A N-terminal intranucleosome cross-linking and that the cross-linking nearly completely inhibits the movement of nucleosomes to the endmost position on the 601 DNA fragment.
To further analyze the extent of remodeling in the un-cross-linked and cross-linked nucleosomes, time course experiments were performed with increased amounts of enzyme. Nucleosomes were incubated with SWI/SNF and ATP, and reactions were stopped by adding excess calf thymus DNA. Results were analyzed on native translational gels as above. Remodeling of un-cross-linked nucleosomes resulted in accumulation of end positions with increasing time such that almost 95% of un-cross-linked nucleosomes were mobilized to the DNA ends in 60 min, with the vast majority of nucleosomes moved to the most rapidly migrating species, corresponding to the endmost position on the DNA fragment (Fig. 5A, left). In contrast, cross-linked nucleosomes were mobilized to a reduced extent and, strikingly, accumulated at the interior position near the end of the fragment but did not advance to the extreme endmost location (Fig. 5A, right). A plot of the amount of nucleosomes in the original, unmobilized position shows that, while about 5% of the un-cross-linked nucleosomes remained at the central location after 60 min of remodeling, ∼20% of the cross-linked nucleosomes remained at the central location (Fig. 5C).
Fig. 5.
Cross-linking inhibits remodeling-dependent nucleosome mobilization to a position off the end of the DNA fragment. (A) Nucleosomes were reconstituted on the 343-bp 601 DNA fragment with H2A G2C, and un-cross-linked and cross-linked nucleosomes (0.5 nM) were remodeled with 3 nM SWI/SNF. Reactions were stopped at various times by addition of competitor DNA, and products were analyzed on nondenaturing 4% polyacrylamide translational gels. Assigned translational positions relative to the DNA are shown on the right of the gels. (B) As in panel A except that remodeling was carried out with 0.1 nM RSC complex. (C and D) The bands corresponding to the original centrally located nucleosomes in panels A and B, respectively, were quantified and plotted versus remodeling time. (E) Remodeling of un-cross-linked and cross-linked nucleosomes containing H2A G2C with saturating RSC. Reaction mixtures containing 0.5 nM RSC were incubated at 30°C for 5 min, and the remodeling reactions were initiated by addition of 0.8 μM ATP. Reactions were stopped at the indicated times, and samples were analyzed as described for panel A.
To assess whether cross-linking would similarly restrict the nucleosome mobilization activity of RSC, similar time course experiments were performed. Compared to remodeling by SWI/SNF, nucleosomes were remodeled by RSC more rapidly in the first 10 min (Fig. 5B, lanes 2 to 4), but both remodelers mobilized nucleosomes to apparently the same positions, as shown in the translational gels (Fig. 5, compare panels A and B). Importantly, we found that cross-linking has the same effect on RSC nucleosome mobilization activity as that observed with SWI/SNF, whereby cross-linking results in almost a complete inhibition of mobilization to the extreme end position (Fig. 5B, compare lanes 9). Likewise, a similar effect on the amount of nucleosomes mobilized from the original central position is observed (Fig. 5D). Interestingly, the effect of cross-linking on remodeling-dependent mobilization to the extreme end position was also detected by Exo III analysis. Mapping of cross-linked nucleosome locations after remodeling revealed a diminution of the extreme end position (Fig. 4C, lane 8, open circle), with the majority of nucleosomes mapping to the more interior position (Fig. 4C, lane 8, solid circle). These results indicate that both complexes are similarly affected by intranucleosome cross-linking and are consistent with the idea that both employ similar mechanisms to induce nucleosome movement.
To determine whether the incomplete mobilization of cross-linked nucleosomes to the extreme end position was due to slower kinetics or a physical block to mobilization, we examined nucleosome mobilization with saturating amounts of RSC. Under such conditions, nearly complete movement of cross-linked and un-cross-linked nucleosomes was achieved at very early time points (Fig. 5E). Significantly, remodeled un-cross-linked nucleosomes exhibited the two translational positions close to the DNA ends, as discussed above (Fig. 5E, left). However, the cross-linked nucleosomes were not moved to the extreme end position, even with saturating amounts of RSC, after long incubation times (Fig. 5E, right).
Likewise, experiments with 601 and 5S nucleosome H2As cross-linked via the C-terminal tail domains show that a constraining loop around the DNA near the dyad axis also restricts nucleosome mobilization in a manner similar to that observed with nucleosomes containing N-terminally cross-linked H2A. Cross-linked nucleosomes are mobilized from the central position to end positions on the 601 fragment at a slower rate, and cross-linking restricts mobilization to the extreme end position (data not shown).
Inhibition of nucleosome mobilization is not due to addition of a bulky adduct to the H2A tail domain.
An alternative interpretation of the differences in mobility between remodeled cross-linked and un-cross-linked nucleosomes is that cross-linking with BM[PEO]4 results in restricted dimer eviction or histone dissociation. To address this possibility, nucleosomes were analyzed on 0.7% agarose nucleoprotein gels before and after RSC remodeling reactions. The tetramer, hexamer, and other subnucleosomal species exhibit distinct migrations on these gels, but nucleosomes with different translational positions have similar mobilities on agarose gels. Samples of remodeled and unremodeled nucleosomes were split and loaded onto both acrylamide and agarose gels after addition of CT DNA to dissociate SWI/SNF (Fig. 6A and B), and the results were analyzed by phosphorimaging. While remodeled and unremodeled nucleosomes exhibit distinct migration patterns on the acrylamide gel, as observed in previous experiments, no difference is detected on the agarose gel. Moreover, remodeled un-cross-linked and cross-linked nucleosomes exhibited identical electrophoretic mobilities, even compared to the unremodeled particles on the agarose gel (Fig. 6B). These results indicate that remodeling does not result in significant dissociation of the histone octamer, consistent with a recent report that such activity is minimal on strong nucleosome positioning sequences (54). Moreover, cross-linking clearly does not result in a reduction in histone dissociation from the remodeled species.
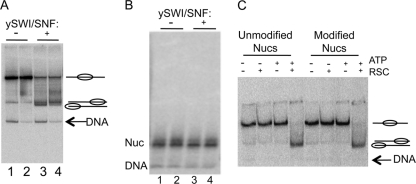
Fig. 6.
Differences in remodeling products are not due to differential histone loss. (A and B) N-terminally cross-linked and un-cross-linked nucleosomes were remodeled with 3 nM SWI/SNF for 60 min. Reactions were terminated by addition of competitor DNA, and samples were split and run on the native acrylamide (A) and agarose (B) gels. Lanes 1 and 3, un-cross-linked nucleosomes; lanes 2 and 4, cross-linked nucleosomes. Nucleosomes in lanes 3 and 4 were remodeled by SWI/SNF, as indicated. (C) Modification without cross-linking of nucleosomes containing H2A G2C does not inhibit nucleosome mobilization. Nucleosomes were modified with fluorescein-maleimide and then subjected to RSC remodeling as indicated. Products were analyzed on 4% acrylamide nucleoprotein gels as described in Materials and Methods.
A second possibility is that the altered movement of cross-linked nucleosomes is due to the presence of the bulky cross-linking modification itself. To investigate this possibility, H2A G2C in reconstituted nucleosomes was site-specifically modified by reacting with fluorescein-5-maleimide (FM). FM efficiently reacts with free sulfhydryl groups within proteins and has a molecular mass of 427 kDa, similar to that of the cross-linker BM[PEO]4. Thus, modification with FM mimics the addition of BM[PEO]4 but without resulting in H2A-H2A cross-linking. Nucleosomes containing H2A G2C were modified with FM and then radiolabeled with [γ-32P]ATP, and the labeled nucleosomes were purified by 5 to 30% glycerol gradients as described above. The alterations of nucleosome translational positions induced by RSC remodeling were characterized by electrophoresis on 4% native translational gels. As shown in Fig. 6C, FM-labeled and unlabeled nucleosomes are efficiently mobilized by RSC and show exactly the same migration properties on the gel, indicating that the site-specific modification does not interfere with the nucleosome mobilization activity of RSC. Thus, the apparent inability of SWI/SNF and RSC to completely mobilize nucleosomes to the very end position on the DNA fragment is unlikely to be due to simple inhibition by the presence of a bulky modification on the H2A tail domains, but rather appears to be due to the presence of a bona fide cross-link.
DISCUSSION
We find that nucleosomes containing protein loops that restrict DNA from separating from the surface of the histone octamer at two locations on the nucleosome do not significantly interfere with exposure of internal DNA sites, as measured by restriction enzyme accessibility assays, but do impair mobilization of the bulk of nucleosomes by SWI/SNF or RSC. When considered with recent evidence that these enzymes translocate DNA within the nucleosome and form internal DNA loops, our results place strict requirements on models for the mechanism of nucleosome remodeling by these enzymes.
Recent work has demonstrated that DNA is likely drawn into the nucleosome by a translocase activity that contacts nucleosomal DNA near superhelix position +2.5 (40, 42, 58) and that RSC nucleosome remodeling intermediates with protruding DNA loops that are resolved in some manner to yield mobilized nucleosomes can be observed (43). Loop formation is believed to result in increased accessibility to restriction enzymes (34) and can occur prior to mobilization (43). A simple model posits that internal DNA loops move around the nucleosome via a wavelike motion to effect advancement of the histone octamer along the DNA (14, 26). However, cross-linking the N- or C-terminal tail domains together results in constraints at superhelix location (SHL) positions ±4 and ∼0 (dyad), respectively, that apparently hinder but do not block nucleosome mobilization. Thus, our results indicate that free diffusion or directed motion of a DNA loop or bubble is not a primary mechanism by which DNA loops progress about the histone octamer surface, as the cross-linked H2A tail domains would prohibit loops from freely passing these restriction points.
Interestingly, our REA analyses indicate that the rate of initial loop formation is likely not affected by the cross-linking, suggesting that the inhibition we observe occurs at a step after initial loop formation. Moreover, the fact that we observe a reduction in the rate of mobilization but not a total block to movement suggests that cross-linking does not grossly alter the remodeling pathway. We therefore propose that loops do not diffuse about the exterior of the nucleosome but rather feed through specific restriction points by threading past fixed constrictions (Fig. 7). An important aspect of this model is that initial loops feed formation of secondary loops by sliding or threading of the DNA through constrictions on the surface of nucleosomes. We propose that such constrictions are a natural feature of nucleosome remodeling and are perhaps due to sites of association of the native tail domains with the DNA on the exterior of the nucleosome.
Fig. 7.
Threading model for movement of DNA vis-à-vis the histone octamer during SWI/SNF or RSC nucleosome remodeling. The model indicates that the remodeling complex induces formation of an internal loop, perhaps by unwrapping and rewrapping of DNA beyond the original register. The loop then feeds into a second loop by shuttling along the surface of the nucleosome at a constriction point (red lines) provided by the H2A tail domains. Further threading, perhaps driven by the remodeling complex, works the extra DNA in the loop through the body of the nucleosome. Superhelix locations 0 to 7 are indicated in the first cartoon, with the dyad axis of the nucleosome oriented horizontally. Primary contacts of the remodeler with nucleosome DNA are indicated in purple and orange (58). DBD, DNA binding domain.
For example, both N-terminal tail domains are intimately associated with nucleosomal DNA at SHL position ±4 and involve contacts to the DNA backbone by four basic residues (two arginines and two lysines) within the 12-residue domain (33). Thus, it is possible that the tail domains serve to maintain tight association of the nucleosome DNA with the surface of the histone octamer, while loops form between tail domain contact points. It is unclear how many of the tail domains provide such constraint, but it is interesting to note that, of these domains, the H4 and H2A N-terminal tails exit the nucleosome over/under the two superhelical gyres, while the H3 and H2B tail domains exit between the gyres, perhaps resulting in less constraint. In addition it is possible that association of the ATP-dependent translocase activity with nucleosome DNA near superhelix location ±2 also provides a constraint (Fig. 7, orange bar), forcing the loop to extrude at about SHL position ±3.5.
We also note that the effect of cross-linked loops on nucleosome mobilization to the most extreme end position observed in the control reactions is not simply due to the presence of a bulky modification on the H2A tail domains or a lack of H2A/H2B dimer dissociation from the remodeled nucleosomes. This result is consistent with a recent study, which showed that octamer ejection or dimer displacement activities of remodelers are minimal on nucleosomes containing robust nucleosome position elements (44, 54). The effects of cross-linking on nucleosome mobilization to the extreme end location may be related to the threading mechanism we propose. It is possible that a larger threshold amount of DNA contained within a primary loop is required to thread past the enhanced restriction provided by the cross-linked loops than is required for the un-cross-linked tail domains. Thus, we envision that, as the nucleosome reaches the extreme end of the fragment, insufficient DNA remains to accumulate to the threshold loop size, thereby resulting in the nucleosome remaining at the penultimate position on the fragment. Alternatively, the presence of the cross-link may emphasize a requirement for remodeler-dependent lifting of the proximal linker DNA, consistent with an unwinding-recapture mechanism for initial loop formation (15). Finally, it is important to note that Exo III trimming denotes the most exterior DNA location in contact with the histone octamer, and therefore an alternative possibility is that cross-linking affects retention of an internal DNA loop (43).
As mentioned above, we also observe that H2A tail-H2A tail cross-linking does not alter remodeling-dependent REA. In prior work, we demonstrated that a single covalent cross-link between nucleosome DNA and a core histone was sufficient to block remodeling-dependent increases in REA at distant sites in the nucleosome, suggesting that initial internal loop formation involves concerted reorganization of histone-DNA interactions throughout the nucleosome (2). While histone-DNA cross-linking restricts both separation of DNA from the histone surface and lateral movement of the DNA with respect to the histone protein, the cross-linking in the current work provides only the former impediment. Moreover, a recent careful analysis of the activity of a minimal RSC translocase complex on naked DNA indicates a distinct rate-limiting initiation phase, which may involve multiple small steps of translocation leading to initial DNA loop formation (45), while ACF translation may involve step sizes as small as 3 bp (6). Thus, remodelers may be designed to thread small segments of DNA past constriction points on the nucleosome surface.
ACKNOWLEDGMENTS
We thank Blaine Bartholomew for the kind gift of purified RSC complex.
This work was supported by NIH grants GM-52426 to J.J.H. and GM-49650 to C.L.P.
Footnotes
Published ahead of print on 22 August 2011.
REFERENCES
- 1. Aoyagi S., Hayes J. J. 2002. hSWI/SNF-catalyzed nucleosome sliding does not occur solely via a twist diffusion mechanism. Mol. Cell. Biol. 22:7484–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aoyagi S., et al. 2002. Nucleosome remodeling by the human SWI/SNF complex requires transient global disruption of histone-DNA interactions. Mol. Cell. Biol. 22:3653–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aoyagi S., Wade P. A., Hayes J. J. 2003. Nucleosome sliding catalyzed by the xMi-2 complex does not occur exclusively via a simple twist-diffusion mechanism. J. Biol. Chem. 278:30562–30568 [DOI] [PubMed] [Google Scholar]
- 4. Bazett-Jones D. P., Côté J., Landel C. C., Peterson C. L., Workman J. L. 1999. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol. Cell. Biol. 19:1470–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker P. B. 2002. Nucleosome sliding: facts and fiction. EMBO J. 21:4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blosser T. R., Yang J. G., Stone M. D., Narlikar G. J., Zhuang X. 2009. Dynamics of nucleosome remodelling by individual ACF complexes. Nature 462:1022–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cairns B. R., et al. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249–1260 [DOI] [PubMed] [Google Scholar]
- 8. Chai B., Huang J., Cairns B. R., Laurent B. C. 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19:1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clapier C. R., Cairns B. R. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273–304 [DOI] [PubMed] [Google Scholar]
- 10. Côté J., Quinn J., Workman J. L., Peterson C. L. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53–60 [DOI] [PubMed] [Google Scholar]
- 11. Damelin M., et al. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 9:563–573 [DOI] [PubMed] [Google Scholar]
- 12. Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319:1097–1113 [DOI] [PubMed] [Google Scholar]
- 13. Flaus A., Martin D. M., Barton G. J., Owen-Hughes T. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34:2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flaus A., Owen-Hughes T. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14:165–173 [DOI] [PubMed] [Google Scholar]
- 15. Flaus A., Owen-Hughes T. 2003. Mechanisms for nucleosome mobilization. Biopolymers 68:563–578 [DOI] [PubMed] [Google Scholar]
- 16. Gangaraju V. K., Bartholomew B. 2007. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 618:3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guschin D., Wade P. A., Kikyo N., Wolffe A. P. 2000. ATP-Dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry 39:5238–5245 [DOI] [PubMed] [Google Scholar]
- 18. Hamiche A., Sandaltzopoulos R., Gdula D. A., Wu C. 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97:833–842 [DOI] [PubMed] [Google Scholar]
- 19. Holstege F. C., et al. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717–728 [DOI] [PubMed] [Google Scholar]
- 20. Jaskelioff M., Gavin I. M., Peterson C. L., Logie C. 2000. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 20:3058–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kassabov S. R., Zhang B., Persinger J., Bartholomew B. 2003. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell 11:391–403 [DOI] [PubMed] [Google Scholar]
- 22. Kent N. A., Chambers A. L., Downs J. A. 2007. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J. Biol. Chem. 282:27693–27701 [DOI] [PubMed] [Google Scholar]
- 23. Kingston R. E., Narlikar G. J. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339–2352 [DOI] [PubMed] [Google Scholar]
- 24. Krebs J. E., Fry C. J., Samuels M. L., Peterson C. L. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587–598 [DOI] [PubMed] [Google Scholar]
- 25. Kulic I. M., Schiessel H. 2003. Chromatin dynamics: nucleosomes go mobile through twist defects. Phys. Rev. Lett. 91:148103. [DOI] [PubMed] [Google Scholar]
- 26. Kulic I. M., Schiessel H. 2003. Nucleosome repositioning via loop formation. Biophys. J. 84:3197–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee K. M., Hayes J. J. 1998. Linker DNA and H1-dependent reorganization of histone-DNA interactions within the nucleosome. Biochemistry 37:8622–8628 [DOI] [PubMed] [Google Scholar]
- 28. Lee K. M., Hayes J. J. 1997. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proc. Natl. Acad. Sci. U. S. A. 94:8959–8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lia G., et al. 2006. Direct observation of DNA distortion by the RSC complex. Mol. Cell 21:417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu N., Balliano A., Hayes J. J. 2011. Mechanism(s) of SWI/SNF-induced nucleosome mobilization. Chembiochem 12:196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu N., Hayes J. J. Preparation of nucleosomes containing a specific H2A-H2A crosslink forming a DNA-constraining loop structure. Methods Mol. Biol., in press [DOI] [PubMed] [Google Scholar]
- 32. Logie C., Peterson C. L. 1997. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 16:6772–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 34. Narlikar G. J., Phelan M. L., Kingston R. E. 2001. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell 8:1219–1230 [DOI] [PubMed] [Google Scholar]
- 35. Peterson C. L. 2000. ATP-dependent chromatin remodeling: going mobile. FEBS Lett. 476:68–72 [DOI] [PubMed] [Google Scholar]
- 36. Peterson C. L., Herskowitz I. 1992. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68:573–583 [DOI] [PubMed] [Google Scholar]
- 37. Peterson C. L., Laniel M. A. 2004. Histones and histone modifications. Curr. Biol. 14:R546–R551 [DOI] [PubMed] [Google Scholar]
- 38. Polach K. J., Widom J. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130–149 [DOI] [PubMed] [Google Scholar]
- 39. Saha A., Wittmeyer J., Cairns B. R. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16:2120–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saha A., Wittmeyer J., Cairns B. R. 2005. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 12:747–755 [DOI] [PubMed] [Google Scholar]
- 41. Schnitzler G. R., et al. 2001. Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips. Mol. Cell. Biol. 21:8504–8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwanbeck R., Xiao H., Wu C. 2004. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J. Biol. Chem. 279:39933–39941 [DOI] [PubMed] [Google Scholar]
- 43. Shukla M. S., et al. 2010. Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer. Proc. Natl. Acad. Sci. U. S. A. 107:1936–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shundrovsky A., Smith C. L., Lis J. T., Peterson C. L., Wang M. D. 2006. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat. Struct. Mol. Biol. 13:549–554 [DOI] [PubMed] [Google Scholar]
- 45. Sirinakis G., et al. 2011. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 30:2364–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith C. L., Horowitz-Scherer R., Flanagan J. F., Woodcock C. L., Peterson C. L. 2003. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 10:141–145 [DOI] [PubMed] [Google Scholar]
- 47. Smith C. L., Peterson C. L. 2005. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65:115–148 [DOI] [PubMed] [Google Scholar]
- 48. Strahl B. D., Allis C. D. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 49. Strohner R., et al. 2005. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat. Struct. Mol. Biol. 12:683–690 [DOI] [PubMed] [Google Scholar]
- 50. van Holde K., Yager T. 2003. Models for chromatin remodeling: a critical comparison. Biochem. Cell Biol. 81:169–172 [DOI] [PubMed] [Google Scholar]
- 51. Whitehouse I., et al. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784–787 [DOI] [PubMed] [Google Scholar]
- 52. Whitehouse I., Stockdale C., Flaus A., Szczelkun M. D., Owen-Hughes T. 2003. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell. Biol. 23:1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Winston F., Carlson M. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387–391 [DOI] [PubMed] [Google Scholar]
- 54. Yang X., Zaurin R., Beato M., Peterson C. L. 2007. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat. Struct. Mol. Biol. 14:540–547 [DOI] [PubMed] [Google Scholar]
- 55. Yang Z., Zheng C., Hayes J. J. 2007. The core histone tail domains contribute to sequence-dependent nucleosome positioning. J. Biol. Chem. 282:7930–7938 [DOI] [PubMed] [Google Scholar]
- 56. Zhang Y., Reinberg D. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343–2360 [DOI] [PubMed] [Google Scholar]
- 57. Zhang Y., et al. 2006. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell 24:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zofall M., Persinger J., Kassabov S. R., Bartholomew B. 2006. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 13:339–346 [DOI] [PubMed] [Google Scholar]