Abstract
The histone variant macroH2A1 contains a carboxyl-terminal ∼30-kDa domain called a macro domain. MacroH2A1 is produced as one of two alternatively spliced forms, macroH2A1.1 and macroH2A1.2. While the macro domain of macroH2A1.1 can interact with NAD+-derived small molecules, such as poly(ADP-ribose), macroH2A1.2's macro domain cannot. Here, we show that changes in the alternative splicing of macroH2A1 pre-mRNA, which lead to a decrease in macroH2A1.1 expression, occur in a variety of cancers, including testicular, lung, bladder, cervical, breast, colon, ovarian, and endometrial. Furthermore, reintroduction of macroH2A1.1 suppresses the proliferation of lung and cervical cancer cells in a manner that requires the ability of macroH2A1.1 to bind NAD+-derived metabolites. MacroH2A1.1-mediated suppression of proliferation occurs, at least in part, through the reduction of poly(ADP-ribose) polymerase 1 (PARP-1) protein levels. By analyzing publically available expression and splicing microarray data, we identified splicing factors that correlate with alterations in macroH2A1 splicing. Using RNA interference, we demonstrate that one of these factors, QKI, regulates the alternative splicing of macroH2A1 pre-mRNA, resulting in increased levels of macroH2A1.1. Finally, we demonstrate that QKI expression is significantly reduced in many of the same cancer types that demonstrate a reduction in macroH2A1.1 splicing.
INTRODUCTION
Similar to the covalent modification of histones, the replacement of canonical histones by histone variants specifies functional differences between chromatin domains (46). A group of H2A-type histone variants (macroH2A1.1, macroH2A1.2, and macroH2A2), collectively referred to as macroH2As, contain an amino-terminal histone-like region and a carboxyl-terminal ∼30-kDa globular “macro” domain. MacroH2A1 is the founding member of a large family of macro domain-containing proteins, members of which can be found from bacteria to humans and in a family of RNA viruses called coronaviruses. In the human genome, 10 genes encode macro domains (33). Often, these domains occur together with other conserved domains and enzymatic activities. Specifically, macro domains are found on histone variants, SNF2-like ATP-dependent chromatin-remodeling enzymes, poly(ADP-ribose) polymerases (PARPs), and sirtuin-type lysine deacetylases. While certain macro domains have been shown to have ADP-ribose (ADPR)-1″-monophosphate phosphatase activity (36), most macro domains are considered ligand-binding domains for NAD+-derived second messengers, including poly(ADP-ribose) (PAR), ADPR, and O-acetyl-ADPR, which are produced by poly(ADP-ribose) polymerases, poly(ADP-ribose) glycohydrolase, and the sirtuin family of lysine deacetylases, respectively (30, 47). Through their ability to bind poly(ADP-ribose), a posttranslational modification, macro domain-containing proteins can be recruited to sites of poly(ADP-ribos)ylated proteins in the nucleus (2, 26, 47).
Even though the histone-like region of macroH2A has only ∼64% identity with canonical H2A, it participates in the formation of a nucleosome core particle with only subtle structural differences from canonical nucleosomes (1, 9, 10). Indeed, what makes macroH2A-containing nucleosomes unique is the large globular macro domain that emerges from the nucleosome near the dyad (4). In vitro biochemical ex- periments have demonstrated that the incorporation of macroH2A1 into nucleosomes affects the ability of some transcription factors to bind their sequences and alters the propensity and determinants for remodeling by ATP-dependent chromatin-remodeling enzymes (3, 4, 11).
The in vivo functions of macroH2A variants have been more difficult to characterize. While early work focused on the role macroH2A plays in X inactivation (18, 28, 49), work from our group and others demonstrated that macroH2A1 is not only enriched on the Xi but is also found in large chromatin domains on all autosomes (6, 12, 13, 23). These macroH2A1-containing domains can be hundreds of kilobases long and occupy roughly a quarter of the human genome (23). Furthermore, while most studies on macroH2A1 have focused on its role in transcriptional repression, our recent work has demonstrated that genes present in macroH2A1-containing domains can be either positively or negatively regulated by macroH2A1 in a context-specific manner (23, 24).
The expression of macroH2A variants is regulated during development. Embryonic stem cells and the early embryo exclusively express macroH2A1.2. But, as differentiation continues during development, macroH2A1.1 and macroH2A2 expression are upregulated (19, 38). The developmental regulation of macroH2A variant expression is consistent with reports implicating macroH2A in the regulation of developmental stage and tissue-specific gene expression (6, 12, 23). However, the specific contribution of individual macroH2A variants in regulating developmental or tissue-specific patterns of gene expression is currently unknown.
One interesting feature that distinguishes the three macroH2A subtypes is their differential ability to bind ADPR-based ligands. Both macroH2A1.2 and macroH2A2 are incapable of binding ADPR and related molecules, leaving macroH2A1.1 as the only macroH2A capable of interacting with these small molecules (35, 47). MacroH2A1.1 and macroH2A1.2 are produced by alternative splicing from the same gene, H2AFY, in a process termed mutually exclusive splicing where each transcript contains a unique exon (Fig. 1A and B). The macroH2A1.1-specific exon encodes part of the macro domain ligand-binding pocket critical for the binding of ADPR-based ligands (35). Substitution of this exon for the macroH2A1.2-specific exon leads to changes in the ligand-binding pocket of the macro domain that are incompatible with binding ADPR. While alternative splicing regulates the ability of macroH2A1 to interact with ADPR-based molecules, the factors that regulate the splicing of macroH2A1 and the biological role of ligand binding to macroH2A1.1 are largely unknown.
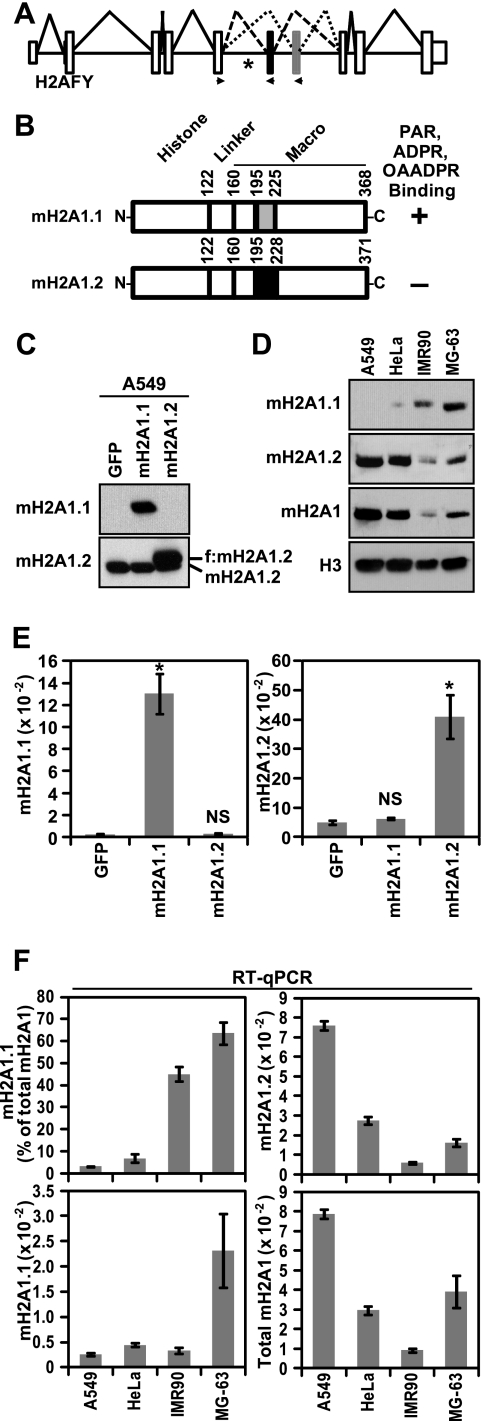
Fig. 1.
Differences in macroH2A1 alternative splicing and protein levels across four cell lines. (A) Diagram depicting the structure and splicing of the gene encoding macroH2A1, H2AFY. The small white boxes, the large white boxes and the solid horizontal lines represent untranslated regions, coding exons and introns, respectively. The macroH2A1.2- and macroH2A1.1-specific exons are black and gray, respectively. Solid, dashed, and dotted lines depict constitutive and macroH2A1.2 or macroH2A1.1 alternative splicing events, respectively. Locations of expression primers used for splice variant-specific qPCR are indicated with arrowheads. The location of a QKI binding site is indicated by an asterisk (27). (B) Schematic depicting the two alternatively spliced macroH2A1 variants. The locations of amino acids encoded by the variable exons are indicated by gray and black boxes for macroH2A1.1 (mH2A1.1) and macroH2A1.2 (mH2A1.2), respectively. Amino acid positions marking the boundaries of the histone-like domain, the linker region, the variable exons, and the macro domain are indicated. The plus and minus signs indicate the affinity or lack of affinity for ADPR-based ligands, respectively. OAADPR, O-acetyl-ADPR. (C) The specificity of anti-macroH2A1.1 and anti-macroH2A1.2 antibodies was determined by immunoblots of A549 cells expressing GFP, Flag-tagged macroH2A1.1, or Flag-tagged macroH2A1.2 (f:mH2A1.2). The location of endogenous and ectopic macroH2A1 is indicated. (D) Immunoblots demonstrating the differences in macroH2A1.1, macroH2A1.2, and total macroH2A protein levels in four cell lines. Anti-histone H3 was used as a loading control. (E) Histograms depicting the specificity of the macroH2A1.1 and macroH2A1.2 primer sets in the RT-qPCR assay with A549 cells expressing the indicated factors as described for panel C. Error bars represent standard errors of the means (SEMs) for four biological replicates. P values are the result of a two-tailed Student's t test. *, P < 0.001; NS, not significant. (F) Histograms depicting the macroH2A1.1 and macroH2A1.2 levels across four cell lines. The data are expressed as either ACTB normalized (mH2A1.1 and mH2A1.2), the sum of macroH2A1.1 and macroH2A1.2 (Total mH2A1), or the percentage of total macroH2A1 that has been spliced as macroH2A1.1 [mH2A1.1 (% of total mH2A)] as indicated. Error bars represent the SEMs for three biological replicates.
Two recent reports have implicated altered expression of macroH2A variants in oncogenesis. The first demonstrates that reduction of macroH2A1.1 protein levels is negatively associated with lung cancer recurrence (43). The second shows that macroH2A1 and macroH2A2 expression is often silenced in malignant melanoma (29). While the functional role of macroH2A expression changes in lung cancer was not determined, in melanoma cells, the loss of expression of macroH2A isoforms leads to increased proliferation and metastatic capacity (29, 43).
In this report, we demonstrate that the expression of macroH2A1.1 is reduced in several types of cancer due to changes in the alternative splicing of macroH2A1 pre-mRNA. Furthermore, we demonstrate that macroH2A1.1 regulates the proliferation of lung and cervical cancer cells, at least in part, by reducing the levels of PARP-1. Bioinformatics analysis of available microarray splicing and expression data allowed us to identify putative macroH2A1 splicing regulators. We demonstrate that one of these splicing factors, QKI, enhances the splicing of macroH2A1.1. Furthermore, we show that QKI expression is repressed in many of the same cancer types in which macroH2A1.1 splicing is downregulated.
MATERIALS AND METHODS
Cell lines and antibodies.
A549 human lung cancer cells and HeLa cervical adenocarcinoma cells were obtained from Susan B. Horwitz and Hayley M. McDaid. MG-63 (CRL-1427) human osteosarcoma cells and IMR90 (CCL-186) primary human fetal lung fibroblast cells were obtained from ATCC. A549 and HeLa cells were maintained in RPMI supplemented with 10% fetal bovine serum. MG-63 and IMR90 cells were maintained in minimal essential medium (MEM) supplemented with 10% fetal bovine serum. A549 and HeLa cell lines expressing either green fluorescent protein (GFP) (as a control), macroH2A1.2, macroH2A1.1, or macroH2A1.1 point mutants G224E or G314E were generated by retroviral-mediated gene transfer using the pQCXIP expression system (Clontech) followed by puromycin drug selection. The GFP-m1.1 construct was made by replacing the histone region of macroH2A1.1 (amino acids 1 to 120) with GFP. All macroH2A1 constructs, including GFP-m1.1, harbor a carboxyl-terminal Flag tag. IMR90 cells were immortalized by using retroviral-mediated gene transfer of the hTERT gene (Addgene plasmid 1773) (20) using the pBABE expression system and hygromycin selection. QKI-depleted IMR90-hTERT and MG-63 cells were created using retroviral-mediated gene transfer of either or both of two small hairpin RNA (shRNA) sequences targeting distinct regions of the QKI transcript common to all known QKI isoforms by using the pSUPER retro system (Oligoengine) and selection with puromycin and/or G418. The targeting sequences used for the QKI knockdown constructs were 5′-GCTCAGAACAGAGCAGAAATC-3′ and 5′-GCACCTACAGAGATGCCAACA-3′. G418-resistant macroH2A1-depleted IMR90-hTERT cells were created similarly with the targeting sequence 5′-GCAATGCAGCGAGAGACAACA-3′. Control cells expressing shRNA targeting luciferase were generated in parallel (34). The ectopic expression and depletion of factors in these cell lines were tested with immunoblots using antibodies against macroH2A1 (07-219, recognizing both macroH2A1.1 and macroH2A1.2; Millipore), macroH2A1.1 (4160S; Cell Signaling), macroH2A1.2 (4827S; Cell Signaling), Flag M2 (F3165; Sigma), QKI (A300-183A; Bethyl), caspase-3 (9665; Cell Signaling Technology), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (2118; Cell Signaling Technology), and tubulin (E7; Developmental Studies Hybridoma Bank [DSHB], University of Iowa). Acid extraction was used to solubilize histone proteins (e.g., histone H3 and macroH2A1) as described in reference 23.
Separation of soluble and chromatin fractions.
An 80 to 90% confluent 10-cm dish of A549 cells stably expressing ectopic GFP or macroH2A1 variants as described above was lysed in 100 μl of a detergent lysis buffer (10 mM Tris-HCl, pH 7.9, 0.1% Triton X-100, 100 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol [DTT], 1× protease inhibitor cocktail [Roche]). After a 15-min incubation on ice, the lysate was cleared with centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was collected as the soluble fraction. To liberate chromatin-incorporated proteins from the resulting pellet, it was resuspended in 85 μl of micrococcal nuclease (MNase) digestion buffer (50 mM Tris-HCl, pH 7.9, 25 mM KCl, 10 mM CaCl2, 4 mM MgCl2, 12.5% glycerol, 2,000 gel units of MNase [NEB]) and incubated for 20 min at 37°C, followed by the addition of 15-μl of MNase stop buffer (20 mM Tris-HCl, pH 7.9, 200 mM EDTA, 0.05 mg/ml RNase [Roche]). After a 15-min incubation at room temperature, the samples were spun at 10,000 × g for 10 min at 4°C. The resulting supernatant was used in immunoblots as the chromatin fraction.
Gene expression analysis.
Expression data from cell lines was obtained from RNA isolated using TriPure (Roche) according to the manufacturer's protocol. The RNA was reverse transcribed using Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) and an oligo(dT)18 primer. The resulting first-strand cDNA was used as the template for quantitative PCR (qPCR) using gene- or splice variant-specific primers. The relative levels of RNA were determined using the efficiency-corrected threshold cycle (ΔCT) method (40). All expression was normalized to that of β-actin (ACTB). Significant changes in RNA levels and splicing were determined by a two-tailed paired Student's t test. Expression data from the 365 cancer patient samples was generated by qPCR with cDNA from poly(A)-purified and oligo(dT)-reverse-transcribed RNA obtained from Origene (CSRT102). Expression data from 5 additional normal tissue samples was also generated from total RNAs obtained from Origene (CR559621, CR561838, CR560512, CR559978, and CR559889). Additional sample information and pathology reports are available from the Origene website (http://www.origene.com/assets/documents/TissueScan/CSRT102.xls). The percentage of total macroH2A1 transcript spliced as macroH2A1.1 was determined by dividing the efficiency-corrected ACTB-normalized macroH2A1.1 expression value by the corrected and normalized sum of both the macroH2A1.1 and macroH2A1.2 values. The significance of the difference in percent macroH2A1.1 and QKI levels in normal and cancer patient samples was determined using a nonparametric two-tailed Mann-Whitney test. Primer sequences used for qPCR are as follows and are listed in the 5′ to 3′ direction: ACTB forward, AGCTACGAGCTGCCTGAC; ACTB reverse, AAGGTAGTTTCGTGGATGC; macroH2A1 forward, GGCTTCACAGTCCTCTCCAC; macroH2A1.1 reverse, GGTGAACGACAGCATCACTG; macroH2A1.2 reverse, GGATTGATTATGGCCTCCAC; QKI forward, ATTAAACGGTCCCCTGAAGC; and QKI reverse, ATCAACAGCCCAAG TGTGAC.
Cell proliferation assays.
A549, HeLa, IMR90, or MG-63 cells were infected with equivalent amounts of retrovirus encoding GFP (as a control), macroH2A1.1, macroH2A1.2, macroH2A1.1 G224E, macroH2A1.1 G314E, GFP fused to the macro domain of macroH2A1.1, shRNA against macroH2A, or shRNA against QKI. The cells were placed under puromycin selection 24 h postinfection. After 5 to 8 days of drug selection, the cell-counting experiments were performed by plating 5 × 104 cells in 6-well dishes and counting 1 well of each cell line with a hemacytometer (Hausser Scientific) on the days indicated below.
Analysis of Rosetta splicing microarray expression data.
Previously analyzed microarray data for 24,426 alternative splicing events and 18,093 genes across 48 different tissues and cell lines was downloaded from (http://rulai.cshl.edu/Rosetta_AS_supp) (8). The raw data are available from the NCBI's Gene Expression Omnibus (GEO accession number GSE11863). The mutually exclusive splicing that generates macroH2A1.1 and macroH2A1.2 is represented in the original data analysis as “H2AFY_MUTEXE_1” in the alternative splicing data sets. The Spearman correlation and associated P values of the relative changes in macroH2A1.1 splicing to the expression of each gene across the 48 tissues were then determined using scripts custom generated with the Python programming language and the Scipy scientific computing module (script available upon request). Gene Ontology (www.geneontology.org) was then used to identify the significant positively and negatively correlated factors that have a known role in regulating alternative splicing (GO accession number GO.0008380).
RESULTS
MacroH2A1 variant protein levels vary with changes in alternative splicing.
In lung cancer, the macroH2A1.1 protein levels are significantly and specifically reduced compared to the levels of macroH2A1.2 (43). Several factors may contribute to these changes, including altered pre-mRNA alternative splicing and altered stability of the two macroH2A1 variant proteins. To examine the contribution alternative splicing plays in specifying macroH2A1 variant protein levels, we determined the relative levels of macroH2A1.1 and macroH2A1.2 proteins and mRNA across four cell lines, including A549 lung cancer cells, HeLa cervical adenocarcinoma cells, IMR90 primary lung fibroblasts, and MG-63 osteosarcoma cells. First, we determined that the macroH2A1.1 and macroH2A1.2 antibodies were highly specific by immunoblotting protein lysates from A549 cells expressing Flag-tagged macroH2A1.1 or macroH2A1.2 (Fig. 1C). The protein levels of macroH2A1.1 varied from undetectable in A549 cells to highly expressed in MG-63 cells (Fig. 1D). To determine the relative levels of macroH2A1 alternatively spliced mRNAs, we used reverse transcription coupled to quantitative PCR (RT-qPCR) with macroH2A1.1 and macroH2A1.2 splice variant-specific primers (23). To confirm the specificity of the macroH2A1 variant RT-qPCR assay, we tested A549 cells expressing ectopic macroH2A1.1 or macroH2A1.2 (Fig. 1E). We observed that macroH2A1.1 and macroH2A1.2 mRNA levels also varied across the four cell lines tested (Fig. 1F). By calculating the percentage of total macroH2A1 transcript that was contributed by macroH2A1.1, we were able to monitor how macroH2A1 alternative splicing changes across the four cell lines. Comparing the macroH2A1.1 immunoblot (Fig. 1D) with the level of macroH2A1.1 splicing (Fig. 1F, top left), we conclude that monitoring changes in macroH2A1 alternative splicing is a reasonable surrogate for changes in macroH2A1.1 protein levels.
MacroH2A1 splicing is perturbed in several types of cancer.
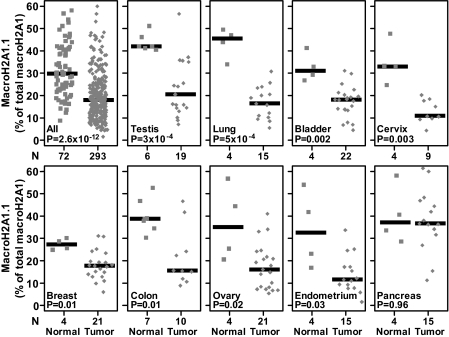
The data presented above led us to hypothesize that the changes in macroH2A1.1 protein levels observed in lung cancer samples (43) were due to changes in alternative splicing of macroH2A1 pre-mRNA. To directly test this hypothesis, we examined samples from both lung cancer patient tumors and normal lung tissue controls (Fig. 2). Similar to the analysis of the cell lines presented in Fig. 1, we determined the macroH2A1.1 and macroH2A1.2 expression and calculated the percentage of total macroH2A1 that exists as macroH2A1.1 as a measure of macroH2A1 alternative splicing. The percentage of macroH2A1 transcript in which the macroH2A1.1-specific exon is included is significantly reduced in the primary lung tumor biopsy specimens compared to that in normal tissue controls (P=5 × 10−4). We expanded the analysis and tested a total of 365 patient samples (293 tumor samples and 72 normal samples) from 17 different tissue types. The combined analysis indicated that the percentage of macroH2A1.1 compared to total macroH2A1 was significantly reduced in the cancer samples compared to the percentage in the normal controls (P=2.6 × 10−12). When each cancer type was analyzed independently, our analysis indicated that splicing of macroH2A1 pre-mRNA was similarly and significantly altered in testicular, lung, urinary bladder, cervical, breast, colon, ovarian, and endometrial cancers compared to that in normal tissue controls (Fig. 2). Not all cancers, however, demonstrated a significant reduction in the macroH2A1.1 alternative splicing (Fig. 2 and Table 1).
Fig. 2.
MacroH2A1 splicing is altered in several cancer types. Splice variant-specific primers were used to determine the percentage of total macroH2A1 transcript that exists as the macroH2A1.1 splice variant in 365 cancer patient samples by RT-qPCR. The squares and diamonds represent data from normal and cancer patient samples, respectively. The x axis represents the number of samples in each group. The solid horizontal bars indicate the median values of particular groups. The reported P values are the results of a two-tailed Mann-Whitney test.
Table 1.
Summary of mean macroH2A1 pre-mRNA alternative splicing and macroH2A1 and QKI mRNA expression in normal and tumor samples from 17 tissue typesa
| Tissue (no. of normal samples, no. of tumor samples) | macroH2A1.1 mRNA |
macroH2A1.2 mRNA [log2(macroH2A1.1/ACTB)] |
QKI mRNA [log2(QKI/ACTB)] |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % of total macroH2A1 |
log2(macroH2A1.1/ACTB) |
|||||||||||
| Normal | Cancer | P value | Normal | Cancer | P value | Normal | Cancer | P value | Normal | Cancer | P value | |
| Esophagus (3, 19) | 18.71 | 17.94 | 1 | −6.09 | −7.51 | 0.52 | −4.87 | −5.35 | 0.26 | −8.96 | −9.83 | 0.29 |
| Thyroid gland (3, 18) | 21.61 | 20.47 | 0.96 | −8.80 | −6.82 | 0.001 | −6.98 | −4.88 | 0.002 | −9.40 | −9.11 | 0.69 |
| Pancreas (4, 15) | 37.10 | 36.61 | 0.96 | −6.63 | −4.49 | 0.08 | −5.48 | −3.55 | 0.05 | −8.44 | −8.38 | 0.89 |
| Liver (3, 15) | 28.23 | 32.14 | 0.65 | −7.2 | −7.12 | 0.82 | −5.87 | −5.98 | 0.65 | −6.59 | −8.27 | 0.02 |
| Lymphoid tissue (3, 31) | 13.68 | 12.28 | 0.49 | −9.28 | −8.41 | 0.15 | −6.45 | −5.57 | 0.10 | −8.76 | −9.34 | 0.30 |
| Stomach (5, 14) | 25.64 | 17.53 | 0.26 | −7.79 | −7.59 | 0.50 | −6.14 | −5.05 | 0.30 | −8.57 | −8.59 | 1.00 |
| Prostate (4, 21) | 23.98 | 18.65 | 0.13 | −6.73 | −7.01 | 0.41 | −4.99 | −4.66 | 0.59 | −8.19 | −8.91 | 0.02 |
| Adrenal gland (5, 10) | 22.77 | 16.79 | 0.13 | −7.23 | −7.43 | 0.95 | −5.28 | −5.09 | 0.13 | −8.51 | −9.08 | 0.13 |
| Kidney (5, 18) | 22.22 | 29.72 | 0.11 | −7.37 | −6.87 | 0.45 | −5.24 | −5.73 | 0.29 | −7.55 | −8.12 | 0.26 |
| Endometrium (4, 15) | 32.46 | 11.54 | 0.03 | −7.25 | −6.49 | 0.47 | −5.86 | −4.20 | 0.0005 | −8.40 | −8.15 | 0.81 |
| Ovary (4, 21) | 34.92 | 16.06 | 0.02 | −7.43 | −7.13 | 0.54 | −6.62 | −4.75 | 0.02 | −7.70 | −9.26 | 0.006 |
| Colon (7, 10) | 38.78 | 15.51 | 0.01 | −7.03 | −6.58 | 0.09 | −6.46 | −4.57 | 0.0001 | −9.34 | −9.99 | 0.04 |
| Breast (4, 21) | 27.18 | 17.69 | 0.01 | −8.09 | −7.28 | 0.30 | −6.69 | −5.10 | 0.03 | −7.61 | −9.39 | 0.0003 |
| Cervix (4, 9) | 33.04 | 10.77 | 0.002 | −6.82 | −7.75 | 0.03 | −5.61 | −4.78 | 0.08 | −8.05 | −9.58 | 0.03 |
| Urinary bladder (4, 22) | 31.08 | 18.08 | 0.001 | −8.64 | −7.58 | 0.10 | −7.66 | −5.15 | 0.0005 | −7.99 | −9.91 | 0.003 |
| Lung (4, 15) | 45.42 | 16.38 | 5.2E−4 | −6.80 | −7.40 | 0.15 | −6.44 | −4.49 | 0.001 | −6.77 | −8.80 | 0.006 |
| Testis (6, 19) | 42.00 | 20.44 | 3.4E−4 | −5.73 | −8.76 | 4.5E−05 | −5.22 | −6.68 | 0.009 | −5.35 | −9.23 | 4.5E−05 |
| All (72, 293) | 29.66 | 18.02 | 2.6E−12 | −7.11 | −7.31 | 0.39 | −6.06 | −5.13 | 3.7E−09 | −8.01 | −9.10 | 7.9E−12 |
The relative alternative splicing of macroH2A1 and the expression of macroH2A1 and QKI were determined as described in the main text. The reported P values are the results of a two-tailed Mann-Whitney test. The gray shading indicates comparisons that are significant (P < 0.05).
MacroH2A1.1 suppresses the growth of lung cancer cells.
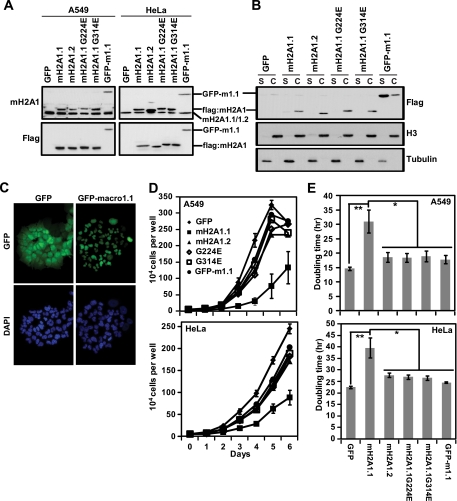
The data presented above demonstrate that reduction of macroH2A1.1 resulting from changes in macroH2A1 pre-mRNA alternative splicing is a hallmark of several types of cancer. Furthermore, these results suggested that macroH2A1.1 may specifically participate in a tumor-suppressive mechanism that is subverted in these cancers. To test this hypothesis, we restored macroH2A1.1 expression in A549 lung cancer cells and HeLa cervical carcinoma cells which normally only express macroH2A1.2. Using stable retrovirus-mediated transduction of both A549 and HeLa cells, we expressed GFP (as a control), flag-tagged macroH2A1.1, macroH2A1.2, two macroH2A1.1 point mutants which prevent binding to NAD+-derived ligands (G224E or G314E), or GFP fused to the macro domain of macroH2A1.1 (GFP-m1.1) (Fig. 3A). To confirm that the ectopic macroH2A1.1, macroH2A1.2, and point mutants were incorporated into chromatin, we fractionated each of the A549 cell lines into soluble and chromatin fractions and performed Flag immunoblots (Fig. 3B). As expected, the macroH2A1 proteins containing the histone-like region demonstrated clear chromatin incorporation. The GFP-m1.1 protein which lacks the histone-like region of macroH2A1 localized predominantly to the soluble fraction, indicating that it is not incorporated into chromatin. However, fluorescence microscopy for GFP indicated that GFP-m1.1 localizes to the nucleus (Fig. 3C).
Fig. 3.
MacroH2A1.1 suppresses the growth of A549 lung cancer cells and HeLa cervical adenocarcinoma cells in a manner that depends on both its ability to bind ADPR-based ligands and its histone-like domain. (A) Anti-macroH2A1 (upper) and anti-Flag (lower) antibodies were used in immunoblots to demonstrate the ectopic expression of macroH2A1 proteins in A549 cells retrovirally transduced with Flag-tagged macroH2A1.1 (mH2A1.1), macroH2A1.2 (mH2A1.2), one of two macroH2A1.1 point mutants, G224E and G314E, or GFP fused to the macro domain of macroH2A1.1 (GFP-m1.1). GFP was used as a control. Note that the Flag-tagged exogenous macroH2A1 proteins have a slower mobility than endogenous macroH2A1. (B) Anti-Flag immunoblot of A549 cells expressing the indicated factors separated into soluble or chromatin fractions (S or C, respectively). Antitubulin and anti-histone H3 antibodies are used as controls for soluble and chromatin fractions, respectively. (C) Fluorescence microscopy was used to detect GFP in A549 cells ectopically expressing GFP alone or GFP-m1.1. The cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) to confirm nuclear localization. (D) Growth curves of A549 (top) and HeLa (bottom) cells expressing the indicated factors counted every day for 6 days. All groups show a significant difference compared to the growth of cells expressing macroH2A1.1 (P < 0.05, Student's two-tailed t test) (E) Histogram of the doubling time of A549 (top) and HeLa (bottom) cells expressing the indicated factors. P values were determined with a two-tailed Student t test. *, P < 0.05; **, P < 0.005. Error bars in panels D and E represent the SEMs for five biological replicates.
To determine the effects of macroH2A1 variants on cell proliferation, we performed cell counting experiments on the stable cell lines expressing the macroH2A1 variants and mutants described above (Fig. 3D and E). Cells expressing ectopic macroH2A1.1 demonstrated significantly slower proliferation than cells expressing GFP as a control. Specifically, macroH2A1.1 expression led to a 2-fold and 1.76-fold increase in doubling time in A549 cells and HeLa cells, respectively. The ability to suppress the growth of A549 and HeLa cells is a specific property of the macroH2A1.1 isoform, as ectopic expression of flag-tagged macroH2A1.2, which was expressed at levels similar to those of macroH2A1.1 (Fig. 3A), did not lead to a similar change in the rate of proliferation compared to GFP-expressing control cells.
The major biochemical difference between the three macroH2A subtypes is that the macroH2A1.1 macro domain is the only macroH2A macro domain that can interact with ADPR-based small molecules. In the case of macroH2A1, the amino acid differences between macroH2A1.1 and macroH2A1.2 are limited to the exchange of a single exon encoding 10 amino acids in macroH2A1.1 for an alternative exon encoding 11 amino acids in macroH2A1.2 (Fig. 1A and B). The macroH2A1.1-specific exon encodes a region of the protein that is critically important for the macro domain of macroH2A1.1 to bind ADPR-based molecules (35). To determine whether ADPR-like ligand-binding was important for the suppression of cancer cell growth by macroH2A1.1, we made use of two macroH2A1.1 point mutants (G224E or G314E) that have been shown previously to abolish the ability of macroH2A1.1 to interact with its ligands (35). When introduced into A549 and HeLa cells, both of the macroH2A1.1 ligand-binding-defective mutants were also defective in their ability to suppress proliferation compared to wild-type macroH2A1.1 (Fig. 3D and E), suggesting that the ability of macroH2A1.1 to interact with one or more of its ADPR-based ligands is critical for its ability to suppress cancer cell proliferation.
We next sought to determine whether nucleosome incorporation is necessary for the ability of macroH2A1.1 to suppress cancer cell proliferation. By using a chimeric protein in which the histone-like domain of macroH2A1.1 was replaced with GFP (GFP-m1.1), we determined that the histone domain of macroH2A1.1 is required to suppress the growth of A549 and HeLa cells (Fig. 3D and E). Overall, these experiments demonstrate that macroH2A1.1 suppresses the proliferation of A549 lung cancer and HeLa cervical adenocarcinoma cells in a process that requires both its ability to incorporate into nucleosomes and to interact with ADPR-based ligands.
MacroH2A1.1 suppresses proliferation by regulating PARP-1 levels.
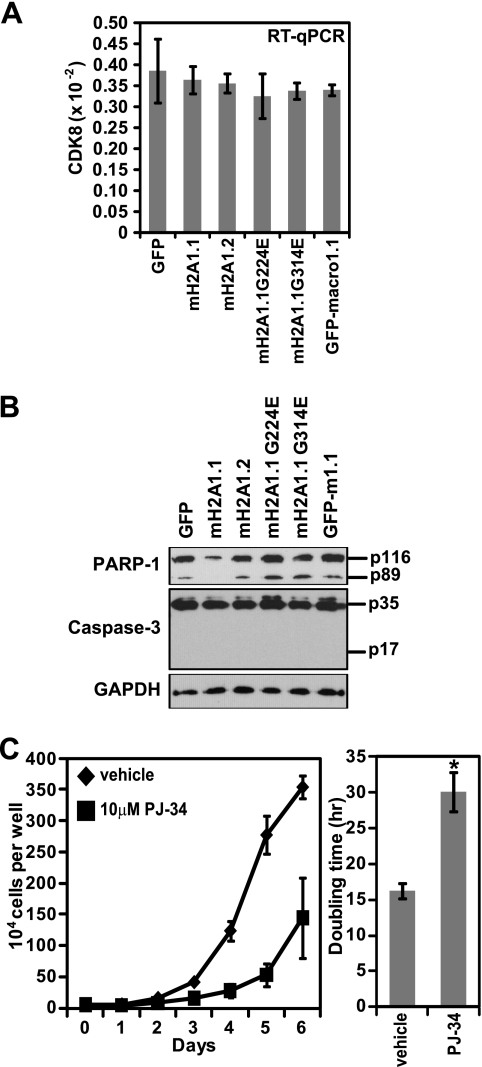
MacroH2A1 and macroH2A2 were previously shown to be silenced at the transcriptional level in a malignant melanoma. Furthermore, macroH2A was shown to suppress malignant melanoma through the transcriptional repression of cyclin-dependent kinase 8 (CDK8) (29). We hypothesized that macroH2A1.1 might be performing a similar function in A549 lung cancer cells. However, when we examined CDK8 mRNA levels in A549 cells expressing macroH2A1.1, we failed to observed a significant change in CDK8 expression (Fig. 4A).
Fig. 4.
MacroH2A1.1 suppresses cancer cell proliferation by downregulating PARP-1 levels. (A) Histogram of CDK8 mRNA levels determined by RT-PCR in A549 cells expressing the indicated factors. Error bars represent the SEMs of four biological replicates. (B) PARP-1, caspase-3, and GAPDH immunoblots of A549 cells expressing the indicated factors. The locations of full-length (p116) and caspase-cleaved (p89) PARP-1 and full-length (p35) and cleaved (p17) caspase-3 are indicated. (C) Growth curve (left) and histogram depicting doubling times of A549 cells in the absence or presence of 10 μM PJ-34. Error bars represent the SEMs of three biological replicates.
So, what is the mechanism by which macroH2A1.1 suppresses cancer cell proliferation? To answer this question, we examined PARP-1 levels in macroH2A1.1-expressing A549 cells for three reasons. First, we wondered if macroH2A1.1-mediated proliferative suppression might be due to enhanced apoptosis, and PARP-1 is cleaved by caspase-3 during apoptosis, yielding an 89-kDa cleavage product (32). Additionally, the ability to bind PARP-1 when automodified with PAR chains distinguishes macroH2A1.1 from macroH2A1.2. Finally, PARP activity has been linked to cell cycle progression through several disparate mechanisms (7, 15-17, 22). Strikingly, we found that PARP-1 levels, both full-length and p89, were specifically reduced in macroH2A1.1-expressing cells (Fig. 4B). Additionally, while we were able to detect a robust signal for pro-caspase-3, we were unable to detect any cleaved (i.e., active) caspase-3 in any of the A549 cells ectopically expressing macroH2A1 variants. From this we conclude that macroH2A1.1 suppresses cell proliferation not by enhancing apoptosis but, rather, by reducing PARP-1 levels. Consistently, pharmacological inhibition of PARP activity suppresses the proliferation of A549 cells in a manner similar to ectopic expression of macroH2A1.1 (Fig. 4C).
Identification of QKI as a factor that regulates the alternative splicing of macroH2A1.
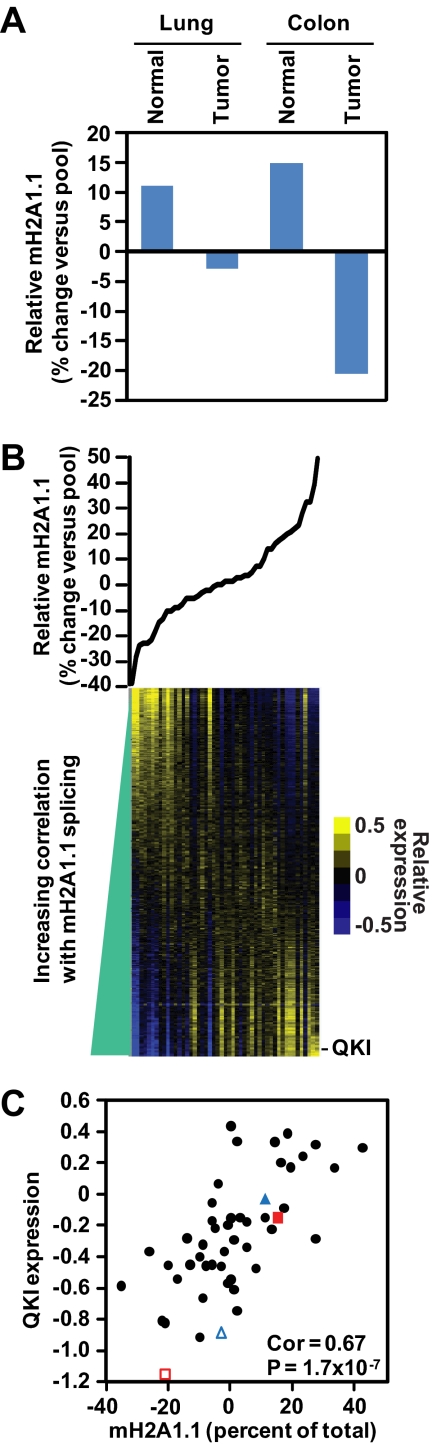
Given the changes in macroH2A1 splicing that occur in several cancers and the role of macroH2A1.1 in suppressing cancer cell growth, we sought to identify the factors that regulate the splicing of macroH2A1. Toward this goal, we undertook a bioinformatics approach using a previously published microarray data set cataloging over 24,000 alternative splicing events while simultaneously monitoring the gene expression of over 18,000 genes across 48 tissues, tumors, and cell lines (8). The microarray data set included splicing data from normal and tumor samples from both lung and colon, two tissues where we observed a cancer-linked reduction in macroH2A1.1 alternative splicing, as shown in Fig. 2. Consistent with our RT-qPCR data, the expression microarray data recapitulate the reduction of macroH2A1.1 splicing in lung and colon tumor samples, confirming the ability of the microarray data to detect differences in macroH2A1 pre-mRNA alternative splicing (Fig. 5A).
Fig. 5.
Identification of putative regulators of macroH2A1 alternative splicing using splicing and expression microarray data from reference 8. (A) Histogram depicting the relative differences in macroH2A1.1 splicing in normal and tumor samples from lung and colon included in the microarray data. (B) Relative levels of macroH2A1.1 splicing (compared to macroH2A1.2) across 48 tissues, tumors, and cell lines (top). Heat map depicting the relative expression of 18,093 genes across the same 48 samples as above (bottom). The tissue samples (i.e., columns) are ordered for the relative level of macroH2A1.1 alternative splicing. See Table S1 at http://www.einstein.yu.edu/docs/labs/matthew-gamble/Novikov-2011-MCB-Supplement.pdf for a list of the 48 samples and the relative levels of macroH2A1.1 exon inclusion. The genes are ordered for increasing correlation to the level of macroH2A1.1 splicing in each tissue. The approximate location of the expression data from the QKI gene is indicated. (C) Scatter plot of the relative macroH2A1.1 splicing in each tissue versus the relative expression of QKI. Correlation (Cor) and P value determined by Spearman's rank correlation test. The location of lung (blue triangles) and colon (red squares) for both normal (filled) and tumor (open) samples are indicated.
Confident in the ability of the microarray data to accurately monitor changes in macroH2A1 splicing, we correlated the observed changes in macroH2A1 splicing across the 48 samples with the expression of each of the 18,093 genes represented on the array. We were able to identify a subset of genes with known or predicted roles in regulating alternative splicing as having a significant correlation (either positive or negative) with the inclusion of the macroH2A1.1 exon (Fig. 5B; also see Tables S1 and S2 at http://www.einstein.yu.edu/docs/labs/matthew-gamble/Novikov-2011-MCB-Supplement.pdf). Among these genes, we focused on QKI for three reasons. First, QKI was the most positively correlated splicing factor identified by the array (Fig. 5B and C; also see Table S2 at the URL above). Second, two recent reports implicate QKI as a potential tumor suppressor (50, 51). Finally, an interaction between QKI and macroH2A1 pre-mRNA was recently reported in a genome-wide screen for site-specific interactions between RNA-binding factors and total RNA called PAR-CLIP (27). These data indicated that QKI binds to macroH2A1 pre-mRNA in the intron upstream of the macroH2A1.2-specific exon (illustrated in Fig. 1A).
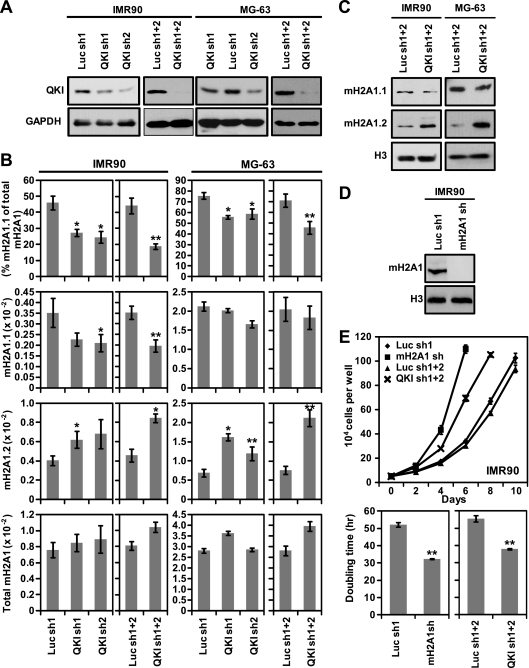
In order to determine whether QKI plays a functional role in regulating macroH2A1 splicing, we used retroviral-mediated shRNA expression of either of two shRNA targeting sequences to deplete QKI from two cell lines that express both macroH2A1.1 and macroH2A1.2 variants, MG-63 osteosarcoma cells and IMR90 primary lung fibroblasts (Fig. 1D). Immunoblotting was used to confirm the depletion of QKI from the cells (Fig. 6 A). While either shRNA targeted against QKI was able to deplete a portion of QKI from the cells, the most robust effect was seen when both shRNA targeting sequences were used simultaneously. The reduction of QKI protein levels led to significant changes in macroH2A1 splicing (Fig. 6B). In the IMR90 cells, where macroH2A1.1 and macroH2A1.2 are expressed at similar levels, QKI knockdown led to a significant reduction of macroH2A1.1 mRNA and an increase in macroH2A1.2 mRNA levels, while total macroH2A1 transcript levels were not significantly altered. In MG-63 cells, we detected a dramatic increase in macroH2A1.2 mRNA levels in the QKI knockdown cells compared to the levels in luciferase knockdown controls. However, we were unable to observe a significant decrease in macroH2A1.1. Our inability to detect a significant decrease in macroH2A1.1 in MG-63 may be due to the fact that the macroH2A1.1 levels are nearly 4-fold greater than those of macroH2A1.2 in MG-63 cells. Thereby, the large increase in macroH2A1.2 levels corresponds to a relatively small decrease in the amount of macroH2A1.1 mRNA. In both cell lines, the percentage of total macroH2A1 transcript that includes the macroH2A1.1-specific exon is significantly reduced due to QKI depletion (Fig. 6B, top). Consistent with a role for QKI in the regulation of the splicing of macroH2A1 transcripts and not transcription of the H2AFY gene, the total level of macroH2A1 mRNA is not significantly altered when QKI is depleted from these cells. Importantly, the changes in macroH2A1 pre-mRNA alternative splicing seen upon QKI depletion are also apparent when examined at the protein level (Fig. 6C).
Fig. 6.
QKI regulates the splicing of macroH2A1 pre-mRNA. (A) Immunoblots with anti-QKI (top) or anti-GAPDH (bottom) antibodies demonstrating the reduced expression of QKI in IMR90 and MG-63 cells harboring one or both of two shRNAs targeting QKI (QKI sh1, QKI sh2, or sh1+2) or one or both of two shRNAs targeting luciferase (Luc sh1, Luc sh2, or Luc sh1+2) as a control. (B) Relative expression of macroH2A1.1, macroH2A1.2, and total macroH2A1 in luciferase and QKI knockdown cells. The percentage of total macroH2A1 mRNA that was spliced as macroH2A1.1 is also indicated (top). Error bars represent the SEMs of four biological replicates. (C) Immunoblots with anti-macroH2A1.1 (mH2A1.1), macroH2A1.2 (mH2A1.2), and anti-histone H3 (H3) antibodies of IMR90 and MG-63 cells expressing two shRNAs against QKI or luciferase as a control. (D) Immunoblot with anti-macroH2A1 (pan-specific) antibody or histone H3 of IMR90 cells expressing an shRNA against macroH2A1 or luciferase as a control. (E) Growth curves (top) and histograms of doubling times (bottom) of IMR90 cells harboring the indicated shRNAs. Error bars represent the SEMs of three biological replicates. P values were determined by a two-tailed Student's t test. *, P < 0.05; **, P < 0.01.
Consistent with a role for macroH2A1.1 in suppressing proliferation, depletion of macroH2A1 from IMR90 cells leads to a significant decrease in doubling time (Fig. 6D and E). The observation that QKI depletion in IMR90 cells also enhances cellular proliferation further supports a specific role for macroH2A1.1 in the regulation of cell proliferation. Overall, our results suggest that QKI is a key determinant of macroH2A1 pre-mRNA splicing, which in turn leads to the regulation of proliferation.
QKI expression is reduced in several types of cancers.
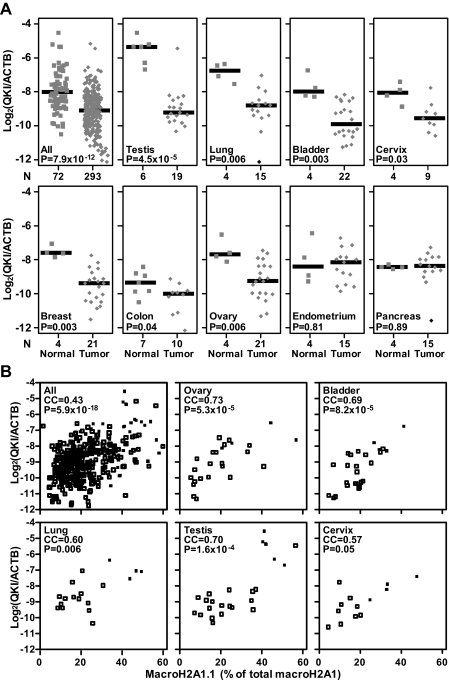
The changes in macroH2A1 alternative splicing caused by QKI depletion in MG-63 and IMR90 cells were reminiscent of the reduction in macroH2A1.1 splicing observed in the tumors in the experiment whose results are shown in Fig. 2. Therefore, we next sought to determine whether changes in the level of QKI could explain the reduced levels of macroH2A1.1 splicing observed in cancer patient tumors. To do this, we determined the relative expression of QKI in the same 365 normal and tumor samples from the experiment whose results are shown in Fig. 2 (Fig. 7). Similar to the observed changes in macroH2A1.1 splicing, QKI mRNA expression was significantly reduced in cancer patient samples compared to its expression in normal controls (P=7.9 × 10−12). A significant decrease in QKI levels was apparent in most cancer types in which macroH2A1.1 splicing is reduced, including testicular, lung, bladder, cervical, breast, colon, and ovarian tumors (Fig. 7A and Table 1). However, a correspondence between the decline of macroH2A1.1 splicing and reduced QKI levels was not observed in endometrial cancer. Additionally, QKI was significantly reduced in prostate cancer, where reductions in macroH2A1.1 splicing were observed but did not satisfy our significance threshold (Table 1). In support of a role for QKI in regulating macroH2A1 splicing in several cancers, we were able to detect a significant positive correlation between macroH2A1.1 splicing and QKI levels when examined for all samples and in several individual tissue types (Fig. 7B). Overall, these findings support our hypothesis that reduction of the level of QKI affects the pre-mRNA splicing of macroH2A1 in several cancers.
Fig. 7.
QKI expression is reduced in many of the same cancer types that demonstrate a reduction in macroH2A1.1 splicing. (A) QKI expression in 365 cancer patient samples was determined by RT-qPCR. The squares and diamonds represent data from normal and cancer patient samples, respectively. The x axis represents the number of samples in each group. The solid horizontal bars represent the median values of particular groups. The reported P values are the results of a two-tailed Mann-Whitney test. (B) Scatter plots demonstrating the correlation between macroH2A1.1 splicing and QKI levels in the indicated tissues. Correlation coefficients (CC) and associated P values were determined using the Spearman rank method.
DISCUSSION
There is an accumulating body of evidence demonstrating that changes in chromatin structure occur during oncogenesis (41). These changes in chromatin structure include alterations in DNA methylation and posttranslational modifications of histones which are thought to collaborate with mutations to alter the expression of oncogenes and tumor suppressors and promote the oncogenic state. Changes in the expression of histone variants have also been observed in cancer and can also lead to important changes in chromatin structure and function. For example, increased expression of the histone variant H2A.Z has been observed in both colon and breast cancers (44). Consistent with a role in oncogenesis, H2A.Z overexpression has been shown to increase breast cancer cell proliferation (45).
Two recent reports have found alterations in macroH2A expression in cancer. The first, from Andreas Ladurner's laboratory, demonstrates that the protein levels of the macroH2A1.1 isoform are reduced relative to the levels of macroH2A1.2 in lung cancer patients (43). The work we present here confirms the reduction of macroH2A1.1 in lung cancer. We also extend this finding by demonstrating that the reduction of macroH2A1.1 is caused, at least in part, by altered regulation of the mutually exclusive splicing of the macroH2A1 transcript, strongly favoring the expression of the macroH2A1.2 variant in this cancer. Furthermore, we demonstrate that a similar alteration of macroH2A1 alternative splicing, leading to a reduction in macroH2A1.1, occurs in a variety of additional cancer types, including testicular, lung, bladder, cervical, breast, colon, ovarian, and endometrial cancer.
The second report documenting a change in macroH2A expression in cancer, from Emily Bernstein's group, demonstrates that the genes encoding macroH2A1 and macroH2A2 are often silenced due to promoter DNA methylation in malignant melanomas (29). Notably, of the 17 cancer types we tested for the expression of macroH2A1 variants, none demonstrated the transcriptional silencing of macroH2A1 observed in malignant melanoma and only testicular cancer exhibited a significant reduction in macroH2A1 total expression (Table 1). Overall, this suggests that silencing of macroH2A expression and perturbed regulation of macroH2A1 alternative splicing represent two independent mechanisms by which macroH2A expression is altered in cancer.
In addition to demonstrating that macroH2A genes are silenced in malignant melanoma, the Bernstein laboratory demonstrated a functional role for macroH2A1 in suppressing the growth of melanoma cells (29). They show that silencing macroH2A leads to increased proliferation of melanoma cells through the derepression of CDK8. Consistent with Bernstein's report, we demonstrate a role for macroH2A1 in suppressing the proliferation of cancer cells. However, our data specifically implicate a role for macroH2A1.1 in this process. Unlike the previous report for malignant melanoma, the suppression of lung cancer proliferation by macroH2A1.1 does not seem to involve transcriptional repression of the CDK8 gene.
It appears that in addition to having different mechanisms by which macroH2A expression is altered in cancers, the mechanism by which macroH2A suppresses proliferation in lung cancer appears to be distinct from its role in suppressing malignant melanoma. As described above, the defining characteristic of macroH2A1.1 is its ability to interact with ADPR-based molecules, such as PARP-1-catalyzed PAR. As our experiments with macroH2A1.1 mutants suggest, the ADPR-binding function is required for macroH2A1.1-mediated suppression of lung cancer cell proliferation (Fig. 3). We have shown that macroH2A1.1 performs this role, at least in part, by downregulating PARP-1 protein levels. In turn, reduced PARP activity leads to decreased proliferation. PARP-1 has been implicated in several processes that promote cellular proliferation, including growth factor-mediated mitogen-activated protein (MAP) kinase cascades (16, 17), cell cycle progression (7), and prevention of the redifferentiation of tumor cells (22). PARP-1 overexpression has been documented in several cancers, including colon (37), breast (5), endometrial (25), liver (42), lymphoma (48), and Ewing's sarcoma (39). In the future it will be important to determine exactly how macroH2A1.1 leads to the reduction of PARP-1 and which pathways important for cellular proliferation are affected.
Most genes in the human genome are regulated by alternative splicing, which can yield proteins with divergent or even antagonistic functions from a single gene. Consequently, alternative splicing represents an important control point that is often leveraged by cancer cells (21). While mutually exclusive inclusion of alternative exons, as seen in macroH2A1 pre-mRNA splicing, is not the most common form of alternative splicing, there are several examples in which dysregulation of mutually exclusive splicing has been shown to occur in cancer. Dysregulation of mutually exclusive splicing of FGFR2 and PKM1/2 pre-mRNA has been implicated in the epithelial-to-mesenchymal transition (31) and the Warburg effect (14), respectively. Both of these examples of cancer-perturbed mutually exclusive splicing require a variety of protein factors and cis-acting elements for appropriate regulation (21). While there is still much work to be done in determining the mechanisms that regulate the mutually exclusive splicing of macroH2A1 transcripts, we have made an important advance by identifying QKI as a splicing factor that plays a significant role in this process. Our results also demonstrate that reduced QKI levels likely cause the reduced macroH2A1.1 levels observed in several types of cancer. Overall, these results suggest that QKI may be an important tumor suppressor. This hypothesis finds additional support in two prior studies. First, the QKI gene is found on a region of chromosome 6 often deleted in glioblastoma multiforme (51). Additionally, QKI suppresses the growth of colon cancer cells in culture, which is exactly the phenotype we would expect from a splicing factor that enhances the splicing of macroH2A1.1 (50).
In summary, our results demonstrate that macroH2A1.1 splicing is reduced in several types of cancer. Additionally, macroH2A1.1 mediates growth suppression of cancer cells which requires the interaction of macroH2A1.1 with ADPR-based ligands and results in a reduction of PARP-1. Shifting the splicing of macroH2A1 pre-mRNA to favor macroH2A1.2 provides a convenient mechanism for cancer cells to bypass macroH2A1.1-mediated growth suppression. In our search for factors that regulate macroH2A1 alternative splicing, we were able to quickly focus on QKI due, in part, to the availability of whole-transcriptome binding data for this factor using a novel high-throughput technique called PAR-CLIP (27). However, given that there is not a complete correspondence between cancers that show decreased macroH2A1.1 splicing and decreased QKI expression (e.g., endometrial cancer), we fully expect a variety of additional splicing factors to be involved in regulating this process.
ACKNOWLEDGMENTS
We thank T. Zhang for critical reading of the manuscript and helpful discussions. We thank H. M. McDaid and S. B. Horwitz for providing A549 and HeLa cells and helpful discussions. We thank C.W. Chow and F. Macian for the antitubulin and anti-caspase-3 antibodies, respectively. We thank C. Rubin and A. Skoultchi for helpful discussions. We thank W. L. Kraus for the GFP retroviral expression construct and the luciferase retroviral shRNA construct.
We declare no competing financial interests.
This work was supported by a grant from the Sidney Kimmel Foundation for Cancer Research to M.J.G.
Footnotes
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Abbott D. W., et al. 2004. Structural characterization of macroH2A containing chromatin. Biochemistry 43:1352–1359 [DOI] [PubMed] [Google Scholar]
- 2. Ahel D., et al. 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325:1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angelov D., et al. 2006. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25:1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angelov D., et al. 2003. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11:1033–1041 [DOI] [PubMed] [Google Scholar]
- 5. Bieche I., de Murcia G., Lidereau R. 1996. Poly(ADP-ribose) polymerase gene expression status and genomic instability in human breast cancer. Clin. Cancer Res. 2:1163–1167 [PubMed] [Google Scholar]
- 6. Buschbeck M., et al. 2009. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 16:1074–1079 [DOI] [PubMed] [Google Scholar]
- 7. Carbone M., et al. 2008. Poly(ADP-ribosyl)ation is implicated in the G0-G1 transition of resting cells. Oncogene 27:6083–6092 [DOI] [PubMed] [Google Scholar]
- 8. Castle J. C., et al. 2008. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 40:1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakravarthy S., et al. 2005. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 25:7616–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakravarthy S., Luger K. 2006. The histone variant macro-H2A preferentially forms “hybrid nucleosomes.” J. Biol. Chem. 281:25522–25531 [DOI] [PubMed] [Google Scholar]
- 11. Chang E. Y., et al. 2008. MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry 47:13726–13732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Changolkar L. N., et al. 2007. Developmental changes in histone macroH2A1-mediated gene regulation. Mol. Cell. Biol. 27:2758–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Changolkar L. N., et al. 2010. Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol. Cell. Biol. 30:5473–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen M., Zhang J., Manley J. L. 2010. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 70:8977–8980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chevanne M., et al. 2010. Inhibition of PARP activity by PJ-34 leads to growth impairment and cell death associated with aberrant mitotic pattern and nucleolar actin accumulation in M14 melanoma cell line. J. Cell. Physiol. 222:401–410 [DOI] [PubMed] [Google Scholar]
- 16. Cohen-Armon M. 2007. PARP-1 activation in the ERK signaling pathway. Trends Pharmacol. Sci. 28:556–560 [DOI] [PubMed] [Google Scholar]
- 17. Cohen-Armon M., et al. 2007. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol. Cell 25:297–308 [DOI] [PubMed] [Google Scholar]
- 18. Costanzi C., Pehrson J. R. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599–601 [DOI] [PubMed] [Google Scholar]
- 19. Costanzi C., Pehrson J. R. 2001. MACROH2A2, a new member of the MARCOH2A core histone family. J. Biol. Chem. 276:21776–21784 [DOI] [PubMed] [Google Scholar]
- 20. Counter C. M., et al. 1998. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. U. S. A. 95:14723–14728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. David C. J., Manley J. L. 2010. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24:2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Blasio A., et al. 2005. Differentiative pathway activated by 3-aminobenzamide, an inhibitor of PARP, in human osteosarcoma MG-63 cells. FEBS Lett. 579:615–620 [DOI] [PubMed] [Google Scholar]
- 23. Gamble M. J., Frizzell K. M., Yang C., Krishnakumar R., Kraus W. L. 2010. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 24:21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gamble M. J., Kraus W. L. 2010. Multiple facets of the unique histone variant macroH2A: from genomics to cell biology. Cell Cycle 9:2566–2572 [DOI] [PubMed] [Google Scholar]
- 25. Ghabreau L., et al. 2004. Poly(ADP-ribose) polymerase-1, a novel partner of progesterone receptors in endometrial cancer and its precursors. Int. J. Cancer 109:317–321 [DOI] [PubMed] [Google Scholar]
- 26. Gottschalk A. J., et al. 2009. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. U. S. A. 106:13770–13774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hafner M., et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hernandez-Munoz I., et al. 2005. Stable X chromosome inactivation involves the PRC1 polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. U. S. A. 102:7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kapoor A., et al. 2010. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468:1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karras G. I., et al. 2005. The macro domain is an ADP-ribose binding module. EMBO J. 24:1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katoh M. 2008. Cancer genomics and genetics of FGFR2 (review). Int. J. Oncol. 33:233–237 [PubMed] [Google Scholar]
- 32. Kaufmann S. H., Desnoyers S., Ottaviano Y., Davidson N. E., Poirier G. G. 1993. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 53:3976–3985 [PubMed] [Google Scholar]
- 33. Kraus W. L. 2009. New functions for an ancient domain. Nat. Struct. Mol. Biol. 16:904–907 [DOI] [PubMed] [Google Scholar]
- 34. Krishnakumar R., et al. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319:819–821 [DOI] [PubMed] [Google Scholar]
- 35. Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A. G. 2005. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 12:624–625 [DOI] [PubMed] [Google Scholar]
- 36. Martzen M. R., et al. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286:1153–1155 [DOI] [PubMed] [Google Scholar]
- 37. Nosho K., et al. 2006. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur. J. Cancer. 42:2374–2381 [DOI] [PubMed] [Google Scholar]
- 38. Pehrson J. R., Costanzi C., Dharia C. 1997. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell Biochem. 65:107–113 [DOI] [PubMed] [Google Scholar]
- 39. Prasad S. C., Thraves P. J., Bhatia K. G., Smulson M. E., Dritschilo A. 1990. Enhanced poly(adenosine diphosphate ribose) polymerase activity and gene expression in Ewing's sarcoma cells. Cancer Res. 50:38–43 [PubMed] [Google Scholar]
- 40. Schefe J. H., Lehmann K. E., Buschmann I. R., Unger T., Funke-Kaiser H. 2006. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression's CT difference” formula. J. Mol. Med. 84:901–910 [DOI] [PubMed] [Google Scholar]
- 41. Sharma S., Kelly T. K., Jones P. A. 2010. Epigenetics in cancer. Carcinogenesis 31:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shimizu S., et al. 2004. Expression of poly(ADP-ribose) polymerase in human hepatocellular carcinoma and analysis of biopsy specimens obtained under sonographic guidance. Oncol. Rep. 12:821–825 [PubMed] [Google Scholar]
- 43. Sporn J. C., et al. 2009. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28:3423–3428 [DOI] [PubMed] [Google Scholar]
- 44. Svotelis A., Gevry N., Gaudreau L. 2009. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem. Cell Biol. 87:179–188 [DOI] [PubMed] [Google Scholar]
- 45. Svotelis A., Gevry N., Grondin G., Gaudreau L. 2010. H2A.Z overexpression promotes cellular proliferation of breast cancer cells. Cell Cycle 9:364–370 [DOI] [PubMed] [Google Scholar]
- 46. Talbert P. B., Henikoff S. 2010. Histone variants—ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11:264–275 [DOI] [PubMed] [Google Scholar]
- 47. Timinszky G., et al. 2009. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 16:923–929 [DOI] [PubMed] [Google Scholar]
- 48. Tomoda T., et al. 1991. Enhanced expression of poly(ADP-ribose) synthetase gene in malignant lymphoma. Am. J. Hematol. 37:223–227 [DOI] [PubMed] [Google Scholar]
- 49. Wutz A., Rasmussen T. P., Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30:167–174 [DOI] [PubMed] [Google Scholar]
- 50. Yang G., et al. 2010. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology 138:231–240.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yin D., et al. 2009. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol. Cancer Res. 7:665–677 [DOI] [PubMed] [Google Scholar]