Abstract
Spt6 is a conserved factor, critically required for several transcription- and chromatin-related processes. We now show that Spt6 and its binding partner, Iws1, are required for heterochromatic silencing in Schizosaccharomyces pombe. Our studies demonstrate that Spt6 is required for silencing of all heterochromatic loci and that an spt6 mutant has an unusual combination of heterochromatic phenotypes compared to previously studied silencing mutants. Unexpectedly, we find normal nucleosome positioning over heterochromatin and normal levels of histone H3K9 dimethylation at the endogenous pericentric repeats. However, we also find greatly reduced levels of H3K9 trimethylation, elevated levels of H3K14 acetylation, reduced recruitment of several silencing factors, and defects in heterochromatin spreading. Our evidence suggests that Spt6 plays a role at both the transcriptional and posttranscriptional levels; in an spt6 mutant, RNA polymerase II (RNAPII) occupancy at the pericentric regions is only modestly increased, while production of small interfering RNAs (siRNAs) is lost. Taken together, our results suggest that Spt6 is required for multiple steps in heterochromatic silencing by controlling chromatin, transcriptional, and posttranscriptional processes.
INTRODUCTION
Spt6 is conserved from prokaryotes (49) to humans (25) and plays many roles in regulation of transcription and chromatin structure. Studies in several organisms, particularly Saccharomyces cerevisiae, have elucidated many of its functions. Spt6 acts as a transcription elongation factor, increasing the rate of RNA polymerase II (RNAPII) progression in vivo (6) and in vitro (35). Spt6 also plays a role in the regulation of transcription initiation (1, 46, 47) and 3′ end formation (16, 50). Spt6 associates with elongating RNAPII (4, 6, 50, 52, 53) and also functions as a histone chaperone (15, 51, 68). Mutation of spt6 produces a more open chromatin structure in specific regions of the genome (46, 51), which is suggested to have a number of consequences. These include increased genomic instability (12), hyperrecombination (62), the failure of certain promoters to repress transcription (1), and the initiation of cryptic transcripts within particular open reading frames (24, 51). Furthermore, Spt6 has been linked to mRNA splicing, processing, and export activities, through both genetic (20) and physical (5, 58, 94) interactions with some of the components. Taken together, these studies indicate that Spt6 plays important roles in many chromatin and transcription-related processes.
Studies have identified several conserved domains in the Spt6 protein (Fig. 1A). It has an N-terminal acidic domain (84) and a tandem SH2 motif at its C-terminal end (29, 30, 32, 59, 61, 83). The core of the protein encodes a domain homologous to the Pseudomonas Tex activator, which contains several nucleic acid-interacting domains, including an RNase H-like domain, an S1 domain, and a helix-hairpin-helix (HhH) domain (33, 76). Recent structural and biochemical studies have begun to address the function of these domains. The tandem SH2 domain binds the phosphorylated C-terminal domain of elongating RNAPII (29, 30, 32, 59, 83, 94). The Tex domain was crystallized, and its S1 domain was shown to have a preference for single-stranded RNA (49). Finally, an N-terminal region of Spt6 that interacts with nucleosomes has been identified (68). Interestingly, this overlaps with the region that binds Iws1, the binding partner of Spt6, and the two interactions appear to be mutually exclusive (31, 68). These conserved domains encode many functions that are important for Spt6 but which are only beginning to be understood with respect to its in vivo function.
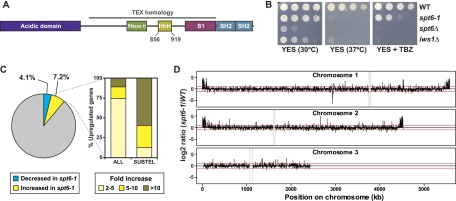
Fig. 1.
Characterization of spt6+ and iws1+ in S. pombe. (A) Domain structure of Spt6. Spt6 encodes a number of conserved domains and motifs, including an acidic domain, a YqgFc RNase H-like domain (RNase H), a helix-hairpin-helix motif (HhH), an S1 domain, and tandem SH2 domains. Homology to the prokaryotic TEX factor is indicated by the gray bar. The boundaries of the HhH domain are at amino acids 856 to 919; these residues are removed in the spt6-1 mutant. Scale is approximate. (B) Phenotypic characterization of S. pombe spt6-1, spt6Δ, and iws1Δ mutants by serial dilution spot tests. These images show 3 days of growth on rich medium (YES) at 30°C and with the microtubule inhibitor thiabendazole (TBZ) and 2 days of growth at 37°C. (C) Transcriptional changes in the spt6-1 mutant by microarray analysis. Transcripts were considered significantly affected if the spt6-1/wild-type (WT) ratio was ≤0.5 (decreased, 198 genes, 4.1%) or ≥2 (increased, 353 genes, 7.2%). The bar graph shows the genes with increased transcripts (all genes and subtelomeric genes) binned by fold increase. (D) spt6-1/wild-type log2 expression ratios for each gene on the array are plotted according to their genomic location. Red lines correspond to the 2-fold cutoffs used in this study. The repetitive centromeric regions (gray bars) and the subtelomeric regions of chromosome 3 containing the rDNA repeats are not represented in this diagram, due to lack of coverage on the microarray.
Iws1 (Spn1/Iws1 in Saccharomyces cerevisiae) binds Spt6 at substoichiometric levels and is required for transcriptional regulation (36, 99), histone modification (95), and mRNA export (94). Interestingly, Spt6 and Iws1 have been shown to play contradictory regulatory roles at certain loci (99), and high-resolution chromatin immunoprecipitation (ChIP) studies have recently shown that while both Spt6 and Iws1 associate with elongating RNAPII, they have distinct binding patterns (67). Consequently, many questions remain about the relationship between Spt6 and Iws1.
Given that Spt6 plays many critical roles in chromatin-related processes, we have initiated the study of Spt6 and Iws1 in the fission yeast, Schizosaccharomyces pombe, which is greatly divergent from the budding yeast, Saccharomyces cerevisiae (91). Notably, S. pombe heterochromatin is similar to heterochromatin in larger eukaryotes in that it requires histone H3 lysine 9 (H3K9) methylation, HP1 homologs, and the RNA interference (RNAi) machinery (reviewed in references 41 and 69). There are three types of heterochromatic loci in S. pombe: the pericentric repeats, the silent mating-type locus, and the subtelomeric regions.
We have found that both Spt6 and Iws1 are required for silencing of heterochromatin in S. pombe. We have further investigated the roles of Spt6 in silencing and found that it controls certain histone modifications; specifically, in an spt6 mutant, we observed a decrease in H3K9 trimethylation and an increase in H3K14 acetylation at the pericentric repeats, which may be partially caused by differential recruitment of the complexes regulating these marks. Spreading of heterochromatin marks over inner and outer centromeric silencing reporters is also defective in the spt6-1 mutant, as these reporters show loss of both H3K9 di- and trimethylation. We also found that Spt6 binding at the pericentric repeats correlates with that of RNAPII, indicating that Spt6 is present during active transcription of these regions. Finally, we observed that the increase in RNAPII binding over the pericentric repeats is modest in an spt6 mutant, indicating that the silencing defect may be at least partly posttranscriptional. This is supported by our observation that an spt6 mutation causes a defect in small interfering RNA (siRNA) production. In summary, spt6 mutants have a distinct pattern of heterochromatin defects compared to other silencing mutants, likely due to both transcriptional and posttranscriptional effects. Taken together, our results reveal previously unknown roles for two generally required transcription factors, Spt6 and Iws1, in heterochromatic silencing in S. pombe.
MATERIALS AND METHODS
S. pombe strains and genetic manipulations.
The S. pombe strains used in this study are listed in Table S1, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm. Unless otherwise indicated, strains were cultured at 30°C, using yeast extract-supplemented (YES) medium, as previously described (38). For defined medium, an adaptation of Edinburgh minimal medium (EMM) was used. Ammonium chloride was replaced with 3.75 g/liter of monosodium glutamate (Sigma) as a nitrogen source, and 2 g/liter of synthetic complete mix, containing all amino acids, inositol, p-aminobenzoic acid, and uracil (79), was added to supplement the medium. This medium was designated pombe glutamate medium with SC (PMGSC). Cells containing the thiamine repressible nmt promoters were grown in PMGSC ± 15 μM thiamine. Crosses and tetrad dissection were done as previously described (38).
Gene deletions, epitope tagging, and construction of nmt-spt6+ fusions were performed by homologous recombination of PCR-generated DNA sequences into the genome. PCR primers contained 80 bases of homology to the genome and 20 bases for amplification from previously described plasmids (9). For gene deletions, a KanMX6 or NatMX6 cassette was targeted to sites flanking the open reading frame. For epitope tagging, cassettes encoding the tag and marked by KanMX6 or NatMX6 were integrated at the 3′ end of the gene, removing the endogenous stop codon. For nmt promoter insertions, cassettes encoding KanMX6 upstream of the nmt promoter were inserted at the 5′ end of the gene, adjacent to the ATG. All deletions, promoter insertions, and tags were verified by PCR analysis. Tags were also verified by Western blot analysis with a peroxidase-antiperoxidase antibody (PAP antibody; Sigma) for TAP tags, an anti-Myc antibody (A14; Santa Cruz Biotechnology), or an anti-Flag antibody (M2; Sigma). Ponceau red staining was used to determine if equal amounts of protein were loaded in each lane. All primers used for strain construction are listed in Table S2, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm.
The spt6-1 allele, which deletes the region of spt6+ encoding the helix-hairpin-helix (HhH) motif (+2566 to +2758), was constructed in a haploid strain by transformation with a PCR product that encodes the deletion. The PCR product was generated using primer FO5965, which encodes the deletion, and primer FO5795, a reverse primer 3′ of the spt6+ coding region. These primers were used to amplify the 3′ end of spt6+ with the HA3-TAP2 tag and a KanMX cassette integrated downstream of spt6+ from the TAP integration strain described below (FWP384). The PCR product was then used to transform strain FWP172 to G418 resistance, thereby constructing the deletion in a single step. Sequencing revealed that these manipulations also introduced two silent mutations, T3402C and T3661C. Subsequently, the epitope tags were deleted by integration of a stop codon at the end of the spt6 ORF, using a downstream NatMX cassette (FO5555/7615).
Spot tests were conducted by growing saturated cultures of each strain in YES, making 10-fold serial dilutions, and spotting them onto the different media to be tested. For phenotypic testing, YES plates were supplemented with 17 μg/ml thiabendazole, 10 mM caffeine, 10 μg/ml cycloheximide, or 2% formamide. For silencing reporter assays, PMGSC medium was supplemented with 0.1% 5-fluoroorotic acid (5-FOA).
Purification of Spt6 and mass spectrometry analysis.
Spt6 was purified by the TAP method, using the method previously described (40). Briefly, Spt6 was tagged with an HA3-TAP2 fusion epitope (pFS209, a generous gift from N. Rhind), generating strain FWP384. TAP preparations were done with 2-liter YES cultures of FWP384 and an untagged control (FWP172) grown to 1 × 107 cells/ml. Purified proteins were visualized by silver staining on a 5% to 20% gradient SDS-polyacrylamide gel. Total protein mixtures were analyzed at the Taplin Mass Spectrometry Facility (https://taplin.med.harvard.edu).
Microarray experiments.
Microarrays were performed on total RNA isolated from strains containing TAP-tagged versions of wild-type spt6+ (FWP384) and spt6-1 (FWP389). Protocols, microarrays, and normalization were previously described (60). Briefly, 10 to 20 μg of RNA was fluorescently labeled by direct incorporation of Cy3- or Cy5-dCTP (GE Healthcare) and hybridized to S. pombe cDNA microarrays. Microarrays were scanned with a GenePix 4000B laser scanner (Axon Instruments), and fluorescence intensity ratios were calculated with GenePix Pro (Axon Instruments). The data were normalized using a script, and two biological repeats of wild-type and spt6-1 strains were analyzed by dye swap and averaged. Genes with ratios ≥2 or ≤0.5 (mutant/wild-type) were called up- or downregulated, respectively.
Northern blot and quantitative real-time PCR analysis.
RNA isolation and Northern analyses were performed as previously described (8). The probes were amplified by PCR from S. pombe genomic DNA with primers for dg (FO7071/FO7072), spt6+ (FO7062/FO7063), and hcs1+ (FO5760/5761) and labeled by random priming with [α-32P]dATP. The dg probe anneals to multiple repetitive sequences across the pericentric regions on all three chromosomes. For measurement of RNA levels by quantitative PCR (qPCR), RNA was treated with DNase (TURBO DNA-free kit; Ambion), and then 100 ng was reverse transcribed with an oligonucleotide dT primer (Superscript III kit; Invitrogen). Each sample was analyzed in triplicate and quantified by comparison to a standard curve, composed of 10-fold serial dilutions of genomic DNA. The qPCRs were denatured at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min. Primers used for qPCR amplification were the following: dg, FO7066/FO7067; dh, FO7064/FO7065; tlh1+, FO7091/FO7092; act1+, FO3812/FO3813; hcs1+, FO5758/FO5759. The signal at regions of interest was normalized to act1+ or hcs1+. For the purposes of graphing, combined data were also normalized to wild-type values.
Isolation and Northern analysis of small RNAs were conducted as previously described (18). Briefly, small RNAs were isolated from total RNA by fractionation in RNeasy Midi columns (Qiagen) and precipitated in isopropanol. The resulting fraction was run on a 17.5% polyacrylamide, 7 M urea gel, transferred to a positively charged membrane (Perkin Elmer) by semidry transfer, UV cross-linked, and baked at 80°C for 2 h. The blot was preincubated at 35°C for 1 h in UltraHyb-Oligo buffer (Ambion), followed by overnight hybridization at 35°C with [γ-32P]dATP end-labeled oligonucleotides. Blots were washed three times in 2× SSC-0.5% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min and exposed to film (Kodak MS) on a SuperScreen. The probe for centromeric siRNAs was comprised of a mix of oligonucleotides derived from sequencing of S. pombe small RNAs (78), and the probe for snoR69 was oligonucleotide FO8016.
Minichromosome loss assays.
Wild-type and spt6-1 strains with the circular minichromosome CM3112 (66) were analyzed and the percentage of divisions resulting in loss was calculated as previously described (2, 54). Briefly, strains were streaked for single colonies on PMGSC lacking adenine. Single colonies were picked, resuspended, and used to seed 10-ml YES cultures with ∼1,000 cells/ml, which were grown for ∼15 generations. At initial and final points, cells were plated on selective and nonselective media (PMGSC with or without ade) to determine cell concentrations and percentages of cells retaining the minichromosome. These values were used to calculate the percentages of divisions resulting in loss of the minichromosome.
ChIP experiments.
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (42). Briefly, 100-ml cultures grown to 1 × 107 to 2 × 107 cells/ml were cross-linked (1% formaldehyde, 30 min) and lysed by bead beating. The chromatin fraction was isolated and sheared to 200- to 500-bp fragments using a Bioruptor sonicator (Diagenode). Immunoprecipitations (IPs) were performed overnight at 4°C with 1 μg of anti-H3 (ab1791; Abcam), 2 μg of anti-H3K9me2 (ab1220; Abcam), 2 μg of anti-H3K9me3 (07-442; Upstate), 3 μg of anti-H3K14ac (07-353; Millipore), 5 μl of anti-Rpb1 (8WG16; Covance), 2 μg of anti-Swi6 antisera (generous gift from Jun-ichi Nakayama), 5 μg of anti-protein A (SPA-27; Sigma), 5 μl of anti-Myc (9E10; Santa Cruz Biotechnology), or 3 μl of anti-Flag (M2; Sigma). The immunoprecipitates were coupled to 50 μl of protein G-Sepharose beads (GE Healthcare Life Sciences) at 4°C for 4 h. The beads were washed and eluted; the eluate was reverse-cross-linked overnight at 65°C and incubated with proteinase K and glycogen for 2 h at 37°C. DNA was purified by phenol-chloroform extraction and precipitated in ethanol overnight at −20°C. ChIP DNA was analyzed by qPCR on the Stratagene MX3000P cycler. Each sample was run in triplicate and quantified by comparison to a standard curve (10-fold serial dilutions of input DNA). Primers used for qPCR amplification were as follows: dg, FO7066/FO7067; dh, FO7064/FO7065; ura4+, FO7089/FO7090 gfr, FO7646/FO7647. Relative percent IP (% IP) values for all experiments were calculated by dividing the % IP (calculated as IP/input) at the regions of interest (dg, dh, and ura4+) by the % IP at a previously described intergenic background locus (gfr) (27). For all histone H3 modifications, the % IP was also normalized to the % IP of H3. Specificity of enrichment was determined by analysis of either an untagged or no-antibody control, which can be found in Fig. S1 to S3, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm, but were not used for the purposes of normalization.
Micrococcal nuclease digestion.
Bulk chromatin analysis was conducted as described previously (55), except with a modified NP buffer (39). Briefly, a midlog YES culture was spheroplasted and partially digested with MNase (Sigma N5386). Samples were treated with proteinase K, phenol-chloroform extracted in the presence of sodium perchlorate, and precipitated in ethanol. The resulting nucleic acid was resuspended, treated with RNase, reextracted, and precipitated. The remaining DNA was run on a 1% Tris-acetate-EDTA (TAE) agarose gel, transferred to an uncharged membrane, and Southern blotted according to standard protocol (8). Probe labeling was performed as described for Northern blots. Microarray analysis of nucleosome positioning in heterochromatin was described previously (39).
Microarray data accession number.
Microarray accession data are available at ArrayExpress under accession number E-MTAB-738.
RESULTS
S. pombe spt6+ is not essential for viability but is required for normal transcription.
Temperature-sensitive alleles of both spt6+ and iws1+ were previously isolated in a screening procedure (97), but no further characterization has been done in S. pombe. We initiated our analysis of S. pombe Spt6 with both biochemical and genetic approaches. First, we tandem-affinity purified Spt6 to identify interacting proteins. As in Saccharomyces cerevisiae and mammalian cells, we found that Spt6 stably interacts with one protein, Iws1 (see Fig. S4A at http://genepath.med.harvard.edu/∼winston/supplemental.htm). To analyze spt6+ and iws1+ genetically, we constructed a deletion of the entire coding region of each gene to determine their null phenotypes. Briefly, we deleted each gene in a diploid strain, sporulated the heterozygous diploid, and examined the haploid progeny after tetrad dissection. In contrast to Saccharomyces cerevisiae and several other organisms, but like Candida albicans (3), we found that the haploid spt6Δ progeny were viable though too sick to use experimentally; iws1Δ strains were also viable (Fig. 1B; see Fig. S4B at the URL mentioned above). To allow genetic study of Spt6, we constructed two other classes of spt6 mutations. The first was designed to be similar to the well-characterized Saccharomyces cerevisiae spt6-1004 mutation (51). This allele, designated spt6-1 in S. pombe, deletes the region of spt6+ that encodes the helix-hairpin-helix (HhH) motif (33) and causes a modest growth defect at 30°C (Fig. 1A and B). We compared the growth of spt6Δ, spt6-1, and iws1Δ and characterized their phenotypes using serial dilution spot tests (Fig. 1B; see Fig. S4C at the URL mentioned above). We found that the spt6-1 and iws1Δ mutants shared a number of phenotypes, including sensitivity to temperature, thiabendazole, caffeine, cycloheximide, and formamide, as well as poor growth on minimal media. The thiabendazole sensitivity was of particular interest, since many mutants that affect pericentric heterochromatin demonstrate a sensitivity to this microtubule inhibitor in S. pombe (34).
Given the established role of Spt6 in transcriptional regulation, we conducted microarray experiments with the spt6-1 mutant. Our results provided data on 4,873 transcripts, although the microarray did not extensively cover ribosomal DNA (rDNA), mating type, or centromeric loci. We found that levels of 11.3% of the transcripts were changed more than 2-fold in the mutant, with 4.1% (198) decreased and 7.2% (353) increased compared to wild-type transcripts (Fig. 1C; see Table S3 in the supplemental material). Using an alternative microarray normalization protocol based on spiked-in external controls, we did not detect signs of coordinated global increase or decrease in transcript levels in the spt6 mutant. To determine whether the affected genes fall into any particular functional category, they were analyzed for enrichment of Gene Ontology categories (7) and several categories were found (see Table S4, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). Notably, the genes with increased transcript levels in the spt6-1 mutant were enriched in stress response and meiotic genes (both P < 1 × 10−8); genes from both of these sets tend to cluster in subtelomeric regions (23, 65, 90). Further analysis showed that genes with increased transcript levels in spt6-1 are significantly enriched in subtelomeric regions, which are normally silenced (Fig. 1D; see Fig. S5, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). We analyzed the subtelomeric regions of chromosomes 1 and 2 that have H3K9 dimethylation and Swi6 binding (21) and found that most subtelomeric transcripts are increased in the spt6-1 mutant, with 84% of them increased at least 2-fold and more than half of that subset increased 10-fold or greater (Fig. 1C; see Fig. S5A and Table S5, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). We note that the clusters of genes with increased transcript levels extend past these boundaries (Fig. 1D; see Fig. S5 at the URL mentioned above), consistent with recent data indicating that H3K9 dimethylation extends approximately 100 kb from the telomeres (100). Finally, our microarray results showed that a representative transcript from the pericentric repeats is nearly 40-fold higher in the spt6-1 mutant than in wild-type transcripts (data not shown). These findings, in conjunction with the observed thiabendazole sensitivity, indicate that Spt6 is required for heterochromatic silencing.
Spt6 is required for silencing of all heterochromatic loci.
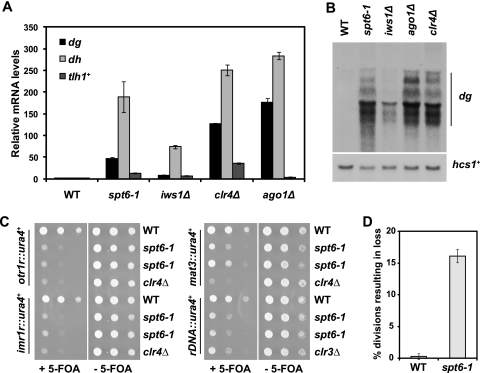
To confirm that Spt6 is required for heterochromatic silencing, we measured transcript levels of the pericentric dg and dh repeats and the subtelomeric gene tlh1+ by reverse transcription followed by quantitative PCR (qPCR). We used deletions of clr4+ and ago1+, which encode the H3K9 methyltransferase and Argonaute, respectively, as positive controls. We found that the spt6-1 mutant displays a strong pericentric silencing defect, with an approximately 50-fold increase in dg transcripts and 190-fold increase in dh transcripts (Fig. 2A; see Fig. S6A, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). The iws1Δ mutant also displayed a silencing defect, albeit with more modest increases in dg and dh levels (Fig. 2A). We confirmed the derepression of the dg repeats by Northern analysis of the same strains (Fig. 2B). To test if the silencing defect in the spt6-1 mutant was allele specific or due to a general loss of function, we analyzed an nmt81-spt6+ transcriptional shutoff strain. The nmt81 promoter (11) allows for repression of spt6+ in the presence of thiamine (see Fig. S6B at the URL mentioned above). Repression of nmt81-spt6+ also leads to a significant increase in pericentric transcripts (see Fig. S6B at the URL mentioned above), suggesting that the silencing defect results from reduced levels or function of Spt6. Both the spt6-1 and iws1Δ mutants also displayed subtelomeric silencing defects, with 13-fold and 7-fold increases in transcript levels of the subtelomeric gene tlh1+, respectively (Fig. 2A). Taken together, these results show that both Spt6 and Iws1 are required for heterochromatic silencing. As the spt6-1 mutant displayed a significantly stronger silencing defect than iws1Δ, we focused on Spt6 for the remainder of our studies.
Fig. 2.
Spt6 is required for silencing of all heterochromatic loci. (A) Measurement of transcript levels of the pericentric dg and dh repeats and the subtelomeric gene tlh1+ by quantitative PCR in spt6-1 and iws1Δ mutants. The clr4Δ and ago1Δ mutants served as positive controls, and act1+ served as a control for normalization across samples. Each column represents the mean normalized value ± standard error (SE) (n = 3 to 6). (B) Northern analysis of transcript levels over the pericentric dg repeats, with hcs1+ serving as a loading control. The dg repeat is present in multiple copies in the genome and consequently produces several transcripts, ranging in size from ∼3.5 to ∼1.5 kb. (C) ura4+ silencing reporters inserted at the outer (otr1R) or inner (imr1R) centromeric repeats, the mating type locus (mat3), and rDNA loci (rDNA) were used to assess silencing in the spt6-1 mutant. Cells were serially diluted and spotted onto media with and without 5-FOA. Failure to grow on 5-FOA indicates defective silencing. The clr4Δ and clr3Δ strains served as positive controls for loss of silencing. The results shown are after incubation at 30°C for 3 days. (D) Genomic stability was assessed by monitoring inheritance of the circular minichromosome CM3112 in wild-type (WT) and spt6-1 strains. Each column represents the mean normalized value ± SE (n = 4).
To determine if Spt6 is required for silencing at other heterochromatic loci, we used established silencing reporter strains containing the ura4+ gene inserted at the inner (imr1R) and outer (otr1R) centromeric repeats, the silent mating type cassette (mat3M), and the rDNA repeats (rDNA) (2, 86). We performed serial dilution spot tests to assay silencing, using either clr4Δ or clr3Δ mutants (clr3+ encodes the H3K14 deacetylase) as positive controls. Failure to grow on 5-FOA containing medium indicates a loss of silencing of the ura4+ reporter. We found that Spt6 is required for silencing at all of these loci, with effects comparable to those for the positive controls at otr1R, imr1R, and rDNA and an intermediate effect at mat3M (Fig. 2C). We observed comparable results on −ura media (see Fig. S6C, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). Taken together, these results indicate that Spt6 is required for silencing of heterochromatin throughout the S. pombe genome.
Finally, we wanted to assess genomic stability in the spt6-1 mutant, as many mutants that destabilize centromeric heterochromatin compromise centromere function (2). We compared inheritance of a circular minichromosome (66) in the wild type and the spt6-1 mutant; in wild-type strains, the minichromosome was lost in less than 1% of all divisions, while the spt6-1 mutant lost the plasmid in more than 15% of divisions (Fig. 2D). This rate of loss is consistent with those of several other silencing mutants, including alleles of swi6, clr4, and rik1 (2). Given that an acentric plasmid is lost in 20 to 50% of divisions in S. pombe (43, 66), it is clear that centromere function is significantly compromised in the spt6-1 mutant.
Spt6 does not cause significant changes in nucleosome positioning in heterochromatin.
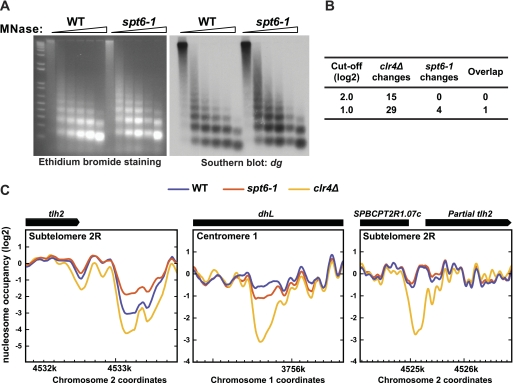
As Spt6 has been shown to control chromatin structure, we tested whether an spt6-1 mutation causes changes in nucleosome position over heterochromatin. First, we examined the sensitivity of both total chromatin and pericentric chromatin to micrococcal nuclease (MNase) digestion, comparing wild-type and spt6-1 strains. We saw a comparable laddering pattern in both strains by ethidium bromide staining and after Southern analysis of the endogenous dg repeats (Fig. 3A) and the outer centromeric reporter (otr1R::ura4+; see Fig. S7A, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). These results indicate that no detectable change in nucleosome level or position occurs over these regions in the spt6-1 mutant.
Fig. 3.
Spt6 does not affect nucleosome positioning or occupancy over heterochromatin. (A) Bulk analysis of nucleosomes was conducted by partial MNase digestion and electrophoresis on an ethidium-stained agarose gel (left), followed by Southern blotting for the endogenous dg repeats (right). (B) The number of sites with changes in nucleosome occupancy in the clr4Δ and spt6-1 mutants, using 2.0 and 1.0 cutoffs for significance. (C) Nucleosome occupancy over three clr4Δ NFRs in wild-type (WT), spt6-1, and clr4Δ strains, displayed as the moving average of normalized log2 scores over 19 probes. The site of overlap from panel B is depicted at left, with the clr4Δ and spt6-1 mutants showing opposing effects. The other clr4Δ NFRs showed minimal (center) or no (right) change in the spt6-1 mutant.
To examine nucleosome positioning at higher resolution, we isolated mononucleosomes from wild-type and spt6-1 strains after MNase digestion and annealed the recovered DNA to a tiling array covering representative regions of heterochromatin, including centromere 1, subtelomere 2R, and the mating type locus (39). From this analysis, we did not observe any significant effects (4-fold or greater) on nucleosome positioning or occupancy in heterochromatin in the spt6-1 mutant (Fig. 3B); at this same threshold, there are 15 nucleosome-free regions (NFRs) in a clr4Δ mutant (39). When we relaxed the threshold 2-fold, we found only four changes in the spt6-1 mutant: one site with nucleosome loss and three with increased occupancy. We compared the sites of these changes to those in a clr4Δ mutant using the same cutoff (29 in total) and found that the one site of overlap showed opposing effects, with decreased occupancy in clr4Δ and increased occupancy in spt6-1 (Fig. 3B and C). The remaining 28 clr4Δ NFRs were unaffected by this cutoff in the spt6-1 mutant (Fig. 3C). In contrast, we did observe a significant decrease in nucleosome occupancy over the control euchromatic act1+ locus in the spt6-1 mutant (Fig. S7B, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm); this is consistent with a previous study in Saccharomyces cerevisiae that showed that many highly transcribed genes display nucleosome loss in an spt6 mutant (46). Overall, these results suggest that Spt6 does not play a significant role in nucleosome positioning or occupancy in heterochromatin and that this is not the cause of the silencing defect in spt6 mutants.
Analysis of histone H3 lysine 9 dimethylation and trimethylation levels in an spt6-1 mutant.
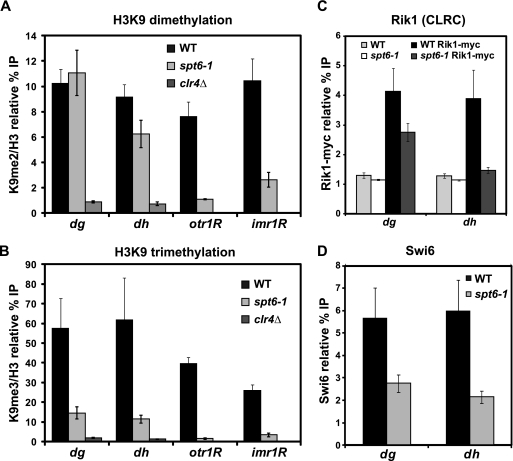
Many S. pombe silencing mutants have severely reduced levels of H3K9 dimethylation over heterochromatic regions (reviewed in references 41 and 69). To test if this loss occurs in the spt6-1 mutant, we measured both H3K9 di- and trimethylation levels at dg and dh, as well as the ura4+ reporter gene inserted at otr1R and imr1R, by chromatin immunoprecipitation (ChIP) in wild-type and spt6-1 strains, using a clr4Δ mutant as a control. Unexpectedly, the level of H3K9 dimethylation was not affected at dg and dh (Fig. 4A; see Fig. S1, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm), while in contrast, we observed a large decrease in the level of trimethylation in the spt6-1 mutant (Fig. 4B; see Fig. S1 at the URL mentioned above). The low level of H3K9 trimethylation still present in the spt6-1 mutant, compared to that in a clr4Δ mutant, might reflect either residual trimethylation, possibly due to a low level of Spt6 activity or a low level of cross-reactivity of the antibody with H3K9 dimethylation. We saw a different pattern at the otr1R and imr1R reporters, which demonstrated a significant loss of both H3K9 di- and trimethylation in the spt6-1 mutant. These results indicate that the spt6-1 mutant has a defect in heterochromatin spreading, like RNAi mutants, such as ago1Δ (45).
Fig. 4.
Analysis of histone H3 lysine 9 dimethylation and trimethylation levels at pericentric repeats and silencing reporters. (A, B) ChIP analysis was used to measure H3K9 dimethylation and trimethylation levels over the pericentric dg and dh repeats, as well as the ura4+ reporter gene inserted at otr1R and imr1R, in both wild-type (WT) and spt6-1 strains. ChIP was conducted with antibodies specific to H3K9me2, H3K9me3, or total H3. In all cases, binding was assessed by qPCR. Columns represent the mean normalized value ± standard error (SE) (n = 3 to 13). Unnormalized data and no-antiserum controls are shown in Fig. S1, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm. (C) ChIP analysis of the Rik1 subunit of the CLRC H3K9 methyltransferase complex. Rik1 was tagged with 13-myc at the endogenous locus, and binding was measured by immunoprecipitation with an anti-myc antiserum. Columns represent the mean normalized value ± SE (n = 6). Untagged controls indicate background levels of Rik1 binding. (D) ChIP analysis of the recruitment of Swi6, using a Swi6-specific antiserum (72). Columns represent the mean normalized value ± SE (n = 6), and no-antiserum controls are shown in Fig. S2, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm.
We next addressed two questions raised by the finding that H3K9 trimethylation is decreased at the endogenous pericentric repeats in spt6-1 mutants. First, we asked if the decreased level of H3K9 trimethylation might be caused by reduced recruitment of the CLRC complex, which contains Clr4 and is responsible for H3K9 methylation (20, 44, 48, 57). To this end, we used ChIP to measure the level of recruitment of a Myc-tagged version of Rik1, a component of CLRC, in wild-type and spt6-1 strains. In the spt6-1 mutant, CLRC occupancy is decreased at both the dg and dh repeats, although there is still significant binding at dg (Fig. 4C). Second, we investigated the functional consequences of loss of H3K9 trimethylation by examining the recruitment of Swi6, an HP1 homolog that binds H3K9 methyl marks (10, 73). Again using ChIP, we found that Swi6 occupancy persists but is decreased 2- to 3-fold at dg and dh (Fig. 4D; see Fig. S2C at http://genepath.med.harvard.edu/∼winston/supplemental.htm). Taken together, these results indicate that at the pericentric repeats, decreased recruitment of the CLRC complex in spt6-1 mutants may lead to loss of H3K9 trimethylation, which subsequently leads to a decrease in Swi6 binding.
Histone H3 lysine 14 acetylation is increased in spt6-1 mutants.
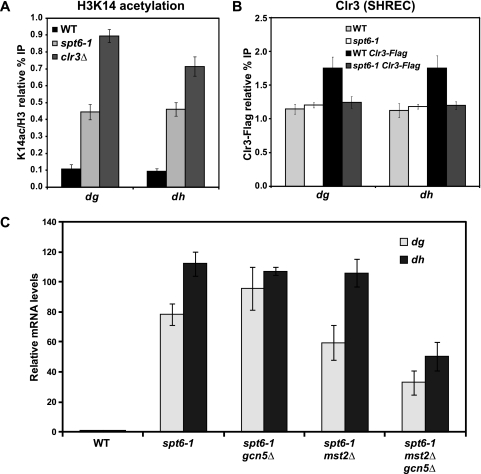
Like the spt6-1 mutant, loss of the histone H3 lysine 14 deacetylase complex, SHREC, has been previously demonstrated to decrease heterochromatic silencing while leaving H3K9 dimethylation intact (70, 81, 82). Therefore, we measured H3K14 acetylation levels in the wild-type strain and the spt6-1 mutant by ChIP, using a clr3Δ strain as a control. We observed a 4-fold increase in H3K14 acetylation levels at dg and dh in the spt6-1 mutant, compared to that in the wild type (Fig. 5A; see Fig. S2, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). Consistent with these findings, ChIP analysis of a Flag-tagged version of Clr3 revealed that its recruitment to dg and dh is at background levels in an spt6-1 mutant, even though its recruitment was modest in wild-type cells (Fig. 5B).
Fig. 5.
Measurement of histone H3 lysine 14 acetylation levels over the pericentric repeats in wild-type (WT) and spt6-1 strains. (A) ChIP analysis was performed to measure the level of H3K14 acetylation over the pericentric dg and dh repeats using antisera specific to H3K14ac and unmodified H3. In all cases, binding was assessed by quantitative real-time PCR. Columns represent the mean normalized value ± standard error (SE) (n = 6). Unnormalized data and no-antiserum controls are shown in Fig. S2, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm. (B) Recruitment of the SHREC deacetylase complex to the pericentric repeats was measured by ChIP analysis of the subunit Clr3, which was C-terminally Flag tagged at its endogenous locus. Columns represent the mean normalized value ± SE (n = 6). Untagged controls indicate background levels of Clr3 binding. (C) Isogenic strains containing specific combinations of spt6-1 with gcn5Δ and mst2Δ were constructed by cross and tetrad analysis. Expression of the dg and dh transcripts were assessed by qPCR. Columns represent the mean normalized value ± SE (n = 3 to 4).
To investigate the possibility that this increase in H3K14 acetylation contributes to the silencing defect, we tested whether deletion of the genes encoding the histone acetyltransferases primarily responsible for H3K14 acetylation, gcn5+ and mst2+ (75), might suppress the spt6-1 silencing defect. Our results show that neither gcn5Δ nor mst2Δ suppresses the silencing defect of spt6-1 (Fig. 5C). However, combining the two deletions conferred a partial suppression of the silencing defect, with a 2-fold decrease of dg and dh transcript levels in a gcn5Δ mst2Δ spt6-1 triple mutant, compared to levels in a single spt6-1 mutant (Fig. 5C). These results suggest that increased acetylation contributes to, but is not the sole reason for, the silencing defect in spt6-1 mutants.
Spt6 binding correlates with that of RNAPII at heterochromatin.
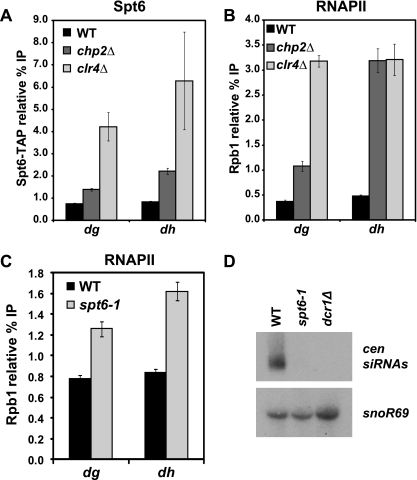
Genome-wide studies in Saccharomyces cerevisiae have shown that Spt6 associates with transcribed regions at a level proportional to that of RNAPII (46, 67). As a low level of transcription is known to be required for heterochromatic silencing in S. pombe, we asked if the level of Spt6 occupancy correlates with that of RNAPII at silenced regions. To do this, we used ChIP to measure the occupancy of RNAPII and of a TAP-tagged version of Spt6 at dg and dh. In wild-type cells, neither Spt6 nor RNAPII is detectable at these regions (Fig. 6A and B; see Fig. S3A and B, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). In contrast, we see a 50-fold enrichment of both factors at the highly expressed gene pma1+ (see Fig. S3D at the URL mentioned above). However, there is a detectable ChIP signal for both Spt6 and RNAPII in a chp2Δ mutant, which derepresses transcription, while leaving heterochromatin intact (37, 70, 80, 86), and a stronger ChIP signal in a clr4Δ mutant, which abolishes heterochromatin altogether (Fig. 6A and B; see Fig. S3A and B at the URL mentioned above). Our results, then, show that Spt6 occupancy mimics that of RNAPII. Taken together with the known physical interaction between Spt6 and RNAPII (32, 83, 94), it is reasonable to conclude that Spt6 is present during transcription of the pericentric repeats and, like RNAPII, may contribute to silencing via a direct mechanism.
Fig. 6.
Spt6 may play a role in transcriptional and posttranscriptional silencing. (A) ChIP analysis of Spt6 in a wild-type (WT) strain, in a chp2Δ strain, where transcription is derepressed, and in a clr4Δ strain, where heterochromatin is lost (n = 6). Spt6 was C-terminally TAP tagged at its endogenous locus and immunoprecipitated with an anti-protein A antibody. Controls using untagged Spt6 are shown in Fig. S3A, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm. (B) ChIP analysis of RNAPII, using an antibody directed against the Rpb1 subunit, demonstrates a similar pattern to Spt6 (n = 6). No-antiserum controls are shown in Fig. S3B. (C) ChIP analysis of RNAPII in wild-type and spt6-1 strains, conducted as for panel B. No-antiserum controls are shown in Fig. S3C at the URL mentioned above. (D) Northern analysis of pericentric siRNA (cen siRNA) levels in wild-type and spt6-1 strains. The dcr1Δ mutant served as a positive control for loss of siRNAs, and snoR69 served as a loading control.
Spt6 may contribute transcriptionally and posttranscriptionally to silencing.
Finally, we wanted to assess the transcriptional contribution to the silencing defect in the spt6-1 mutant; we used ChIP to measure RNAPII occupancy of dg and dh in the spt6-1 mutant and saw only a modest increase in comparison to wild-type occupancy (Fig. 6C; see Fig. S3C, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). Compared to the larger increases in RNAPII occupancy seen in the chp2Δ and clr4Δ mutants (Fig. 6B), it seems unlikely that the observed increase in the spt6-1 mutant would entirely account for the high level of dg and dh transcripts. Although ChIP may not detect some types of transcription defects, our results suggest that Spt6 may play a role in posttranscriptional silencing.
To assess posttranscriptional defects in the spt6-1 mutant, we measured the level of centromeric siRNAs. RNAi has been shown to play a role in the transcriptional and posttranscriptional silencing of heterochromatin in S. pombe, particularly at the pericentric repeats (71, 74, 87, 88). Many silencing mutants in S. pombe are defective for siRNA biogenesis. These mutants include deletions of the genes encoding components of the RNAi machinery, such as dicer (dcr1+) (87) and the RNA-dependent RNA polymerase (rdp1+) (71), as well as clr4+ (71, 74). However, some mutants that disrupt silencing do not show any defect in siRNA production; these include deletions of genes encoding components of the SHREC complex (70) and the chromatin remodeler Pob3 (56). To assess siRNA biogenesis in the spt6-1 mutant, we measured the level of pericentric siRNAs by Northern analysis. Our results show that siRNAs are effectively undetectable in the spt6-1 mutant, with a defect comparable to that in the dcr1Δ mutant (Fig. 6D). Taken together with the RNAPII ChIP results, this provides evidence that Spt6 may regulate posttranscriptional silencing.
DISCUSSION
In this study, we have provided the first characterization of Spt6 in S. pombe, leading to the discovery that both Spt6 and its binding partner, Iws1, are required for heterochromatic silencing. While Spt6 is known to play a number of critical roles in regulation of transcription and chromatin in diverse organisms, this is the first time it has been shown to be important for the formation of heterochromatin. There is no evidence that Saccharomyces cerevisiae spt6 mutants are defective for silencing (C. Fung and F. Winston, unpublished data), although heterochromatin structure in Saccharomyces cerevisiae is markedly different from that in S. pombe. Our results suggest that the heterochromatic silencing defect in S. pombe spt6 mutants is not caused by changes in nucleosome occupancy or positioning (Fig. 3). We show, however, that the silencing defect at the pericentric dg and dh repeats is at least partly due to changes in histone modifications, specifically a decreased level of H3K9 trimethylation, without a decrease in dimethylation, and an increased level of H3K14 acetylation (Fig. 4 and 5). We also observe a defect in spreading of H3K9 di- and trimethylation over the otr1R and imr1R reporters (Fig. 4). Additional results suggest that there are both transcriptional and posttranscriptional defects in spt6 mutants (Fig. 6). Taken together, our results have shown that a broadly required transcription factor, Spt6, is critical for heterochromatin formation in S. pombe.
Other S. pombe mutants have been identified that alleviate silencing at the pericentric repeats without substantially affecting H3K9 dimethylation. One example is a deletion of clr3+, which encodes the histone H3K14 deacetylase and a component of the SHREC complex. Several studies have shown that H3K14 deacetylation is required for transcriptional silencing but not for H3K9 dimethylation (70, 81, 82, 92). While we have shown that H3K14 acetylation levels are increased over pericentric heterochromatin, this is unlikely to be the sole cause of the spt6-1 silencing defect, as loss of both Mst2 and Gcn5, the acetyltransferases primarily responsible for H3K14 acetylation (75), only weakly suppresses the spt6-1 silencing defect. Furthermore, spt6 mutants have some phenotypes that are inconsistent with the loss of the SHREC complex, such as loss of centromeric siRNA production and modest, if any, changes in nucleosome occupancy in heterochromatin. We cannot rule out that other changes in histone acetylation in spt6-1 mutants may contribute to the silencing defect.
There is precedence for Spt6 to control the degree of histone methylation in other contexts. Previous studies have shown that Spt6 controls histone H3K36 methylation in both HeLa cells (95) and Saccharomyces cerevisiae (22, 28, 96). In both cases, some specificity has been seen for effects on H3K36 trimethylation. In HeLa cells, there is a specific defect in H3K36 trimethylation due to a failure to recruit Iws1, which subsequently recruits Set2D, the methyltransferase responsible for the trimethyl mark. In Saccharomyces cerevisiae, certain mutant alleles of SPT6, including spt6-1004, which is analogous to our S. pombe spt6-1 allele, have defects in both H3K36 di- and trimethylation (22, 28, 96). However, only the H3K36 dimethylation defect can be rescued by overexpression of SET2, which encodes the H3K36 methyltransferase, and recombinant Set2 was able to in vitro dimethylate but not trimethylate chromatin isolated from the spt6 mutant, suggesting that Spt6 regulates some property of chromatin that affects its ability to be trimethylated at H3K36 (96). By extension, a similar mechanism may control H3K9 trimethylation in an S. pombe spt6-1 mutant, as binding of the CLRC complex persists at the dg repeats (Fig. 4C), but trimethylation is nearly lost (Fig. 4A).
The spt6-1 mutation causes many changes in transcription; consequently, the silencing defect could be indirect, by altered transcription of a gene encoding a silencing factor. To address this possibility, we have mined our microarray data for altered mRNA levels of genes known to be involved in silencing (see Table S6, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). Using a 2-fold cutoff for regulation, we found that one or more of the genes encoding histone H3 had increased mRNA levels in an spt6-1 mutant, though cross-hybridization of the array probes did not allow us to determine which H3 locus had altered expression. However, protein levels of H3 were similar between spt6-1 and wild-type strains, based on Western analysis (see Fig. S8A, available at http://genepath.med.harvard.edu/∼winston/supplemental.htm). We also found three genes, encoding factors required for silencing, that have modestly reduced mRNA levels: arb1+, stc1+, and raf1+. We were able to rule out two of them as causative based on phenotypic differences with spt6-1 mutants. arb1Δ mutants have reduced H3K9 dimethylation (19), and stc1Δ mutants do not impair silencing of a mating type reporter (14). For raf1+, we found reduced transcript levels (see Fig. S8B at the URL mentioned above) but normal Raf1 protein levels (see Fig. S8C at the URL mentioned above) in the spt6-1 mutant. This result suggests posttranscriptional regulation of Raf1 but also indicates that it is unlikely to be the source of the silencing defect in spt6-1. Furthermore, like arb1Δ, raf1Δ leads to a loss of H3K9 dimethylation (44, 57, 85). Taken together, these results indicate that the silencing defects in spt6 mutants are not caused by altered transcription of another gene required for silencing.
Given previous studies linking Spt6 to the control of chromatin structure, we were surprised that an spt6-1 mutant does not alter nucleosome occupancy or position over heterochromatin. A recent study showed that mutations in genes encoding a number of silencing factors cause nucleosome-free regions to form at specific sites in heterochromatin (39). The lack of such changes in an spt6-1 mutant indicates that changes in nucleosome position are not the cause of the silencing defect we observe, and it also suggests that Spt6 silences heterochromatin in a manner distinct from that of many other silencing factors. Similar to spt6-1, deletion of a nonessential component of the FACT histone chaperone complex, pob3+, also causes a silencing defect (56) but fails to display large changes in nucleosome positioning in heterochromatin (39). The FACT complex contains Spt16, and given some of the phenotypic similarities between spt6 and spt16 mutants in Saccharomyces cerevisiae (24, 51, 56, 63, 64), it is possible that these two factors control silencing in a similar way in S. pombe, perhaps by altering heterochromatin structure in a way not detectable by conventional assays. In contrast, a recent study demonstrated that the Asf1/HIRA histone chaperone complex is required for silencing of pericentric and mating type heterochromatin and is important for regulation of nucleosome positioning over these regions (93). Asf1/HIRA mutants do not affect production of siRNAs or lead to substantial loss of K9 dimethylation at the otr1R::ura4+ reporter (93), suggesting that the mechanism by which it contributes to silencing is distinct from that of Spt6.
Spt6 is known to be involved in many processes, and our studies have revealed a combination of silencing defects in the spt6-1 mutant that is distinct from the phenotypes observed in other silencing mutants. Taken together, our data suggest that Spt6 contributes to silencing in multiple ways, at both the transcriptional and posttranscriptional levels. The modest increase in RNAPII occupancy in the spt6-1 mutant, compared to the stronger increases in the chp2Δ and clr4Δ mutants, shows that the mechanism is not solely transcriptional. On the other hand, the failure of the mst2Δ to suppress the spt6-1 silencing defect indicates that it is not solely posttranscriptional either, as a recent study in S. pombe reported that an mst2Δ mutant could suppress the silencing defects of the RNAi machinery but not the chromatin modifiers (77). Spt6 could play many roles at either stage of the process. It could regulate transcription initiation or elongation of pericentric transcripts. The proper balance between RNAPII transcription and occlusion is required for silencing, and it is possible that Spt6 is needed to promote transcription, much like the JmjC factor Epe1 (reviewed in reference 41). Spt6 could also affect splicing of pericentric transcripts, as a number of splicing factors have been shown to play a role in silencing pericentric transcripts (13, 26). Like Spt6, the splicing mutants show only a modest decrease in H3K9 dimethylation. However, they differ from spt6-1 in other respects; for example, these mutants do not alleviate silencing at the mating type reporter mat3M::ura4+ (13). It is also possible that Spt6 interacts with other factors and complexes that affect silencing, such as Mlo3 (98) or the TRAMP complex (17, 89). Finally, it is possible that Spt6 contributes to silencing by a mechanism we have not yet considered. Further studies should focus on the activities of Spt6 that are required for heterochromatic silencing. Going forward, it will be interesting to understand how a generally required transcription factor plays specialized roles in specific parts of the genome.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jun-ichi Nakayama for providing anti-Swi6 antisera, to Janet Partridge and Susan Forsburg for strains, and to Nick Rhind for plasmids. We thank Danesh Moazed and members of his lab, especially Shane Buker, Erica Gerace, and Danny Holoch, for strains, protocols, and advice. We also thank Dom Helmlinger and Steve Doris for helpful comments on the manuscript.
This work was supported by NIH grant GM32967 to F.W., NIH grant GM71801 to H.D.M., and Cancer Research UK (J.B.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Adkins M. W., Tyler J. K. 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21:405–416 [DOI] [PubMed] [Google Scholar]
- 2. Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218–233 [DOI] [PubMed] [Google Scholar]
- 3. Al-Rawi N., Laforce-Nesbitt S. S., Bliss J. M. 2010. Deletion of Candida albicans SPT6 is not lethal but results in defective hyphal growth. Fungal Genet. Biol. 47:288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andrulis E. D., Guzman E., Doring P., Werner J., Lis J. T. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andrulis E. D., et al. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420:837–841 [DOI] [PubMed] [Google Scholar]
- 6. Ardehali M. B., et al. 2009. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 28:1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aslett M., Wood V. 2006. Gene Ontology annotation status of the fission yeast genome: preliminary coverage approaches 100%. Yeast 23:913–919 [DOI] [PubMed] [Google Scholar]
- 8. Ausubel F. M., et al. 1991. Current protocols in molecular biology. Green Publishing Associates and Wiley-Interscience, New York, NY [Google Scholar]
- 9. Bähler J., et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951 [DOI] [PubMed] [Google Scholar]
- 10. Bannister A. J., et al. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124 [DOI] [PubMed] [Google Scholar]
- 11. Basi G., Schmid E., Maundrell K. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131–136 [DOI] [PubMed] [Google Scholar]
- 12. Basrai M. A., Kingsbury J., Koshland D., Spencer F., Hieter P. 1996. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2838–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bayne E. H., et al. 2008. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science 322:602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bayne E. H., et al. 2010. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140:666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bortvin A., Winston F. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473–1476 [DOI] [PubMed] [Google Scholar]
- 16. Bucheli M. E., Buratowski S. 2005. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 24:2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buhler M., Haas W., Gygi S. P., Moazed D. 2007. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129:707–721 [DOI] [PubMed] [Google Scholar]
- 18. Buhler M., Verdel A., Moazed D. 2006. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125:873–886 [DOI] [PubMed] [Google Scholar]
- 19. Buker S. M., et al. 2007. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 14:200–207 [DOI] [PubMed] [Google Scholar]
- 20. Burckin T., et al. 2005. Exploring functional relationships between components of the gene expression machinery. Nat. Struct. Mol. Biol. 12:175–182 [DOI] [PubMed] [Google Scholar]
- 21. Cam H. P., et al. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37:809–819 [DOI] [PubMed] [Google Scholar]
- 22. Carrozza M. J., et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592 [DOI] [PubMed] [Google Scholar]
- 23. Chen D., et al. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14:214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheung V., et al. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiang P. W., et al. 1996. Identification and analysis of the human and murine putative chromatin structure regulator SUPT6H and Supt6h. Genomics 34:328–333 [DOI] [PubMed] [Google Scholar]
- 26. Chinen M., Morita M., Fukumura K., Tani T. 2010. Involvement of the spliceosomal U4 small nuclear RNA in heterochromatic gene silencing at fission yeast centromeres. J. Biol. Chem. 285:5630–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi E. S., Shin J. A., Kim H. S., Jang Y. K. 2005. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res. 33:7102–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chu Y., Sutton A., Sternglanz R., Prelich G. 2006. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 26:3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Close D., et al. 2011. Crystal structures of the S. cerevisiae Spt6 core and C-terminal tandem SH2 domain. J. Mol. Biol. 408:697–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dengl S., Mayer A., Sun M., Cramer P. 2009. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J. Mol. Biol. 389:211–225 [DOI] [PubMed] [Google Scholar]
- 31. Diebold M. L., et al. 2010. The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26. EMBO J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diebold M. L., et al. 2010. Noncanonical tandem SH2 enables interaction of elongation factor Spt6 with RNA polymerase II. J. Biol. Chem. 285:38389–38398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doherty A. J., Serpell L. C., Ponting C. P. 1996. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 24:2488–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ekwall K., et al. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109:2637–2648 [DOI] [PubMed] [Google Scholar]
- 35. Endoh M., et al. 2004. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 24:3324–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fischbeck J. A., Kraemer S. M., Stargell L. A. 2002. SPN1, a conserved gene identified by suppression of a postrecruitment-defective yeast TATA-binding protein mutant. Genetics 162:1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fischer T., et al. 2009. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 106:8998–9003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Forsburg S. L., Rhind N. 2006. Basic methods for fission yeast. Yeast 23:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia J. F., Dumesic P. A., Hartley P. D., El-Samad H., Madhani H. D. 2010. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 24:1758–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gould K. L., Ren L., Feoktistova A. S., Jennings J. L., Link A. J. 2004. Tandem affinity purification and identification of protein complex components. Methods 33:239–244 [DOI] [PubMed] [Google Scholar]
- 41. Grewal S. I. 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 20:134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Helmlinger D., et al. 2008. The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev. 22:3184–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heyer W. D., Sipiczki M., Kohli J. 1986. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol. Cell. Biol. 6:80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Horn P. J., Bastie J. N., Peterson C. L. 2005. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 19:1705–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Irvine D. V., et al. 2006. Argonaute slicing is required for heterochromatic silencing and spreading. Science 313:1134–1137 [DOI] [PubMed] [Google Scholar]
- 46. Ivanovska I., Jacques P. E., Rando O. J., Robert F., Winston F. 2011. Control of chromatin structure by spt6: different consequences in coding and regulatory regions. Mol. Cell. Biol. 31:531–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jensen M. M., Christensen M. S., Bonven B., Jensen T. H. 2008. Requirements for chromatin reassembly during transcriptional downregulation of a heat shock gene in Saccharomyces cerevisiae. FEBS J. 275:2956–2964 [DOI] [PubMed] [Google Scholar]
- 48. Jia S., Kobayashi R., Grewal S. I. 2005. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 7:1007–1013 [DOI] [PubMed] [Google Scholar]
- 49. Johnson S. J., et al. 2008. Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J. Mol. Biol. 377:1460–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaplan C. D., Holland M. J., Winston F. 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280:913–922 [DOI] [PubMed] [Google Scholar]
- 51. Kaplan C. D., Laprade L., Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096–1099 [DOI] [PubMed] [Google Scholar]
- 52. Kaplan C. D., Morris J. R., Wu C., Winston F. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim M., Ahn S. H., Krogan N. J., Greenblatt J. F., Buratowski S. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kipling D., Kearsey S. E. 1990. Reversion of autonomously replicating sequence mutations in Saccharomyces cerevisiae: creation of a eucaryotic replication origin within procaryotic vector DNA. Mol. Cell. Biol. 10:265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lantermann A., Stralfors A., Fagerstrom-Billai F., Korber P., Ekwall K. 2009. Genome-wide mapping of nucleosome positions in Schizosaccharomyces pombe. Methods 48:218–225 [DOI] [PubMed] [Google Scholar]
- 56. Lejeune E., et al. 2007. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr. Biol. 17:1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li F., et al. 2005. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr. Biol. 15:1448–1457 [DOI] [PubMed] [Google Scholar]
- 58. Lindstrom D. L., et al. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu J., et al. 2011. Solution structure of the tandem SH2 domains from Spt6 and their binding to the phosphorylated RNA polymerase II C-terminal domain. J. Biol. Chem. 286:29218–29226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lyne R., et al. 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maclennan A. J., Shaw G. 1993. A yeast SH2 domain. Trends Biochem. Sci. 18:464–465 [DOI] [PubMed] [Google Scholar]
- 62. Malagon F., Aguilera A. 2001. Yeast spt6-140 mutation, affecting chromatin and transcription, preferentially increases recombination in which Rad51p-mediated strand exchange is dispensable. Genetics 158:597–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Malone E. A., Clark C. D., Chiang A., Winston F. 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5710–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mason P. B., Struhl K. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mata J., Lyne R., Burns G., Bähler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32:143–147 [DOI] [PubMed] [Google Scholar]
- 66. Matsumoto T., Murakami S., Niwa O., Yanagida M. 1990. Construction and characterization of centric circular and acentric linear chromosomes in fission yeast. Curr. Genet. 18:323–330 [Google Scholar]
- 67. Mayer A., et al. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17:1272–1278 [DOI] [PubMed] [Google Scholar]
- 68. McDonald S. M., Close D., Xin H., Formosa T., Hill C. P. 2010. Structure and biological importance of the spn1-spt6 interaction, and its regulatory role in nucleosome binding. Mol. Cell 40:725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moazed D., et al. 2006. Studies on the mechanism of RNAi-dependent heterochromatin assembly. Cold Spring Harbor Symp. Quant. Biol. 71:461–471 [DOI] [PubMed] [Google Scholar]
- 70. Motamedi M. R., et al. 2008. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 32:778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Motamedi M. R., et al. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119:789–802 [DOI] [PubMed] [Google Scholar]
- 72. Nakayama J., Klar A. J., Grewal S. I. 2000. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101:307–317 [DOI] [PubMed] [Google Scholar]
- 73. Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113 [DOI] [PubMed] [Google Scholar]
- 74. Noma K., et al. 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 36:1174–1180 [DOI] [PubMed] [Google Scholar]
- 75. Nugent R. L., et al. 2010. Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation. BMC Genomics 11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ponting C. P. 2002. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 30:3643–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Reddy B. D., et al. 2011. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 25:214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Reinhart B. J., Bartel D. P. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831. [DOI] [PubMed] [Google Scholar]
- 79. Rose M. D., Winston F., Hieter P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 80. Sadaie M., et al. 2008. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol. Cell. Biol. 28:6973–6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shimada A., et al. 2009. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 23:18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sugiyama T., et al. 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128:491–504 [DOI] [PubMed] [Google Scholar]
- 83. Sun M., Lariviere L., Dengl S., Mayer A., Cramer P. 2010. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II CTD. J. Biol. Chem. 285:41597–41603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Swanson M. S., Carlson M., Winston F. 1990. SPT6, an essential gene that affects transcription in Saccharomyces cerevisiae, encodes a nuclear protein with an extremely acidic amino terminus. Mol. Cell. Biol. 10:4935–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thon G., et al. 2005. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 171:1583–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thon G., Verhein-Hansen J. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155:551–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Verdel A., et al. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Volpe T. A., et al. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837 [DOI] [PubMed] [Google Scholar]
- 89. Wang S. W., Stevenson A. L., Kearsey S. E., Watt S., Bähler J. 2008. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol. Cell. Biol. 28:656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wiren M., et al. 2005. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 24:2906–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wood V., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880 [DOI] [PubMed] [Google Scholar]
- 92. Yamada T., Fischle W., Sugiyama T., Allis C. D., Grewal S. I. 2005. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell 20:173–185 [DOI] [PubMed] [Google Scholar]
- 93. Yamane K., et al. 2011. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol. Cell 41:56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yoh S. M., Cho H., Pickle L., Evans R. M., Jones K. A. 2007. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 21:160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yoh S. M., Lucas J. S., Jones K. A. 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 22:3422–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Youdell M. L., et al. 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28:4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yuasa T., et al. 2004. An interactive gene network for securin-separase, condensin, cohesin, Dis1/Mtc1 and histones constructed by mass transformation. Genes Cells 9:1069–1082 [DOI] [PubMed] [Google Scholar]
- 98. Zhang K., et al. 2011. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science 331:1624–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang L., Fletcher A. G., Cheung V., Winston F., Stargell L. A. 2008. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol. Cell. Biol. 28:1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zofall M., et al. 2009. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature 461:419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.