Abstract
The RNA-binding protein (RBP) nucleolin promotes the expression of several proliferative proteins. Nucleolin levels are high in cancer cells, but the mechanisms that control nucleolin expression are unknown. Here, we show that nucleolin abundance is controlled posttranscriptionally via factors that associate with its 3′ untranslated region (3′UTR). The RBP HuR was found to interact with the nucleolin (NCL) 3′UTR and specifically promoted nucleolin translation without affecting nucleolin mRNA levels. In human cervical carcinoma HeLa cells, analysis of a traceable NCL 3′UTR bearing MS2 RNA hairpins revealed that NCL RNA was mobilized to processing bodies (PBs) after silencing HuR, suggesting that the repression of nucleolin translation may occur in PBs. Immunoprecipitation of MS2-tagged NCL 3′UTR was used to screen for endogenous repressors of nucleolin synthesis. This search identified miR-494 as a microRNA that potently inhibited nucleolin expression, enhanced NCL mRNA association with argonaute-containing complexes, and induced NCL RNA transport to PBs. Importantly, miR-494 and HuR functionally competed for modulation of nucleolin expression. Moreover, the promotion of cell growth previously attributed to HuR was due in part to the HuR-elicited increase in nucleolin expression. Our collective findings indicate that nucleolin expression is positively regulated by HuR and negatively regulated via competition with miR-494.
INTRODUCTION
Cell proliferation is strongly influenced through changes in the collection of expressed proteins. Their abundance is largely driven by posttranscriptional mechanisms, particularly changes in the stability and translation of mature mRNAs. These processes are controlled by two main types of mRNA-interacting factors: RNA-binding proteins (RBPs) and noncoding RNAs (46, 47). Numerous RBPs that affect the stability and translation of mRNAs encoding proliferative proteins have been described, including elav/Hu proteins (the ubiquitous HuR and the primarily neuronal HuB, HuC, and HuD), AU-binding factor 1 (AUF1), tristetraprolin (TTP), KH domain-containing RBP (KSRP), the T-cell intracellular antigen 1 (TIA-1) and related (TIAR) proteins, nuclear factor 90 (NF90), polypyrimidine tract-binding protein (PTB), and CUG repeat binding protein 1 (CUGBP1) (16, 26, 28, 35, 43, 50, 53, 63, 65). Together, these RBPs govern the expression of cyclins A2, B1, D1, and E, cyclin-dependent kinases (cdk's [e.g., cdk4]), cdk inhibitory proteins (e.g., p21 and p27), and other cell cycle regulatory proteins and transcription factors (e.g., c-myc, p53, c-fos, and c-jun) (reviewed in reference 4).
The RBP nucleolin has also been implicated in controlling cell proliferation (57). Nucleolin interacts with RNA via four RNA-recognition motifs (RRMs) and a C-terminal, glycine-arginine-rich domain (25, 30, 45, 60). Nucleolin is prominently abundant in the nucleolus, where it interacts with precursor rRNA and assists in its maturation and processing (12, 24, 25, 55); accordingly, nucleolin downregulation disrupted nucleolar function, impairing cell cycle progression and centrosome duplication (57). In the cytoplasm, nucleolin interacts with mature mammalian mRNAs, typically at the 3′ untranslated region (3′UTR), but sometimes at the 5′UTR and coding region (CR) (7). The actions of nucleolin on target mRNAs vary widely depending on the experimental system and the bound mRNA. Besides a role in nucleocytoplasmic transport, nucleolin was shown to promote the stability of mRNAs encoding β-globin, amyloid precursor protein (APP), gastrin, B-cell leukemia/lymphoma 2 (Bcl-2), Bcl-xL, interleukin 2 (IL-2), and growth arrest and DNA damage-inducible 45 (Gadd45α) (18, 29, 39, 49, 52, 66). It also reduced the translation of p53 and prostaglandin endoperoxide H synthase 1 (PGHS1) (14, 59). Recently, nucleolin was shown to associate with dozens of mRNAs encoding proteins with roles in cell growth and proliferation as well as in cancer, including Bcl-2, p53, cyclin I, and Akt1 (7). Additionally, nucleolin was found to promote the translation of matrix metalloprotease 9 (MMP9), several selenoproteins, and a subset of mRNAs bearing a G-rich element (7, 22, 44). Nucleolin was also found on the cell surface in many cancer cells and thus serves as a cancer marker (19, 31, 54). As a DNA-binding protein, nucleolin induces chromatin decondensation by the remodeling complex SWI/SNF (switch/sucrose nonfermentable in yeast), functions as a histone chaperone, facilitates transcription by RNA polymerases I and II, and modulates DNA replication (8, 45, 61).
While nucleolin is expressed ubiquitously, its levels are significantly elevated in many cancer cells. Given its influence on the expression of cancer proteins (e.g., Bcl-2, Bcl-xL, p53, and MMP9), nucleolin has become an important target of anticancer therapy in recent years. In addition, nucleolin's involvement in other pathologies, such as viral infections, autoimmune diseases, graft-versus-host reaction, and Alzheimer's and Parkinson's diseases (9, 15, 17, 20, 58, 61), has increased its diagnostic and therapeutic value. However, very little is known about the regulation of nucleolin expression.
Here, we sought to investigate the molecular mechanisms that govern nucleolin expression. We previously identified nucleolin as a putative target of the RBP HuR (5, 6, 41). In this study, we obtained evidence that HuR associated with the 3′UTR of the nucleolin (NCL) mRNA and that this interaction promoted nucleolin translation without affecting NCL mRNA half-life. Further insight into the enhancement of nucleolin expression by HuR came through the identification of a microRNA, miR-494, which lowered nucleolin expression by competing with HuR. MicroRNAs (∼22-nucleotide [nt]-long noncoding RNAs) are integral components of argonaute (Ago)-containing RNA-induced silencing complexes (RISC) and typically function as repressors of gene expression. As part of RISC/Ago, microRNAs interact with mRNAs with partial complementarity and generally reduce mRNA stability and/or translation (34); these processes can occur in specialized cytoplasmic regions, including processing bodies (PBs) (10, 40). Our results indicate that HuR competed with miR-494 for modulation of nucleolin production, with HuR preventing the recruitment of NCL mRNA to PBs and miR-494 favoring it. The competing actions of HuR and miR-494 influenced nucleolin expression, in turn modulating cell proliferation and survival.
MATERIALS AND METHODS
Cell culture, transfection, small RNAs, and plasmids.
HeLa cells were cultured in Dulbecco's modified essential medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum and antibiotics. miRNAs (Ambion), control small interfering RNA (Ctrl siRNA; Qiagen), HuR siRNA (Qiagen), and enhanced green fluorescent protein (EGFP) reporter plasmids were transfected with Lipofectamine-RNAiMAX or Lipofectamine 2000 (Invitrogen). Plasmid DNAs were transfected at 25 ng/ml [pEGFP, pEGFP-NCL(3′), and pEGFP-NCL(3′mut)], 0.5 μg/ml [pEGFP, pEGFP-NCL(CR), and pMS2-YFP (expressing yellow fluorescent protein)], or 1 μg/ml [pMS2-RL and pMS2-RL-NCL(3′)]. The Renilla luciferase coding region (RL), was added in order to provide a reporter open reading frame to the MS2 RNA-containing constructs. NCL 3′UTR reporter constructs were made by inserting cDNA corresponding to the NCL 3′UTR into pEGFP-C1 or pMS2. pMS2, or pMS2-YFP plasmids were described previously (38). EGFP reporters were cloned by inserting specific fragments from the NCL 3′UTR into pEGFP-C1 (BD Bioscience). The seed region of the miR-494 binding site on the NCL 3′UTR in pEGFP-NCL(3′mut) was disrupted by site-directed mutagenesis (Stratagene).
Western blot analysis.
Whole-cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS, 1 mM dithiothreitol), separated by electrophoresis in SDS-containing polyacrylamide gels, and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). Incubations with primary antibodies to detect nucleolin, HuR, or EGFP (Santa Cruz Biotech) or to detect β-actin (Abcam) were followed by incubations with the appropriate secondary antibodies conjugated with horseradish peroxidase (HRP) (GE Healthcare) and by detection using enhanced luminescence (GE Healthcare).
RNA analysis.
Total RNA was prepared from whole cells, gradient fractions, or RNP immunoprecipitation (IP) samples using TRIzol (Invitrogen). After reverse transcription (RT) using random hexamers and SSII reverse transcriptase (Invitrogen) for mRNA or using a QuantiMir RT kit (System Biosciences) for mature miRNA and U6 snRNA, the abundance of transcripts was assessed by quantitative PCR (qPCR) analysis using the SYBR green PCR master mix (Kapa Biosystems) and gene-specific primer sets (below). RT-qPCR analysis was performed on Applied Biosystems model 7300 and 7900 instruments. The forward and reverse primers used were TGCACCACCAACTGCTTAGC and GGCATGGACTGTGGTCATGAG for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), GAAGGAAATGGCCAAACAGA and ACGCTTTCTCCAGGTCTTCA for nucleolin, CGCAGAGATTCAGGTTCTCC and CCAAACCCTTTGCACTTGTT for HuR, and TAAACGGCCACAAGTTCAGCGT and AAGTCGTGCTGCTTCATGTGGT for EGFP. To measure the abundance of mature microRNAs, the forward primers were TGAAACATACACGGGAAACCTC for miR-494 and CACCACGTTTATACGCCGGTG for U6 snRNA (QuantiMir detection assay; System Biosciences). miR-494 miRNA was from Applied Biosystems.
RNP IP analysis was performed using as primary antibodies anti-HuR, anti-GFP, or control IgG (Santa Cruz Biotech.) or anti-pan Ago antibody (Millipore), as explained in reference 33. RNA in the IP samples was extracted and further analyzed by RT-qPCR using the primers listed above.
Biotin pulldown analysis.
The primers used to prepare biotinylated transcripts spanning the NCL mRNA (NM_005381.2) are listed below. After purification of the template PCR products, biotinylated transcripts were synthesized using the MaxiScript T7 kit (Ambion), whole-cell lysates (200 μg per sample) were incubated with 3 μg of purified biotinylated transcripts for 30 min at room temperature, and then complexes were isolated with streptavidin-coupled Dynabeads (Invitrogen). Proteins present in the pulldown material were studied by Western blot analysis as described previously (7). To synthesize biotinylated transcripts, PCR fragments were prepared using forward primers that contained the T7 RNA polymerase promoter sequence: CCAAGCTTCTAATACGACTCACTATAGGGAGA [presented as “(T7)” below]. Primers to amplify PCR templates for the synthesis of NCL 5′UTR (forward and reverse, respectively) were (T7)CTTTCGCCTCAGTCTCGAGCTCT and TGATGGCGGCGGAGTGTGAAGCGG, and primers to synthesize the NCL coding region (CR) were (T7)ATGGTGAAGCTCGCGAAG and CTATTCAAACTTCGTCTTCTTTC. PCR templates to synthesize the NCL 3′UTR fragments were prepared using primers (T7)CTTCTGTCCCTCTGCTTT and AACCAACACGGTATTGCC for fragment a, (T7)TCCGTCTAGTTAACATTTCAAG and CAAAACCCAAACTATTTGTAGG for fragment b, and (T7)TGTACAAAACCATTTTTTCCTAC and CTGGCACCAGAGTTGACT for fragment c. The biotinylated GAPDH gene (GAPDH) 3′UTR was synthesized using the primer pair (T7)CCTCAACGACCACTTTGTCA and GGTTGAGCACAGGGTACTTTATT.
Biotin pulldown analysis to narrow down the HuR binding site in the NCL 3′UTR was performed by synthesizing small RNA fragments using as templates the short DNA segments obtained after annealing the following oligomer pairs: (T7)TCCGTCTAGTTAACATTTCAAGGGCAAT and ATTGCCCTTGAAATGTTAACTAGACGGA for b-1, (T7)CAATACCGTGTTGGTTTTGACTGGATAT and ATATCCAGTCAAAACCAACACGGTATTG for b-2, (T7)ATATTCATATAAACTTTTTAAAGAGTTG and CAACTCTTTAAAAAGTTTATATGAATAT for b-3, (T7)GTTGAGTGATAGAGCTAACCCTTATCTG and CAGATAAGGGTTAGCTCTATCACTCAAC for b-4, (T7)TCTGTAAGTTTTGAATTTATATTGTTTC and GAAACAATATAAATTCAAAACTTACAGA for b-5, (T7)CAATACCGTGTTGGTTTTGACTGGATAT and AAAAAATGGTTTTGTACATGGGATGAAA for b-6, (T7)TTTTCCTACAAATAGTTTGGGTTTTGTT and AACAAAACCCAAACTATTTGTAGGAAAA for b-7, (T7)TGTTGTTGTTTCTTTTTTTTGTTTTGTT and AACAAAACAAAAAAAAGAAACAACAACA for c-1, (T7)TGTTTTTGTTTTTTTTTTTTTTGCGTT and AACGCAAAAAAAAAAAAAACAAAAACA for c-2, (T7)CGTTCGTGGGGTTGTAAAAGAAAAGAAA and TTTCTTTTCTTTTACAACCCCACGAACG for c-3, (T7)GAAAGCAGAATGTTTTATCATGGTTTTT and AAAAACCATGATAAAACATTCTGCTTTC for c-4, (T7)TTTTGCTTCAGCGGCTTTAGGACAAATT and AATTTGTCCTAAAGCCGCTGAAGCAAAA for c-5, and (T7)AATTAAAAGTCAACTCTGGTGCCAG and CTGGCACCAGAGTTGACTTTTAATT for c-6.
Translation assays.
Polyribosome fractionation assays were carried out as explained in reference 23. In short, cells were incubated with cycloheximide (Sigma) (100 μg/ml, 15 min), and cytoplasmic lysates (500 μl) were fractionated by centrifugation through 15 to 60% linear sucrose gradients and divided into 20 fractions for RT-qPCR analysis to determine the distribution of NCL mRNA and GAPDH mRNAs.
For nascent translation assays, HeLa cells were incubated with 1 mCi l-[35S]methionine and l-[35S]cysteine (NEN/Perkin Elmer) per 60-mm plate for 15 min. After lysis, IP reactions were carried out (16 h, 4°C) using IgG (Santa Cruz) or anti-GAPDH or antinucleolin antibodies, and the reaction products were resolved by SDS-PAGE, transferred to PVDF filters, and visualized using a PhosphorImager (Molecular Dynamics).
Interaction between mRNA and microRNA.
The interaction of the indicated microRNAs was tested by RNP IP analysis using anti-GFP antibody. HeLa cells were cotransfected with plasmid pMS2-RL or pMS2-RL-NCL(3′) and pMS2-YFP. Sixteen hours later, candidate microRNAs enriched in GFP IP samples were assessed by RT-qPCR analysis and by the enrichment in microRNAs in pMS2-RL-NCL(3′)-transfected cells relative to that in pMS2-RL-transfected cells.
Immunocytochemistry.
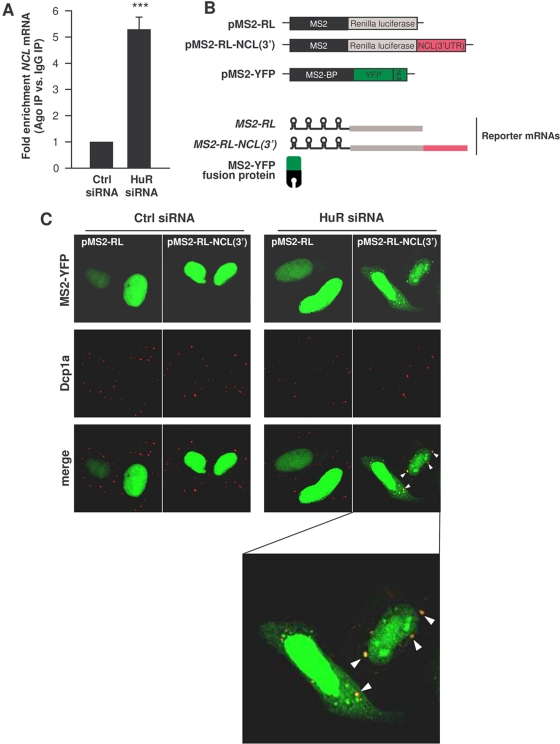
HeLa cells were fixed with 2% formaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 5% bovine serum albumin (BSA). After incubation with a primary antibody recognizing Dcp1a (Abcam), an Alexa Fluor 568-conjugated secondary antibody (Invitrogen) was used to detect primary antibody-antigen complexes (red). YFP fluorescence was green. Images were acquired using Axio Observer microscope (Zeiss) with AxioVision 4.7 Zeiss image processing software or with LSM 510 Meta (Zeiss). Confocal microscopy images were acquired with the Z-sectioning mode with 15 slices and 0.4-μm spacing and merged using maximum intensity.
RESULTS
RNA-binding protein HuR interacts with NCL mRNA.
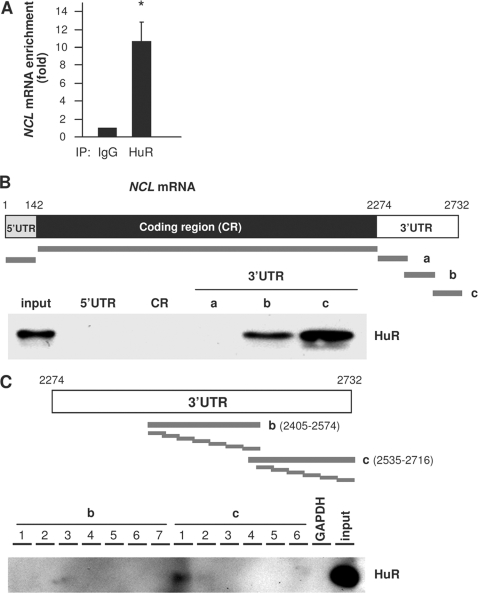
Earlier en masse searches identified NCL mRNA as a putative target of HuR (5, 6, 41). To investigate if HuR regulated nucleolin expression, we began by examining the interaction of HuR with the NCL mRNA in human cervical carcinoma (HeLa) cells. First, we performed ribonucleoprotein (RNP) immunoprecipitation (IP) assays using anti-HuR antibodies under conditions that preserved RNP integrity. After isolating RNA from the IP material and subjecting it to reverse transcription (RT) and quantitative real-time PCR (qPCR) analysis, we monitored the association of NCL mRNA with HuR. As shown in Fig. 1A, NCL mRNA was enriched more than 10-fold in HuR IP samples compared with IgG IP samples, supporting the existence of NCL mRNA-HuR complexes. Second, we studied if endogenous HuR bound to ectopic NCL mRNAs by using the biotin pulldown assay. Biotinylated RNAs spanning the NCL 5′UTR, CR, and 3′UTR were synthesized (Fig. 1B, top), and their interaction with HuR was studied. We did not detect any association of HuR with biotinylated RNAs containing the NCL 5′UTR, CR, or proximal 3′UTR. However, HuR associated prominently with 3′UTR fragments b and c. No binding was observed between HuR and a negative control transcript spanning the 3′UTR of GAPDH mRNA. Further subdivision of the 3′UTR revealed that HuR interacted more strongly with segment c-1, spanning positions 2573 to 2600 (Fig. 1C).
Fig. 1.
HuR binds NCL mRNA. (A) HeLa cell lysates were subjected to RNP IP followed by RT-qPCR analysis to measure the enrichment of NCL mRNA in HuR IP compared with control IgG IP. (B) (Top) schematic depiction of the NCL 5′UTR, CR, and 3′UTR, as well as the biotinylated RNAs (thick gray lines) synthesized for use in biotin pulldown analysis. (Bottom) Biotinylated RNA was incubated with HeLa cell lysates, and the interaction of HuR with the biotinylated segments was assessed by Western blot analysis. The input represents 5 μg of HeLa whole-cell lysate. (C) (Top) Biotinylated RNAs spanning short segments of the 3′UTR transcripts b and c were incubated with cell lysates, and the association of HuR with these RNAs was detected by Western blot analysis.
HuR regulates nucleolin expression.
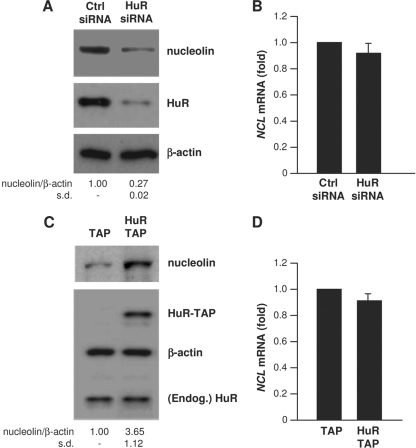
To test the functional consequences of the association between HuR and NCL mRNA, we studied if modulation of HuR abundance in HeLa cells affected nucleolin expression. As shown, silencing HuR by using specific HuR-directed small interfering RNA (siRNA) markedly decreased nucleolin protein levels but did not significantly lower NCL mRNA abundance (Fig. 2A and B), nor did it affect NCL mRNA stability (not shown). Conversely, overexpression of HuR (as the fusion protein HuR-TAP) significantly increased nucleolin levels but did not affect the NCL mRNA concentration (Fig. 2C and D).
Fig. 2.
HuR increases nucleolin expression, not NCL mRNA levels. (A and B) Forty-eight hours after transfection of HeLa cells with either control (Ctrl) siRNA or HuR-directed siRNA, lysates were prepared to assess the levels of nucleolin, HuR, and loading control β-actin by Western blot analysis (A), and the levels of NCL mRNA were measured by RT-qPCR using GAPDH mRNA for normalization (B). (C and D) Twenty-four hours after transfection with a control plasmid expressing TAP or a plasmid overexpressing HuR as a fusion protein (TAP-HuR), the levels of nucleolin, HuR-TAP, endogenous (Endog.) HuR, and β-actin were assessed by Western blot analysis (C), and the levels of NCL mRNA were assessed by RT-qPCR (D), as explained in panel B. Western blotting signals in panels A and C were quantified by densitometry, and the relative abundance of nucleolin was calculated. Data in panels A to D represent the means of 3 independent experiments. s.d., standard deviation.
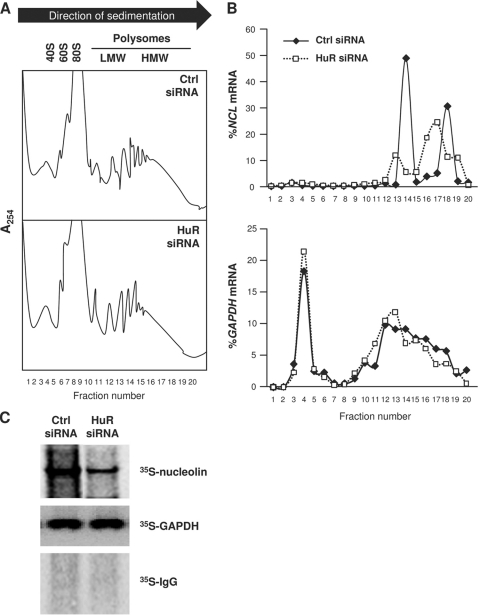
HuR promoted the translation of several target mRNAs without affecting their stability, as reported for mRNAs encoding prothymosin α, p53, cytochrome c, and hypoxia-inducible factor 1α (HIF-1α) (23, 32, 36, 42). To test if HuR similarly enhanced the translation of NCL mRNA, we tested the sizes of polysomes associated with NCL mRNA in HeLa cells expressing either normal HuR levels (Ctrl siRNA) or reduced HuR levels (HuR siRNA). Following the fractionation of cytoplasmic lysates through sucrose gradients (15 to 60%), the lightest components sedimented at the top (fractions 1 to 5), small (40S) and large (60S) ribosomal subunits and monosomes (80S) appeared in fractions 6 to 10, and progressively larger polysomes, ranging from low to high molecular weight (LMWP and HMWP, respectively) appeared in fractions 12 to 20 (Fig. 3A). Forty-eight hours after siRNA transfection, the distribution of NCL mRNA peaked at fractions 14 and 18 in control (Ctrl siRNA) cells, but these peaks shifted down and leftward in HuR siRNA cells, indicating that when HuR levels were lower, the sizes of NCL mRNA polysomes were also reduced, consistent with a decline in nucleolin translation (Fig. 3B, top). The distributions of the GAPDH mRNA, encoding a housekeeping protein, largely overlapped between the two groups (Fig. 3B, bottom). According to these results, HuR appears to contribute to enhancing nucleolin translation by increasing the initiation of NCL mRNA translation.
Fig. 3.
HuR increased nucleolin translation. (A) Lysates were prepared from HeLa cells 48 h after transfection with either Ctrl siRNA or HuR siRNA and fractionated through linear sucrose gradients (15 to 60%) in order to fractionate cytoplasmic components according to their molecular weight. The arrow indicates the direction of sedimentation; fractions 1 to 4 had no ribosomal components, fractions 6 to 10 contained 40S, 60S, and monosomes (80S), and fractions 11 to 20 contained polysomes of increasingly larger sizes (low-molecular weight [LMW] and high-molecular-weight [HMW]). (B) The relative distributions (percentages) of NCL mRNA (top) and GAPDH mRNA (bottom) on the sucrose gradients were quantified by RT-qPCR analysis of RNA in each of 20 gradient fractions. (C) Following siRNA transfections as explained in panel A, nascent nucleolin biosynthesis was monitored by incubating HeLa cells with l-[35S]methionine and l-[35S]cysteine for 15 min, whereupon cell lysates were prepared, and the levels of newly synthesized nucleolin and GAPDH were assessed by immunoprecipitation with antinucleolin and anti-GAPDH antibodies, respectively, followed by separation by SDS-PAGE and visualization using a PhosphorImager. Data in panels A to C are representative of three independent experiments.
Additional evidence that HuR promoted nucleolin translation came from studies to assess de novo nucleolin biosynthesis. Forty-eight hours after silencing HuR in HeLa cells, nascent translation of nucleolin was measured by culturing cells in the presence of l-[35S]methionine and l-[35S]cysteine for 15 min and immediately lysing cells for IP. This assay revealed that de novo translation of nucleolin in HuR-silenced HeLa cells was significantly decreased, while de novo translation of GAPDH was unaffected (Fig. 3C). Taking into consideration that HuR silencing did not affect NCL mRNA levels (Fig. 2B) or nucleolin protein stability (not shown), these data indicate that HuR promotes nucleolin translation.
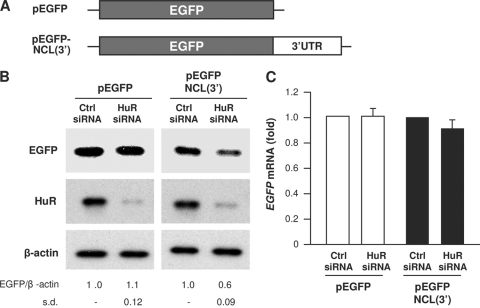
Further experiments revealed that HuR influenced nucleolin translation through the NCL 3′UTR. A heterologous reporter construct expressing a chimeric RNA that spanned the EGFP gene (EGFP) CR and the NCL 3′UTR was constructed and studied (Fig. 4A). As shown, HuR silencing decreased significantly the expression of EGFP (Fig. 4B) from the reporter chimeric plasmid pEGFP-NCL(3′) but not from the parent control plasmid, pEGFP. This reduction was due to changes in translation and not due to changes in the levels or stability of the chimeric RNA, since the levels of the ectopic reporter RNAs [EGFP mRNA and EGFP-NCL(3′) mRNA] did not significantly change after silencing HuR (Fig. 4C). Taken together, these data indicate that HuR promotes nucleolin translation via the NCL 3′UTR.
Fig. 4.
HuR regulates nucleolin expression through the NCL 3′UTR. (A) Schematic representation of the parent reporter plasmid pEGFP and the pEGFP-NCL(3′) reporter plasmid bearing the NCL 3′UTR. (B and C) HeLa cells were transfected with each of reporter constructs, together with either Ctrl siRNA or HuR siRNA. Forty-eight hours after transfection, EGFP expression levels, as well as the levels of HuR and β-actin were assessed by Western blot analysis, and quantified (B), and the levels of EGFP mRNA were examined by RT-qPCR analysis (C). The data are representative of 3 independent experiments. Standard deviations (s.d.) are indicated.
HuR excludes NCL mRNA from RISC/Ago complexes and from PBs.
We sought to further examine the mechanisms whereby HuR promotes nucleolin translation. We hypothesized that the absence of HuR may facilitate the binding of a translational repressor of NCL mRNA, resulting in reduced translation of nucleolin. A plausible candidate repressor was the microRNA/RISC/Ago (RNA-induced silencing complex/Argonaute) machinery. To measure if NCL mRNA associated with RISC/Ago, we performed RNP IP analysis using an antibody that recognized all isoforms of the protein Ago, a critical component of the RISC. As shown, silencing of HuR increased the interaction of Ago with NCL mRNA (Fig. 5A). These results suggested that HuR might prevent microRNA/RISC-mediated translational repression.
Fig. 5.
Silencing HuR increases the association of NCL mRNA with Ago and PBs. (A) By 48 h after transfection of either Ctrl siRNA or HuR siRNA, the interaction of NCL mRNA with Ago-containing complexes was assessed by RNP IP using anti-pan-Ago antibody followed by RT-qPCR analysis. (B) Schematic of the plasmids used for tracking the NCL mRNA intracellularly. pMS2-RL and pMS2-RL-NCL(3′) were derived from pSL-MS2(24X), and each expressed the Renilla luciferase (RL) coding region and 24 tandem MS2 RNA hairpins; pMS2-RL-NCL(3′) additionally contained the NCL 3′UTR. The plasmid pMS2-YFP expressed a fusion fluorescent protein (MS2-YFP) capable of binding MS2-containing RNA. NLS, nuclear localization signal. (C) Cells were transfected with either Ctrl or HuR siRNAs, together with pMS2-YFP and either pMS2-RL and pMS2-RL-NCL(3′). Forty-eight hours after transfection, the subcellular localization of the MS2-tagged RNAs was monitored by confocal microscopy. PBs were visualized by staining with an antibody that recognizes the PB marker Dcp1a. The “merge” panels show colocalization of MS2-tagged RNA and PBs. (The bottom panel shows an enlarged merged image in the HuR siRNA group to visualize colocalized [yellow] signals [arrowheads].)
To identify which microRNA(s) interacted with NCL mRNA, we employed a recently reported method based on the tagging of the RNA of interest (NCL mRNA in this case) with bacteriophage MS2 RNA hairpins (56). Plasmids pMS2-RL (containing the Renilla luciferase gene [RL] CR) and pMS2-RL-NCL(3′) were constructed to express reporter (MS2-tagged) MS2-RL and MS2-RL-NCL(3′) mRNAs. Each culture was also transfected with plasmid pMS2-YFP, which expresses a fusion protein that has a strong nuclear localization signal (NLS), is fluorescent (via the YFP domain), and recognizes the MS2 RNA hairpins (via the MS2-binding protein region) (Fig. 5B). The plasmids expressing reporter RNAs were transfected along with pMS2-YFP; 24 h later, the localization of the reporter RNAs was studied in cells that expressed different levels of HuR. As shown in Fig. 5C, the YFP fluorescent signal (green) was almost exclusively nuclear in all of the cells, due to the NLS present in the fusion MS2-YFP protein; however, in HuR siRNA cells, there was a sizeable amount of fluorescence in the cytoplasm, and some of the cytoplasmic fluorescent signal overlapped with the signals of Dcp1a, a PB marker (overlapping signals are displayed yellow in the “merge” images). These findings revealed that silencing HuR permitted the colocalization of ectopic NCL RNA in PBs, while the NCL RNA was excluded from PBs in cells that expressed normal HuR levels (Ctrl siRNA). Given that microRNA/RISC can elicit translational repression at PBs (40), we postulated that a microRNA might be implicated in recruiting NCL mRNA into PBs when the cellular HuR levels are low.
miR-494 represses nucleolin expression by competing with HuR, recruiting NCL mRNA to PBs.
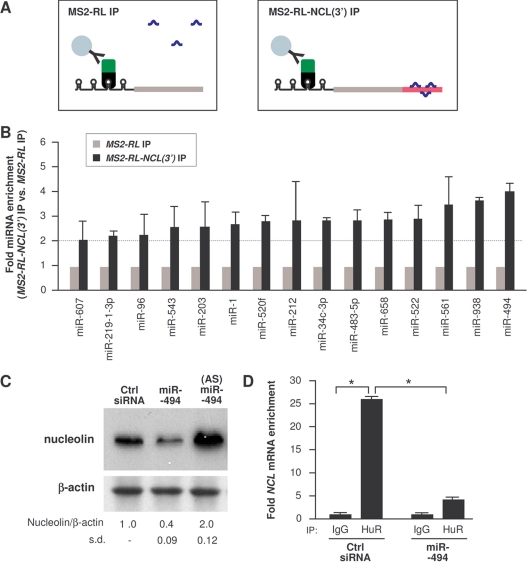
This MS2 RNA-tagging system was further used in order to identify the microRNAs that interact with the NCL 3′UTR. First, cells were transfected with pMS2-YFP and with either pMS2-RL or pMS2-RL-NCL(3′), as explained in Fig. 5C. The MS2-YFP fusion protein (bound to the MS2-tagged RNA) was then subjected to IP using anti-GFP antibody, which selectively detected RNP complexes containing MS2-YFP, MS2-tagged RNA, and any microRNAs bound to the tagged mRNA (shown schematically in Fig. 6A). As previously reported (56), this IP selectively enriched in microRNAs associated with an RNA of interest. Second, we predicted computationally all of the possible microRNAs that could interact with the NCL 3′UTR, using the programs TargetScan, microRNA org, and miRDB. Among the 46 microRNAs that were predicted to target the NCL 3′UTR, 20 were screened for their enrichment in the MS2-YFP IP samples from cells expressing MS2-RL-NCL(3′) RNA relative to samples from cells expressing MS2-RL RNA. As shown in Fig. 6B, 15 of these microRNAs were enriched more than 2-fold in MS2-RL-NCL(3′) relative to MS2-RL RNAs; among the most enriched microRNAs was miR-494. Subsequent studies indicated that miR-494 did lower nucleolin expression, while other microRNAs tested did not (not shown). Overexpression of miR-494 reduced nucleolin abundance, while transfection of an antisense RNA complementary to miR-494 [the antagomir (AS)-miR-494] increased nucleolin abundance, as assessed by Western blot analysis (Fig. 6C). Importantly, HuR binding to NCL mRNA was potently reduced when miR-494 was overexpressed, suggesting that miR-494 was capable of competing with HuR, displacing it from the NCL mRNA (Fig. 6D).
Fig. 6.
miR-494 targets the NCL 3′UTR. (A) Schematic of the detection of microRNAs interacting with an RNA of interest. HeLa cells were transfected with the plasmids indicated in Fig. 5B. (B) Forty-eight hours after transfection of plasmids, RNP IP analysis was performed using anti-EGFP antibody, and the levels of microRNAs present in the IP samples were assessed by RT-qPCR analysis. The microRNAs enriched greater than 2-fold in the MS2-RL-NCL(3′) mRNA IP group compared with those in the MS2-RL mRNA IP group are shown. (C) Forty-eight hours after transfection of miR-494 or an antisense miR-494 antagomir [(AS)miR-494], the levels of nucleolin and loading control β-actin were assessed by Western blot analysis and quantified. (D) Forty-eight hours after transfection with either Ctrl siRNA or miR-494, the enrichment of NCL mRNA in HuR IP samples was tested by RNP IP followed by RT-qPCR analysis. Data in panels B to D are the means and standard deviations (s.d.) from three experiments. *, P < 0.05.
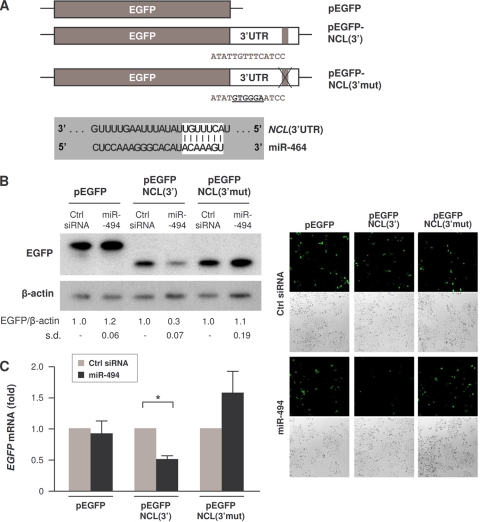
We tested if miR-494 directly affected nucleolin expression by studying the expression of a reporter construct carrying the intact NCL 3′UTR [pEGFP-NCL(3′)] relative to the expression of a reporter construct in which the region of complementarity with miR-494 in the NCL 3′UTR was mutated [pEGFP-NCL(3′mut)] (Fig. 7A). As shown, EGFP expressed from the parent vector (pEGFP) in transfected HeLa cells did not change after overexpressing miR-494; in contrast, expression of EGFP from cells transfected with pEGFP-NCL(3′) declined markedly (Fig. 7B). Importantly, however, destroying the site of interaction with miR-494 rendered the reporter refractory to miR-494-mediated reduction of EGFP levels (Fig. 7B). Overexpression of miR-494 lowered EGFP-NCL(3′) mRNA levels significantly (Fig. 7C), suggesting that miR-494, besides displacing HuR, may also affect NCL mRNA stability.
Fig. 7.
miR-494 directly represses the NCL 3′UTR. (A) Schematic of plasmids pEGFP, pEGFP-NCL(3′), and pEGFP-NCL(3′mut), the latter bearing 6 mutant nucleotides in the NCL 3′UTR site corresponding to the miR-494 seed region. (B) HeLa cells were cotransfected with the plasmids shown in panel A and with either Ctrl siRNA or HuR siRNA, as indicated. Forty-eight hours after transfection, EGFP expression levels were assessed by Western blot analysis and quantified (left) and assessed by green fluorescence (right). The reporter EGFP protein expressed from pEGFP-NCL(3′) and pEGFP-NCL(3′mut) was slightly shorter because cloning of the 3′UTR regulatory sites required the removal of 16 amino acids from pEGFP and the introduction of a new stop codon. (C) The levels of EGFP mRNA in cells transfected as described in panel B were assessed by RT-qPCR analysis and normalized to GAPDH mRNA abundance. The data in panels B and C represent the means and standard deviations (s.d.) from 3 independent experiments. *, P < 0.05.
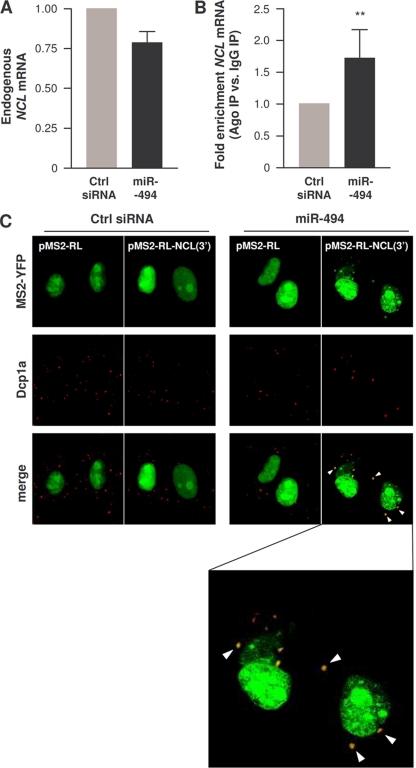
Overexpression of miR-494 in HeLa cells modestly lowered NCL mRNA levels (by close to 25% [Fig. 8A]) and significantly enriched the association of NCL mRNA with Ago-containing complexes (Fig. 8B). In agreement with the notion that miR-494 competed with HuR functionally, miR-494 overexpression in cells transfected with the reporters described in Fig. 5 triggered an increase in localization of the chimeric MS2-RL-NCL(3′) mRNA into PBs, while it did not affect the subcytoplasmic distribution of control MS2-RL mRNA (Fig. 8C).
Fig. 8.
miR-494 recruits NCL 3′UTR to Ago-containing complexes and to PBs. (A) Forty-eight hours after transfecting cells with either Ctrl siRNA or miR-494, the levels of endogenous NCL mRNA were assessed by RT-qPCR. (B) The levels of NCL mRNA in IgG IP relative to pan-Ago IP were quantified by RNP IP followed by RT-qPCR analysis in cells that had been transfected 48 h earlier with either Ctrl siRNA or miR-494. (C) HeLa cells were transfected with either Ctrl siRNA or miR-494, together with pMS2-YFP and either pMS2-RL and pMS2-RL-NCL(3′), as explained in the legend to Fig. 5. Forty-eight hours after transfection, the subcellular localization of the MS2-tagged RNAs was monitored by confocal microscopy. PBs were visualized by staining with an antibody that recognizes the PB marker Dcp1a. The “merge” panels show colocalization of MS2-tagged RNA and PBs. The bottom panel is an enlarged merged image within the miR-494 group to visualize colocalized (yellow) signals.
HuR enhances cell proliferation in part by promoting nucleolin expression.
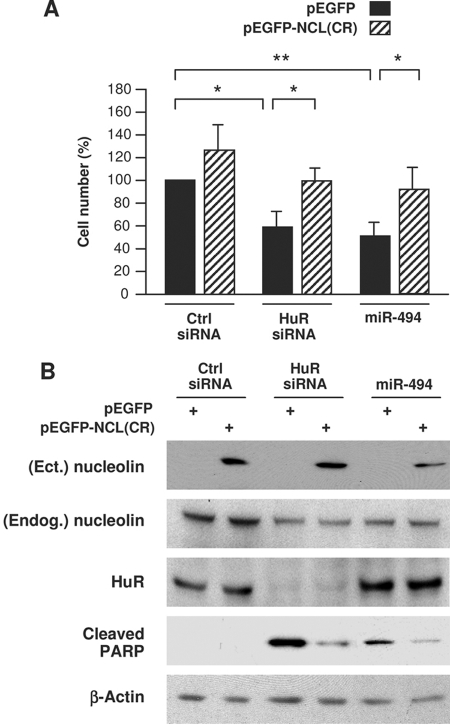
As reported previously (5), silencing HuR significantly reduced the proliferation of HeLa cells (Fig. 9A). Interestingly, overexpression of nucleolin in HuR siRNA cells using a vector that transcribed only the NCL CR significantly restored cell numbers, indicating that HuR promoted cell proliferation in part by elevating nucleolin levels. Overexpression of miR-494 similarly lowered the number of cells in the population, and this effect was likewise reversed if nucleolin levels were ectopically increased using a vector [pEGFP-NCL(CR)] that lacked the regulatory NCL 3′UTR (Fig. 9A). The changes in cell number elicited by HuR, nucleolin, and miR-494 were at least partly due to alterations in apoptosis, since the levels of cleaved PARP (a marker of apoptosis) increased after HuR silencing and after miR-494 overexpression, but in both instances, overexpression of nucleolin partially rescued apoptosis (Fig. 9B). Taken together, these findings indicate that HuR promotes cell proliferation and prevents apoptosis at least in part by promoting nucleolin expression and that these effects are antagonized by miR-494.
Fig. 9.
HuR-enhanced cell proliferation and survival depend partly on upregulation of nucleolin. HeLa cells were transfected with Ctrl siRNA, HuR siRNA, or miR-494 together with either a control plasmid (pEGFP) or with a plasmid expressing the nucleolin CR [pEGFP-NCL(CR)], and 48 h later, cells were analyzed. (A) The cells remaining in each transfection group were counted using a hemacytometer. (B) Whole-cell lysates prepared from each transfection group were analyzed by Western blotting to assess the levels of ectopic (Ect.) nucleolin, endogenous (Endog.) nucleolin, HuR, loading control β- actin, and cleaved PARP, an apoptotic marker. The data shown are means and standard deviations (A) or representative results (B) from three independent experiments.
DISCUSSION
Nucleolin is known to participate in important cellular processes, such as chromatin dynamics, ribosome biogenesis, DNA replication, cell proliferation, and cell survival. Its role is also recognized in pathologies like cancer, autoimmunity, and neurodegeneration. Although nucleolin has these critical functions, the mechanisms that control nucleolin expression have not been reported. Here we have identified two potent posttranscriptional regulators of nucleolin expression: the RBP HuR and the microRNA miR-494. Both of these molecules interact with the NCL 3′UTR [NCL(3′)] and influence nucleolin biosynthesis: HuR enhances it, and miR-494 represses it. HuR and miR-494 bind the NCL 3′UTR competitively, and each has an opposite effect on the localization of NCL mRNA in PBs, cytoplasmic foci where mRNAs are subject to translational repression and mRNA decay. Our data indicate that HuR excludes the NCL mRNA from PBs, while miR-494 recruits NCL mRNA to PBs. Although controversy remains regarding the role of PBs in gene regulation, our findings are consistent with HuR precluding the localization of NCL mRNA within a cellular domain where mRNAs can be degraded and translationally repressed, while miR-494 has the opposite influence.
The discovery that the levels of the RBP nucleolin are controlled by another RBP (HuR) and by a microRNA (miR-494) adds to a growing body of evidence that expression of posttranscriptional regulatory factors is controlled by other posttranscriptional regulatory factors. For example, RBPs HuR, AUF1, TIA-1, TIAR, KSRP, NF90, and TTP were shown to associate with their cognate mRNAs, and in several instances, they auto- and cross-regulated their expression (13, 51, 64). Similarly, several microRNAs have been found to modulate the expression of several RBPs; for example, miR-519 and miR-125 lowered HuR levels and miR-375 reduced HuD production (2, 5, 27). It appears that in instances in which the cellular fate depends critically on posttranscriptional gene regulation (for example, after severe DNA damage that shuts down de novo transcription), posttranscriptional regulators rely on each other and on preexisting mRNAs in order to properly adapt the patterns of expressed proteins.
Examples of posttranscriptional regulation of a target of HuR via the joint influence of HuR and a specific microRNA are also on the rise. For example, HuR antagonizes miR-122 to regulate expression of CAT-1 mRNA (11), antagonizes miR-548c-3p to regulate the expression of TOP2A (56), and cooperates with let-7 in repressing c-Myc expression (33). MicroRNA sites are often present near HuR sites (37, 48), suggesting that in some cases HuR and microRNA could compete via their physical interaction with mRNAs (possibly via steric hindrance). However, in the above examples (11, 33, 56), HuR and the coregulatory microRNAs bind at distances up to several hundreds of bases apart; likewise in this report, the miR-494 site and the main interaction site for HuR (c-1) are 44 bases apart on the NCL 3′UTR. RNA structure and folding analyses will be needed to investigate systematically how binding of HuR in one area of the 3′UTR affects association of a microRNA/RISC in a remote site and vice versa.
It was interesting to find that the HuR-elicited promotion of cell proliferation and inhibition of apoptosis was significantly dependent on its upregulation of nucleolin. Nucleolin inhibits apoptosis by enhancing the expression of antiapoptotic proteins such as Bcl-2 and Bcl-xL and increases cell division by elevating expression of many cancer-, cell growth-, and proliferation-associated proteins (e.g., cyclin I) (7) and by promoting processes like DNA replication and transcription (58). HuR helps to implement a malignant phenotype by controlling the production of subsets of cancer-related proteins, including proteins that promote cell division (e.g., cyclins), degrade the extracellular matrix (e.g., matrix metalloproteases), induce angiogenesis (e.g., HIF-1α, vascular endothelial growth factor [VEGF], and cyclooxygenase 2), inhibit apoptosis (e.g., Bcl-2, prothymosin α, and sirtuin 1), and evade immune recognition (e.g., transforming growth factor β) (1). In light of the findings reported here, HuR is an upstream regulator of nucleolin expression and hence nucleolin's functions as inhibitor of apoptosis and promoter of cell proliferation. Thus, HuR and nucleolin appear to work in tandem to mediate these two cellular functions and thereby favor a pro-oncogenic phenotype. Whether HuR is also involved in other physiologic and pathological facets of nucleolin, such as neurodegeneration and autoimmunity, remains to be studied.
It will also be important to determine if miR-494, a strong inhibitor of nucleolin production, possesses antitumor activities. Overexpression of miR-494 triggered an increase in PARP cleavage (Fig. 9), suggesting that miR-494 is proapoptotic; this effect was rescued by overexpression of nucleolin, indicating that miR-494 enhanced apoptosis specifically by lowering nucleolin levels. Other studies have also shown that miR-494 triggered growth arrest in bronchial epithelial cells (21). Thus, it will be interesting to investigate if miR-494 can function more broadly as a tumor supressor microRNA. For example, miR-519 (a repressor of HuR translation) is much less abundant in cancer tissues than in noncancer tissues, and its overexpression in human carcinoma cells reduces tumorigenesis (3). It will be important to study systematically if miR-494 levels differ between tumor and normal tissues and whether miR-494 recapitulates the antitumorigenic effects of miR-519.
In conclusion, since nucleolin has been linked to numerous pathologies, understanding the mechanisms that control nucleolin is critical to designing specific approaches to modulate nucleolin levels. Here, we have identified two posttranscriptional regulators of nucleolin expression. Interventions directed toward HuR, miR-494, or other regulators of nucleolin expression can become promising therapies to combat diseases in which nucleolin is implicated, including cancer.
ACKNOWLEDGMENT
This work was supported in its entirety by the National Institute on Aging-Intramural Research Program of the National Institutes of Health.
Footnotes
Published ahead of print on 22 August 2011.
REFERENCES
- 1. Abdelmohsen K., Gorospe M. 2010. Post-transcriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 1:214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdelmohsen K., et al. 2010. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol. Cell. Biol. 30:4197–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abdelmohsen K., et al. 2010. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 9:1354–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abdelmohsen K., Kuwano Y., Kim H. H., Gorospe M. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 389:243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdelmohsen K., Srikantan S., Kuwano Y., Gorospe M. 2008. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. U. S. A. 105:20297–20302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abdelmohsen K., et al. 2009. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28:1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abdelmohsen K., et al. 6 July 2011, posting date Enhanced translation by nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Angelov D., et al. 2006. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25:1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bell S. A., Faust H., Mittermüller J., Kolb H. J., Meurer M. 1996. Specificity of antinuclear antibodies in scleroderma-like chronic graft-versus-host disease: clinical correlation and histocompatibility locus antigen association. Br. J. Dermatol. 134:848–854 [PubMed] [Google Scholar]
- 10. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 71:513–521 [DOI] [PubMed] [Google Scholar]
- 11. Bhattacharyya S. N., Habermacher R. R., Martine U., Closs E. I., Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111–1124 [DOI] [PubMed] [Google Scholar]
- 12. Bouvet P., Allain F. H., Finger L. D., Dieckmann T., Feigon J. 2001. Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin. J. Mol. Biol. 309:763–775 [DOI] [PubMed] [Google Scholar]
- 13. Brooks S. A., Connolly J. E., Rigby W. F. 2004. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 172:7263–7271 [DOI] [PubMed] [Google Scholar]
- 14. Bunimov N., Smith J. E., Gosselin D., Laneuville O. 2007. Translational regulation of PGHS-1 mRNA: 5′ untranslated region and first two exons conferring negative regulation. Biochim. Biophys. Acta 1769:92–105 [DOI] [PubMed] [Google Scholar]
- 15. Callé A., et al. 2008. Nucleolin is required for an efficient herpes simplex virus type 1 infection. J. Virol. 82:4762–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carballo E., Lai W. S., Blackshear P. J. 1998. Feedback inhibition of macrophage tumor necrosis factor-a production by tristetraprolin. Science 281:1001–1005 [DOI] [PubMed] [Google Scholar]
- 17. Caudle W. M., Kitsou E., Li J., Bradner J., Zhang J. 2009. A role for a novel protein, nucleolin, in Parkinson's disease. Neurosci. Lett. 459:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen C. Y., et al. 2000. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14:1236–1248 [PMC free article] [PubMed] [Google Scholar]
- 19. Derenzini M. 2000. The AgNORs. Micron 31:117–120 [DOI] [PubMed] [Google Scholar]
- 20. Dranovsky A., et al. 2001. Cdc2 phosphorylation of nucleolin demarcates mitotic stages and Alzheimer's disease pathology. Neurobiol. Aging 22:517–528 [DOI] [PubMed] [Google Scholar]
- 21. Duan H., Jiang Y., Zhang H., Wu Y. 2010. MiR-320 and miR-494 affect cell cycles of primary murine bronchial epithelial cells exposed to benzo[a]pyrene. Toxicol. In Vitro 24:928–935 [DOI] [PubMed] [Google Scholar]
- 22. Fähling M., et al. 2005. Role of nucleolin in posttranscriptional control of MMP-9 expression. Biochim. Biophys. Acta 1731:32–40 [DOI] [PubMed] [Google Scholar]
- 23. Galbán S., et al. 2008. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 28:93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ginisty H., Amalric F., Bouvet P. 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17:1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ginisty H., Sicard H., Roger B., Bouvet P. 1999. Structure and functions of nucleolin. J. Cell Sci. 112:761–772 [DOI] [PubMed] [Google Scholar]
- 26. Glisovic T., Bachorik J. L., Yong J., Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582:1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo X., Wu Y., Hartley R. S. 2009. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 6:575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hinman M. N., Lou H. 2008. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65:3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang Y., Xu X. S., Russell J. E. 2006. A nucleolin-binding 3′ untranslated region element stabilizes beta-globin mRNA in vivo. Mol. Cell. Biol. 26:2419–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johansson C., et al. 2004. Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target. J. Mol. Biol. 337:799–816 [DOI] [PubMed] [Google Scholar]
- 31. Jordan P., Heid H., Kinzel V., Kübler D. 1994. Major cell surface-located protein substrates of an ecto-protein kinase are homologs of known nuclear proteins. Biochemistry 33:14696–14706 [DOI] [PubMed] [Google Scholar]
- 32. Kawai T., et al. 2006. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 26:3295–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim H. H., et al. 2009. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23:1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krol J., Loedige I., Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11:597–610 [DOI] [PubMed] [Google Scholar]
- 35. Kuwano Y., et al. 2010. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 38:225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lal A., Kawai T., Yang X., Mazan-Mamczarz K., Gorospe M. 2005. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 24:1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lebedeva S., et al. 30 June 2011, posting date Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 38. Lee E. K., et al. 2010. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 17:732–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee P. T., Liao P. C., Chang W. C., Tseng J. T. 2007. Epidermal growth factor increases the interaction between nucleolin and heterogeneous nuclear ribonucleoprotein K/poly(C) binding protein 1 complex to regulate the gastrin mRNA turnover. Mol. Biol. Cell 18:5004–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lopez de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U. S. A. 101:2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazan-Mamczarz K., et al. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U. S. A. 100:8354–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mazan-Mamczarz K., Lal A., Martindale J. L., Kawai T., Gorospe M. 2006. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 26:2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miniard A. C., Middleton L. M., Budiman M. E., Gerber C. A., Driscoll D. M. 2010. Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression. Nucleic Acids Res. 38:4807–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mongelard F., Bouvet P. 2007. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 17:80–86 [DOI] [PubMed] [Google Scholar]
- 46. Moore M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514–1518 [DOI] [PubMed] [Google Scholar]
- 47. Morris A. R., Mukherjee N., Keene J. D. 2010. Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip. Rev. Syst. Biol. Med. 2:162–180 [DOI] [PubMed] [Google Scholar]
- 48. Mukherjee N., et al. 30 June 2011, posting date Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Otake Y., et al. 2007. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood 109:3069–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Piecyk M., et al. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19:4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pullmann R., Jr., et al. 2007. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 27:6265–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rajagopalan L. E., Westmark C. J., Jarzembowski J. A., Malter J. S. 1998. hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res. 26:3418–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sawicka K., Bushell M., Spriggs K. A., Willis A. E. 2008. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 36:641–647 [DOI] [PubMed] [Google Scholar]
- 54. Semenkovich C. F., Ostlund R. E., Jr., Olson M. O., Yang J. W. 1990. A protein partially expressed on the surface of HepG2 cells that binds lipoproteins specifically is nucleolin. Biochemistry 29:9708–9713 [DOI] [PubMed] [Google Scholar]
- 55. Serin G., et al. 1996. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie 78:530–538 [DOI] [PubMed] [Google Scholar]
- 56. Srikantan S., et al. 18 July 2011, posting date Translational control of Top2A influences doxorubicin efficacy. Mol. Cell. Biol. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Srivastava M., Pollard H. B. 1999. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 13:1911–1922 [PubMed] [Google Scholar]
- 58. Storck S., Shukla M., Dimitrov S., Bouvet P. 2007. Functions of the histone chaperone nucleolin in diseases. Subcell. Biochem. 41:125–144 [DOI] [PubMed] [Google Scholar]
- 59. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. 2005. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123:49–63 [DOI] [PubMed] [Google Scholar]
- 60. Tuteja R., Tuteja N. 1998. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit. Rev. Biochem. Mol. Biol. 33:407–436 [DOI] [PubMed] [Google Scholar]
- 61. Ugrinova I., et al. 2007. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol. Biol. 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reference deleted.
- 63. Xu Y. H., Grabowski G. A. 1999. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid β-glucosidase and other mRNAs. Mol. Genet. Metab. 68:441–454 [DOI] [PubMed] [Google Scholar]
- 64. Yi J., et al. 2010. Reduced nuclear export of HuR mRNA by HuR is linked to the loss of HuR in replicative senescence. Nucleic Acids Res. 38:1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang W., et al. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y., et al. 2006. Nucleolin links to arsenic-induced stabilization of GADD45alpha mRNA. Nucleic Acids Res. 34:485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]