Abstract
The genotyping of human papillomaviruses (HPV) is essential for the surveillance of HPV vaccines. We describe and validate a low-cost PGMY-based PCR assay (PGMY-CHUV) for the genotyping of 31 HPV by reverse blotting hybridization (RBH). Genotype-specific detection limits were 50 to 500 genome equivalents per reaction. RBH was 100% specific and 98.61% sensitive using DNA sequencing as the gold standard (n = 1,024 samples). PGMY-CHUV was compared to the validated and commercially available linear array (Roche) on 200 samples. Both assays identified the same positive (n = 182) and negative samples (n = 18). Seventy-six percent of the positives were fully concordant after restricting the comparison to the 28 genotypes shared by both assays. At the genotypic level, agreement was 83% (285/344 genotype-sample combinations; κ of 0.987 for single infections and 0.853 for multiple infections). Fifty-seven of the 59 discordant cases were associated with multiple infections and with the weakest genotypes within each sample (P < 0.0001). PGMY-CHUV was significantly more sensitive for HPV56 (P = 0.0026) and could unambiguously identify HPV52 in mixed infections. PGMY-CHUV was reproducible on repeat testing (n = 275 samples; 392 genotype-sample combinations; κ of 0.933) involving different reagents lots and different technicians. Discordant results (n = 47) were significantly associated with the weakest genotypes in samples with multiple infections (P < 0.0001). Successful participation in proficiency testing also supported the robustness of this assay. The PGMY-CHUV reagent costs were estimated at $2.40 per sample using the least expensive yet proficient genotyping algorithm that also included quality control. This assay may be used in low-resource laboratories that have sufficient manpower and PCR expertise.
INTRODUCTION
High-risk genital human papillomaviruses (HPV) are etiologically linked to cervical cancer and other anogenital malignancies. HPV16 and HPV18 together account for 70% of the cases worldwide (25). The infectious nature of cervical cancer raises hopes that vaccines against high-risk HPV will effectively reduce the incidence of this disease. The positive effect of the two vaccine formulations available now, Gardasil (quadrivalent HPV6/11/16/18; Merck & Co.) and Cervarix (bivalent HPV16/18; GlaxoSmithKline), has been demonstrated with phase III efficacy trials to yield significant reductions in the frequency of precursor lesions associated with these viruses (21, 27). Some degree of cross-protection against infections by HPV31, HPV33, HPV45, HPV52, and HPV58 also have been reported with Gardasil (37) and for HPV31, HPV33, and HPV45 with Cervarix (27). Long-term epidemiologic surveillance therefore is required to monitor vaccine efficacy and to monitor HPV genotype-specific disease incidence rates in the vaccination context (14, 30). Continued gynecological and pathological examinations as well as HPV diagnosis also will be required for decades prior to the modification of a patient's care approach, since cervical cancer is a rare outcome of long-term persistent infections (9).
There are several methods to identify HPV infections. While the hybrid capture assay (HCII) has proven its clinical reliability for the colposcopic triage of women based on the high-risk profile of their HPV infection (6, 12, 32), more sensitive PCR-based assays with typing capacity are necessary for epidemiological purposes and vaccine surveillance. Typing also may be desirable clinically to identify persistent genotypes as an adjunct to clinically validated tests, as it has been shown that persistent infections confer an increased risk of high-grade disease in an HPV genotype-specific manner, with HPV16, HPV18, HPV31, and HPV33 being the most carcinogenic (2, 26).
Two widely used HPV genotyping assays that rely on reverse blotting hybridization (RBH) against a panel of HPV genotype-specific probes immobilized on membrane strips are available commercially: Inno-Lipa (Innogenetics), based on the SPF primers (22), and linear array (LA; Roche), based on the PGMY primers (17). Both probe panels address the most frequent high-risk genotypes and several low- or undetermined-risk genotypes that are frequently observed. These probe arrays cannot be reused, as the detection reagents leave a precipitate on the membrane that cannot be removed. The performance of both assays is comparable for cervical smears in liquid transport medium in terms of sensitivity, specificity, and predictive values (5, 28, 32, 35). Other commercial assays based on probe hybridization following broad-range PCR also are available (CLART from Genomica, DNA chip from Biocare, Papillocheck from Greiner Bio-One, PCR Luminex from Multimetrix, and HPV genotyping LQ from Qiagen, to name a few). Unlike the RBH platforms, these assays require additional, potentially expensive instruments. The reagent costs and availability of all commercial assays, regardless of platform, may restrict their use, especially in low-resource settings.
We validated the in-house assay based on the PGMY primers and a reusable probe array (PGMY-CHUV) described in chapter 5 of the WHO HPV Laboratory Manual (33). We have used this assay in our laboratory as an adjunct to cytology as well as for the monitoring of treated patients and for epidemiological purposes for 10 years. The assay allows the genotyping of 40 to 80 samples per day. It was developed under a quality assurance program to ensure its reproducibility and accuracy and was optimized in terms of costs ($2.40 per sample). It has been evaluated within the WHO HPV Laboratory Network after technology transfer to member laboratories and was found to be suitable for HPV genotyping with successful participation in proficiency panels established in this network (13, 15).
MATERIALS AND METHODS
Samples.
A total of 9,408 ThinPrep samples were submitted to our laboratory for HPV testing from March 1999 to April 2010. They were from 5,904 patients (median age, 35 years; interquartile range, 27 to 47 years) who visited the outpatient clinics of the Gynecology and Obstetrics Department of our hospital center and affiliated family planning centers. HPV testing was performed as an adjunct to cytology. Cytology results were atypical squamous cells of undetermined significance (n = 5,017 samples; 53.3%) or had low (n = 3,683; 39.1%), high (n = 112; 1.2%), normal (n = 512; 5.4%), or unspecified cytology grades (n = 84; 1.0%).
Two hundred additional samples were obtained from an external laboratory (MCL Diagnostics) for comparison to the LA.
The use of clinical samples for assay development and comparative analyses was approved by our local ethics committee.
Quality controls.
WHO HPV proficiency panel DNAs were kindly provided by Carina Eklund and Joakim Dillner (WHO HPV LabNet Global Reference Laboratory, Malmö, Sweden) (15, 16). HPV16 and HPV18 plasmid DNA international standards were obtained from the NIBSC (38).
Positive controls.
Nonbiotinylated HPV genotype-specific amplicons were cloned with the pGEM-T (Promega, Dübendorf, Switzerland) or pCR-TOPO (Invitrogen, Basel, Switzerland) cloning kit according to the manufacturer's instructions. Each positive control was verified by DNA sequencing and was kept as a bacterial glycerol stock at −80°C.
SiHa (2 copies of HPV16 per cell), CasKi (600 copies of HPV16 per cell), HeLa (50 copies of HPV18 per cell), and HEK293 (HPV negative) cells were used for the evaluation of the PGMY PCR sensitivity. HPV genome content per cell was confirmed by quantitative real-time PCR against HPV16 and HPV18 DNA international standards (NIBSC). Cells were grown in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal calf serum in an atmosphere of 5% CO2 at 37°C according to standard procedures. Cells were counted and mixed at appropriate ratios of infected to uninfected cells prior to DNA extraction with the Qiagen blood DNA kit according to the manufacturer's instructions (Qiagen, Hombrechtikon, Switzerland).
Clinical sample DNA extraction.
Residual ThinPrep cell suspensions were transferred into 15-ml conical polystyrene tubes and sedimented by low-speed centrifugation (1,000 × g) for 10 min at room temperature. Cells were suspended in 10 pellet volumes or 200 μl, whichever was larger, of ThinPrep supernatant prior to DNA extraction.
DNA for PGMY-CHUV was purified with the Magna Pure DNA isolation kit I on the Magna LC robot (Roche, Rotkreuz, Switzerland) and eluted in 100 μl elution buffer according to the manufacturer's instructions.
DNA for LA was purified with the Nuclisens DNA extraction kit on the EasyMAG robot (bioMérieux, Geneva, Switzerland) and eluted in 110 μl according to the manufacturer's instructions.
Phosphate-buffered saline (PBS)-negative controls systematically accompanied each set of clinical samples during the DNA extraction procedure and throughout all PCR and gel electrophoresis steps to assess contaminations.
PGMY-CHUV assay.
The PGMY-CHUV assay was performed as described in chapter 5 of the WHO HPV Laboratory Manual (33). It is based on the PCR amplification of HPV DNA with biotinylated PGMY primers (17) and the determination of HPV genotypes by RBH against a panel of HPV genotype-specific probes. This panel allows the genotyping of 31 HPV (6, 11, 16, 18, 26, 31, 33, 34, 35, 39, 40, 42, 44, 45, 51, 52, 53, 54 [AE9 subtype], 55, 56, 57, 58, 59, 66, 68 [ME180 subtype], 69, 70, 73, 82 [AE2 subtype], 83, and 84). Key points for data interpretation are provided below.
(i) PCR.
Five μl sample DNA was PCR amplified in 50-μl replicate (up to 3) reaction mixtures with buffer A containing 1.5 mM MgCl2 for the first 8,348 samples (33). The most recent 1,060 samples were PCR amplified in 50-μl duplicate reaction mixtures, one with buffer A and the other with buffer B containing 3 mM MgCl2 (33). Buffer B was found to be necessary to achieve adequate sensitivity for epidemiological purposes and was introduced for routine genotyping only recently. The analytical sensitivity of the PCR was determined to be 50 genome equivalents (GE) for genotypes 6, 11, 16, 18, 33, 35, 39, 45, 51, 52, 58, 59, 66, 68 (ME180 subtype), and it was 500 GE for HPV31 and HPV56 with proficiency panel DNAs, DNAs from HPV16 and HPV18 cell lines (CasKi, SiHa and HeLa), and HPV16 and HPV18 international standards (see File S1 in the supplemental material).
Samples were noninformative if the HPV and the HLA internal controls were negative (33).
(ii) RBH.
After gel electrophoretic analysis, amplicons from doubtful or positive reactions were subjected to RBH in a miniblotter, allowing up to 45 samples and controls to be processed in parallel.
The arrays were produced by the covalent binding of the probes to a negatively charged nylon membrane. This allowed washing the membrane at high temperature in a low-salt solution with minimal loss of sensitivity on repeat reprobing. Enhanced chemiluminescence (ECL) was used to reveal biotinylated hybrids with streptavidin peroxidase after exposure to an autoradiography film. This detection method did not leave an imprint on the membrane and therefore allowed reprobing the arrays. Washing the miniblotter with hydrogen peroxide prevented strong, nonspecific background at further membrane usage (see File S2 in the supplemental material).
(iii) Genotype identification.
RBH positives were identified by the visual inspection of films (see File S2 in the supplemental material). A positive was defined by signal strength superior to that of local background, square shape, and position within a corresponding grid array. Evaluation was done by two of us independently (C. Estrade and R. Sahli) to avoid errors due to misreading. Errors were very rare (less than 0.5%). They were associated mainly with the lack of a genotype being recorded in multiple infections.
For objective evaluation, the film was scanned and stored as a .tiff file with 256 gray levels at 200 dots per inch. Image analysis software was used to measure the gray level of each spot (AlphaEaseFC, v.4.01; Cell Biosciences, Santa Clara, CA). It was expressed as a percentage of 255, with 100% corresponding to saturating black pixels and 0% as white. On-screen display was done with the gamma value set at 0.90. Image enhancement was not allowed. A blank lane parallel to the probes was used as a negative control to calculate the average background gray value and its standard errors. The threshold of positivity was set as a gray level of more than the mean background level plus 5 standard errors, corresponding visually to very faint signals. The specificity of such low signals was confirmed by DNA sequencing.
(iv) DNA sequencing.
Amplicons were purified with the Qiaquick PCR purification kit (Qiagen) and sequenced with 4 μl of the BigDye Terminator chemistry (BDT v.1.1; Applied Biosystems) in 10-μl reaction mixtures containing up to 3 μl purified DNA (10 to 50 ng) and 3 pmol of each PGMY11 primer. Labeled DNA from sequencing reactions was purified either by ethanol precipitation or by the Montage SEQ96 purification kit (Millipore, Zug, Switzerland) according to the manufacturer's instructions, and samples were resolved by capillary electrophoresis on ABI instruments (Applied Biosystems, Rotkreuz, Switzerland).
Sequencing chromatograms were evaluated with Chromas (Technelysium, Tewantin, Australia) or Geneious v5.4 (Biomatters LTD, New Zealand). Sequence comparison to the nonredundant nucleic acid GenBank database was performed with BLAST (1) and is available at http://blast.ncbi.nlm.nih.gov/Blast.cgi; probe mismatches within sequenced amplicons were identified with the find motif tool of Geneious.
(v) Reagents and equipment.
Primers and probes were obtained from three different manufacturers (Microsynth, Balgach, Switzerland; Eurogentec, Seraing, Belgium; and Eurofins MWG Operon, Ebersberg, Germany). All other reagents and equipment were from the manufacturers indicated in chapter 5 of the WHO HPV Laboratory Manual (33).
LA.
LA was performed as recommended by the manufacturer (Roche, Basel, Switzerland) except for the volume of input DNA. Five μl DNA in 50-μl PCR mixture was used, as larger relative volumes of DNA purified with the EasyMAG system frequently inhibited PCR. This assay detects nine additional HPV genotypes not detected by PGMY-CHUV RBH: 61, 62, 64, 67, 71, 72, 81, 82 (IS39 subtype), and 89 (CP6108).
Validation studies. (i) PGMY-CHUV PCR buffer comparison.
Of the 9,408 samples submitted to our laboratory for HPV testing, 827 were prospectively collected to analyze the performance of PGMY-CHUV realized with PCR buffer A or with PCR buffer B (see File S3 in the supplemental material).
(ii) Probe sensitivity and specificity.
PGMY-CHUV genotyping was completed by the DNA sequencing of PCR-positive or doubtful samples if they also were negative or weakly positive by RBH (n = 1,024 out of 9,408 samples). Sequencing was performed for quality-control purposes and included randomly selected RBH-positive samples (see File S4 in the supplemental material).
(iii) Comparison between PGMY-CHUV and LA.
Two hundred samples were collected prospectively at the MCL diagnostic laboratory for several weeks to obtain at least 180 positive samples, covering as much as possible of the entire spectrum of high-risk genotypes, and they were equilibrated in single and multiple infections. This set was supplemented with negative samples selected randomly during this period. Original DNA stored at −20°C was used for PGMY-CHUV testing. Genotypes determined with PGMY-CHUV were recorded before the disclosure of LA results. The number of genotypes per sample was determined after combining PGMY-CHUV and LA results, assuming 100% probe specificity for each assay.
(iv) PGMY-CHUV reproducibility.
Two hundred seventy-nine samples from all women known to have persistent HPV among the 5,904 patients whose samples were submitted to our laboratory were selected for repeat testing. A single sample was evaluated from each patient to increase genotype/subtype diversity. This mode of selection did not exclude negative samples due to the known fluctuation in HPV DNA detection rate in the same patient over time (36). DNA was stored at −20°C until PGMY-CHUV retesting. Samples were renamed prior to repeat analysis. Repeat HPV typing results were recorded before sample decoding and then compared to the original data.
Determination of the occurrence of HPV genotypes in routine HPV testing.
All samples (n = 9,408) submitted for HPV testing in our laboratory were considered for occurrence data (see File S5 in the supplemental material). Occurrence did not necessarily reflect that of the healthy patient population, since it was determined in a population previously screened by cytology with a high proportion of patients presenting with abnormal cytology (as described above). The risk attribution of HPV genotypes was according to Munoz et al. (24, 25). Risk categorization based on phylogeny is controversial for HPV26 and HPV53 (8, 19, 29). The former was considered undetermined and the latter was considered low risk due to its high prevalence in low-grade disease and low prevalence, similarly to HPV6, in cervical cancer cases (18, 29).
Reagent cost evaluation.
Reagent cost per sample was evaluated using a typical analysis setup using 96-well PCR plates (33). This setup consisted of analyzing each sample together with five positive controls (one with 1,000, two with 100, and two with 10 copies of HPV16 DNA) and two negative controls (one DNA extraction control and one no-template PCR control) either with buffer B or with both buffer A and buffer B. One 1,000-copy HPV16 control was placed in each additional row of the plate at increasing column numbers to serve as a unique row identifier when more than five samples were analyzed. Systematic gel electrophoretic analysis in 96-well formats was considered together with the PCR plate and plastic sealing foil as fixed PCR costs (21 Swiss francs). PCR reagents, polymerase, and primers were considered variable PCR costs (0.81 Swiss francs per reaction). The cost calculation for RBH took into consideration membranes, probes, probe cross-linking reagent, the production of quality control amplicons, and the array quality control as fixed costs (103 Swiss francs); hybridization solutions, film, ECL detection reagents, foam pads, and streptavidin peroxidase were variable costs (27 Swiss francs). Filter tips and common labware shared with commercial assays have not been taken into consideration for this calculation, except for gel electrophoresis, which was specific to PGMY-CHUV. DNA sequencing was not included in cost estimates, as it was used only as a quality control for assay design purposes and is not necessary for the routine use of PGMY-CHUV. Cost did not include personnel time for the preparation and quality control of reagents. DNA extraction was not evaluated in this study, therefore it was not included in the calculations which were for genotyping per se. DNA extraction with the Qiagen kit mentioned above would cost 4 Swiss francs ($2.20 in the United States) per sample and should be added to the genotyping cost to estimate the total analysis cost.
Data analysis and statistics.
Tables for statistics were generated with Microsoft Access or Excel, and statistics were performed with GraphPad Prism (GraphPad Software, La Jolla, CA). The agreement of HPV typing results between paired cases was evaluated with the Cohen's kappa (κ) statistic, and their uneven distribution was evaluated with McNemar's test. Agreement was interpreted as weak (0.200 < κ < 0.401), moderate (0.400 < κ < 0.601), strong (0.600 < κ < 0.801), near perfect (0.800 < κ < 1.000), and perfect (κ = 1.000). Statistics for unpaired cases were performed with the two-sided Fisher's exact test using two-by-two contingency tables.
RESULTS
Comparison to LA.
To compare PGMY-CHUV to a similar, commercially available, and thoroughly evaluated assay, we examined 200 samples that were genotyped by LA. Of these 200 samples, 182 were positive and 18 were negative. The attribution of positive and negative results was identical with PGMY-CHUV PCR independently of the RBH results. Both methods agreed on all samples at this level of evaluation (positive versus negative).
The array-specific genotypes observed during this study were excluded from subsequent analysis, as they accounted for only 10% of the observed genotype-sample combinations (HPV34, HPV44, and HPV57 with PGMY-CHUV or HPV genotypes 61, 62, 64, 67, 71, 72, 81, 82 [IS39 subtype], and 89 [CP6108] with LA; n = 38 out of 382). One HPV61 single infection determined by LA was confirmed by DNA sequencing of the PGMY-CHUV amplicon, and one IS39 single infection by LA was reported as unknown (mixed profile) after PGMY-CHUV DNA sequencing. Both samples were excluded from comparison. Thus, 180 of the 182 positive samples contained genotypes represented by both arrays (HPV genotypes 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51 to 56, 58, 59, 66, 68, 69, 70, 73, and 82 to 84). Of 83 single infections reported by LA, 81 (98%) each were attributed to the same genotype by PGMY-CHUV (Table 1). The two discordant cases were RBH negative by PGMY-CHUV and were determined to be HPV59 and HPV84 by LA. DNA sequencing was used with the corresponding PGMY-CHUV amplicons to evaluate discordance. Both samples were HPV positive for a genotype not represented on the array as expected: HPV107 and HPV114, respectively.
Table 1.
Comparison of PGMY-CHUV and LA: summary of sample results according to the number of genotypes per sample
| No. of genotypes per samplea | No. of samples | Full agreementb |
|
|---|---|---|---|
| Yes (%) | No | ||
| 1 | 83 | 81 (98) | 2 |
| 2 | 44 | 32 (70) | 12 |
| 3 | 30 | 17 (57) | 13 |
| 4 | 7 | 4 (57) | 3 |
| 5 | 11 | 2 (18) | 9 |
| 6 | 2 | 0 (0) | 2 |
| 7 | 3 | 1 (33) | 2 |
| Total | 180 | 137 (76) | 43 |
Except for assay-specific single infections, the number of genotypes per sample was calculated after combining PGMY-CHUV and LA results.
A sample was considered discordant whenever one of the genotypes represented on both arrays was discordant within the sample.
In contrast to samples with single infections, full agreement on samples with double infections or other multiple infections was significantly lower (<71%; P < 0.0001 by two-tailed Fisher's exact test). On average, full agreement was 76% (137/180). Agreement at the genotype level was slightly higher. Out of 344 genotype-sample combinations, 285 (83%) were concordant (Table 2). Agreement was lower in multiple infections (<86%; P < 0.0001 by two-tailed Fisher's exact test) than in single infections (98%), and its level was not associated with the number of genotypes per sample (P > 0.05 by two-tailed Fisher's exact test).
Table 2.
Comparison of PGMY-CHUV and LA: summary of genotype-sample combination results according to the number of genotypes per sample
| No. of genotypes per samplea | No. of genotype-sample combinationsb | Agreement |
|
|---|---|---|---|
| Yes (%) | No | ||
| 1 | 83 | 81 (98) | 2 |
| 2 | 81 | 69 (85) | 12 |
| 3 | 75 | 57 (76) | 18 |
| 4 | 27 | 23 (85) | 4 |
| 5 | 51 | 35 (67) | 16 |
| 6 | 12 | 7 (58) | 5 |
| 7 | 15 | 13 (87) | 2 |
| Total | 344 | 285 (83) | 59 |
Except for assay-specific single infections, the number of genotypes per sample was calculated after combining PGMY-CHUV and LA results.
The genotypes were restricted to those represented on both arrays.
Except for HPV69, which could not be evaluated (n = 0), all genotypes were identified by both assays with genotype-specific κ values greater than 0.639 (strong agreement) (Table 3). The overall agreement was 98.95% (κ = 0.901; 95% confidence interval [CI], 0.875 to 0.926; near perfect). Average κ values of 0.987 were observed for single infections and of 0.853 for multiple infections (data not shown). HPV42 (P = 0.0233 by two-sided McNemar's test) and HPV56 (P = 0.0026 by two-sided McNemar's test) were detected significantly more often with PGMY-CHUV, conferring, together with HPV53 (P = 0.0736 by two-sided McNemar's test), a slightly higher sensitivity of PGMY-CHUV compared to that of LA (8%; P = 0.0018 by two-sided McNemar's test) when the combined results obtained with both tests served as a reference.
Table 3.
Comparison of PGMY-CHUV and LA: genotype-specific analysisf
| Genotypea | No. of samples for each resultb |
% Agreement | κ | 95% CI | Intc | |||
|---|---|---|---|---|---|---|---|---|
| −/− | +/− | −/+ | +/+ | |||||
| 6 | 185 | 2 | 0 | 13 | 99.00 | 0.923 | 0.817–1.029 | NP |
| 11 | 196 | 0 | 0 | 4 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 16 | 156 | 3 | 0 | 41 | 98.50 | 0.955 | 0.905–1.006 | NP |
| 18 | 189 | 0 | 1 | 10 | 99.50 | 0.950 | 0.852–1.048 | NP |
| 26 | 199 | 0 | 0 | 1 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 31 | 181 | 1 | 0 | 18 | 99.50 | 0.970 | 0.912–1.028 | NP |
| 33 | 198 | 0 | 0 | 2 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 35 | 196 | 0 | 1 | 3 | 99.50 | 0.855 | 0.570–1.139 | NP |
| 39 | 193 | 0 | 0 | 7 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 40 | 197 | 0 | 0 | 3 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 42e | 173 | 7 | 0 | 20 | 96.50 | 0.832 | 0.709–0.954 | NP |
| 45 | 192 | 1 | 1 | 6 | 99.00 | 0.852 | 0.648–1.056 | NP |
| 51 | 169 | 2 | 0 | 29 | 99.00 | 0.961 | 0.907–1.015 | NP |
| 52 | 179 | 3 | 3 | 15 | 97.00 | 0.817 | 0.673–0.961 | NP |
| 53 | 180 | 5 | 0 | 15 | 97.50 | 0.844 | 0.709–0.979 | NP |
| 54 | 183 | 0 | 1 | 16 | 99.50 | 0.967 | 0.902–1.032 | NP |
| 55 | 197 | 0 | 0 | 3 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 56e | 178 | 11 | 0 | 11 | 94.50 | 0.640 | 0.434–0.847 | S |
| 58 | 180 | 0 | 1 | 19 | 99.50 | 0.972 | 0.916–1.027 | NP |
| 59 | 189 | 1 | 2 | 8 | 98.50 | 0.834 | 0.648–1.020 | NP |
| 66 | 185 | 2 | 3 | 10 | 97.50 | 0.787 | 0.602–0.971 | S |
| 68 | 195 | 0 | 0 | 5 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 69 | 200 | 0 | 0 | 0 | NAd | NA | NA | NA |
| 70 | 196 | 1 | 0 | 3 | 99.50 | 0.855 | 0.570–1.139 | NP |
| 73 | 192 | 0 | 0 | 8 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 82 | 195 | 0 | 0 | 5 | 100.00 | 1.000 | 1.000–1.000 | PE |
| 83 | 193 | 1 | 1 | 5 | 99.00 | 0.828 | 0.591–1.065 | NP |
| 84 | 190 | 2 | 3 | 5 | 97.50 | 0.654 | 0.354–0.953 | S |
| Total | 5256 | 42 | 17 | 285 | 98.95 | 0.901 | 0.875–0.926 | NP |
Only the 28 genotypes shared by both assays were considered for analysis.
−/−, Negative by both assays; −/+, PGMY-CHUV negative and LA positive; +/−, PGMY-CHUV positive and LA negative; +/+, positive with both assays.
Interpretation of the κ values. S, strong; NP, near perfect; PE, perfect.
NA, not applicable.
More often detected with PGMY-CHUV (P < 0.05 by two-sided McNemar's test).
The total number of samples was 200, giving 5,600 possible genotype-sample combinations.
To assess whether discordant results in multiple infections were associated with genotypes representing a minor fraction in each sample, they were reevaluated by taking into account RBH signal strength. A genotype was considered weak when it had the weakest relative RBH signal strength compared to that of all other genotypes within a sample. Of the 59 discordant genotype-sample combinations, 49 (83%) were recorded as weak. In contrast, only 41 (14%) out of 285 concordant combinations were recorded as such. Discordant results in multiple infections therefore were significantly associated with weakly positive genotypes (P < 0.0001 by two-sided Fisher's exact test), which is consistent with intergenotypic competition in the PCRs.
Reproducibility.
To verify the robustness of PGMY-CHUV, one sample per patient from a set of 279 independent patients was reanalyzed with different reagent lots and probe arrays. Laboratory work involved four different technicians during a period of 2 years. The original data were compared to the newly acquired data with κ statistics.
Three samples were noninformative on repeat testing, and one was missing. Of the resulting 275 informative samples (98.6%), 13 were reproducibly negative (4.7%), 10 became negative on retesting (3.6%), and 252 were reproducibly positive (91.6%). Most samples (84.0%) showed complete concordance for all genotypes, even with multiple infections containing up to 8 genotypes (not shown). Results from single and multiple infections were combined for genotype-specific analysis (Table 4). Original testing identified 375 genotype-sample combinations, and retesting identified 362 (95.3%). The global level of genotype-specific agreement reached 99.47%, with a κ value of 0.933 (95% CI, 0.914 to 0.952) interpreted as near perfect. The number of discordant cases of each category was not statistically different (30 versus 17; P = 0.08 by two-sided McNemar's test). Genotype-wise, κ values were interpreted as near perfect for all genotypes but HPV82 (strong). The number of HPV genotype 26, 34, 40, 57, and 69 cases was insufficient for statistical analysis.
Table 4.
PGMY-CHUV reproducibility analysis: genotype-specific analysisa
| Genotype | No. of samples for each resultb |
% Agreement | κ | 95% CI | Intc | |||
|---|---|---|---|---|---|---|---|---|
| −/− | +/− | −/+ | +/+ | |||||
| 6 | 262 | 3 | 0 | 10 | 98.91 | 0.864 | 0.711–1.017 | NP |
| 11 | 272 | 0 | 0 | 3 | 100 | 1.000 | 1.000–1.000 | PE |
| 16 | 236 | 3 | 0 | 36 | 98.91 | 0.954 | 0.902–1.006 | NP |
| 18 | 266 | 0 | 0 | 9 | 100 | 1.000 | 1.000–1.000 | PE |
| 26 | 275 | 0 | 0 | 0 | NAd | NA | NA | NA |
| 31 | 257 | 3 | 0 | 15 | 98.91 | 0.903 | 0.795–1.012 | NP |
| 33 | 267 | 0 | 0 | 8 | 100 | 1.000 | 1.000–1.000 | PE |
| 34 | 274 | 1 | 0 | 0 | 99.64 | 0.000 | −1.956–1.956 | W |
| 35 | 257 | 0 | 0 | 18 | 100 | 1.000 | 1.000–1.000 | PE |
| 39 | 268 | 0 | 0 | 7 | 100 | 1.000 | 1.000–1.000 | PE |
| 40 | 275 | 0 | 0 | 0 | NA | NA | NA | NA |
| 42 | 258 | 2 | 2 | 13 | 98.55 | 0.859 | 0.722–0.996 | NP |
| 44 | 266 | 0 | 1 | 8 | 99.64 | 0.939 | 0.821–1.058 | NP |
| 45 | 264 | 1 | 1 | 9 | 99.27 | 0.896 | 0.753–1.040 | NP |
| 51 | 259 | 1 | 0 | 15 | 99.64 | 0.966 | 0.899–1.033 | NP |
| 52 | 256 | 2 | 2 | 15 | 98.55 | 0.875 | 0.753–0.997 | NP |
| 53 | 242 | 1 | 1 | 31 | 99.27 | 0.965 | 0.916–1.013 | NP |
| 54 | 269 | 0 | 1 | 5 | 99.64 | 0.907 | 0.726–1.089 | NP |
| 55 | 263 | 2 | 1 | 9 | 98.91 | 0.851 | 0.684–1.019 | NP |
| 56 | 257 | 0 | 2 | 16 | 99.27 | 0.937 | 0.851–1.024 | NP |
| 57 | 275 | 0 | 0 | 0 | NA | NA | NA | NA |
| 58 | 245 | 3 | 1 | 26 | 98.55 | 0.920 | 0.843–0.998 | NP |
| 59 | 265 | 0 | 0 | 10 | 100 | 1.000 | 1.000–1.000 | PE |
| 66 | 259 | 1 | 1 | 14 | 99.27 | 0.929 | 0.832–1.027 | NP |
| 68 | 268 | 0 | 0 | 7 | 100 | 1.000 | 1.000–1.000 | PE |
| 69 | 275 | 0 | 0 | 0 | NA | NA | NA | NA |
| 70 | 263 | 1 | 0 | 11 | 99.64 | 0.955 | 0.866–1.043 | NP |
| 73 | 266 | 1 | 1 | 7 | 99.27 | 0.871 | 0.693–1.049 | NP |
| 82 | 270 | 0 | 2 | 3 | 99.27 | 0.747 | 0.397–1.097 | S |
| 83 | 268 | 2 | 0 | 5 | 99.27 | 0.830 | 0.595–1.065 | NP |
| 84 | 269 | 0 | 1 | 5 | 99.64 | 0.907 | 0.726–1.089 | NP |
| Othere | 242 | 3 | 0 | 30 | 98.91 | 0.946 | 0.886–1.007 | NP |
| Total | 8,408 | 30 | 17 | 345 | 99.47 | 0.933 | 0.914–0.952 | NP |
The total number of samples was 275, giving 8,800 possible genotype-sample combinations.
−/−, Negative on original as well as on retesting; −/+, original negative and retesting positive; +/−, original positive and retesting negative; +/+, positive on original as well as on retesting.
Interpretation of the κ values. PE, perfect; NP, near perfect; S, strong; W, weak.
NA, not applicable.
Other HPV genotypes not on the probe array and considered a single group (61, 62, 71, 72, 74, 81, 89, 114, HLT7474S, and unknown).
To assess whether discordant results in multiple infections were associated with genotypes representing a minor fraction within each sample, they were reevaluated taking into consideration RBH signal strength. For single infections, samples were considered weak by the stochastic nature of PCR or by their RBH signal strength being less than the average gray value of the negative control plus 8 times the standard deviations. Of the 47 discordant cases, 27 (57%) were recorded as weak. In contrast, only 38 (11%) out of 345 concordant cases were recorded as such. Discordant results therefore were significantly associated with weakly positive genotypes (P < 0.0001 by two-sided Fisher's exact test), which is consistent with the results of the comparison of PGMY-CHUV to LA.
Cost evaluation.
Cost evaluation was determined after distinguishing fixed and variable costs and taking into consideration the systematic use of at least five positive controls and two negative controls per set of samples.
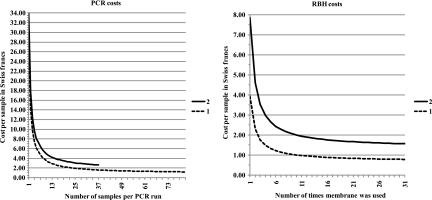
Fixed costs for PCR were estimated at 21 Swiss francs and variable costs at 0.81 Swiss francs per reaction (see Materials and Methods). For PCR, a sharp cost decrease was seen as soon as several samples were PCR amplified in parallel, with a plateau starting at around 16 samples (Fig. 1). Sixteen samples corresponded to 24 wells of a 96-well PCR plate (16 samples plus five positive and two negative controls and one positive control in the second row). Typical sets of PCR performed in 96-well plates with either one (dotted line) or two (solid line) PCR buffers and up to a maximum of 82 or 37 samples, respectively, resulted in a cost per sample of 1.21 to 2.63 Swiss francs.
Fig. 1.
Cost evaluation. Costs in Swiss francs are shown as a function of the number of samples processed in parallel in a 96-well PCR plate with either a single reaction performed with buffer B per sample (PCR costs, line 1) or two reactions performed with buffer A and with buffer B per sample (PCR costs, line 2). Following gel electrophoresis, positive or doubtful samples were processed by RBH by sets of 40 hybridizations (RBH costs). The costs of RBH per sample are shown as a function of membrane reuse and the number of PCRs subjected to RBH: either one reaction per sample (1) or both reactions, in adjacent lanes, per sample (2). The membranes could be used a median of 11 times, with several being used more than 20 times (see File S2 in the supplemental material).
Fixed costs for RBH were estimated at 103 Swiss francs and variable costs at 27 Swiss francs per hybridization run (see Materials and Methods). Filling the miniblotter was necessary to optimize costs. Thus, 40 lanes per hybridization were considered for the calculations presented in Fig. 1. Under this condition, the cost per sample of RBH diminished strongly as soon as the membrane was used more than four times, with a plateau starting at around nine times. The median number of times a membrane could be used was 11 (see File S2 in the supplemental material), well into the plateau. Therefore, the median cost per sample of RBH was 0.97 to 1.94 Swiss francs depending on whether amplicons from 1 (dotted line) or 2 PCRs (solid line) were subjected to RBH. The contribution of RBH to costs was a conservative evaluation. Using membranes more than 11 times was feasible and further reduced costs. For instance, with a membrane used 29 times (see File S2 in the supplemental material), RBH cost per sample would be 0.79 to 1.57 Swiss francs depending on whether amplicons from one or both PCRs are subjected to RBH.
The most expensive algorithm (duplicate PCR, two RBH per sample, median RBH usage) was calculated to be 4.6 Swiss francs ($5.50) per sample. It was used only during the prospective evaluation of the effect of PCR buffer composition on genotyping (see File S3 in the supplemental material). The median cost for routine use with duplicate PCR and one RBH per sample was estimated to be 3.6 Swiss francs ($4.30). This median cost could be reduced to 2.2 Swiss francs ($2.60) with a single PCR (buffer B containing 3 mM MgCl2) and a single RBH per sample. Using the membranes beyond the median usage number allowed further price reduction down to a plateau close to 2 Swiss francs ($2.40).
DISCUSSION
Our data show that we were able to design an in-house HPV genotyping method that compares favorably with the research-grade, commercially available LA. It is reproducible and cost-effective, even taking into account quality-control procedures. Like LA and other HPV genotyping assays, PGMY-CHUV has not been validated for primary cervical cancer screening according to published guidelines (23). It therefore should be used in its present format either as an adjunct to a clinically validated screening test which identifies the risk of progression to high-grade cervical intraepithelial neoplasia, as a follow-up test to monitor patients after treatment, or as an epidemiological or research tool. Unlike a similar assay based on the GP5+/6+ primers (34), PGMY-CHUV relies on gel electrophoresis rather than enzyme immunoassay for sorting positive samples for RBH, hence it is simpler and less expensive.
To confirm its adequacy for genotyping, PGMY-CHUV was compared to LA because both share the same PGMY primers (17) and LA has already been thoroughly evaluated against the original PGMY reverse-line blot array (3, 11) and against other commercial assays with reported good performance (4, 32). PGMY-CHUV agreed with LA for 81 out of 83 assay-specific single infections (98%). Samples were not available for the sequencing of the corresponding discordant LA amplicons. It therefore is impossible to assess the specificity of LA in these two instances. The other discordant results were found in multiple infections, indicating a slightly superior global sensitivity of PGMY-CHUV if the combined results of both assays were used as a reference. At the genotypic level, however, higher sensitivity was significant only for HPV42 and HPV56.
Discordant results were significantly associated with weakly positive genotypes. This may be related not only to the use of duplicate PCRs with PGMY-CHUV versus a single reaction with LA but also to a potentially limiting primer concentration established to minimize competition by dominant targets. The genotyping of high-risk HPVs with both assays therefore is expected to be highly comparable, with discordant results having only a limited impact on epidemiological studies, except possibly for HPV56.
An advantage of PGMY-CHUV over LA is the unambiguous detection of HPV52 regardless of coexisting genotypes in multiple infections. The three samples that were HPV52 positive by PGMY-CHUV and negative by LA corresponded to multiple infections containing HPV58 (n = 2) or HPV35 (n = 1). The simultaneous presence of one of these two genotypes is known to interfere with HPV52 detection by LA (10). Being able to detect HPV52 is important, as its prevalence is high in our patient population (ranking third among the high-risk genotypes and fifth overall; see File S5 in the supplemental material) and also in squamous cell carcinoma in Asia compared to other regions of the world (7, 20). PGMY-CHUV avoids the use of alternative methods like genotype-specific PCR to identify HPV52 in such multiple infections. Another advantage of PGMY-CHUV is the lower number of cases subjected to RBH, since the procedure can be stopped after PCR for PCR-negative samples, hence reducing costs further, especially for studies dealing with low-HPV-prevalence populations.
The intralaboratory reproducibility of PGMY-CHUV was a prerequisite for its technology transfer to members of the WHO HPV laboratory network. Our comparison of genotyping data performed at two different time points showed a very high degree of genotype-specific reproducibility (κ = 0.933), with most samples (84%) being fully concordant for all genotypes. These values are similar to those obtained by Steinau and colleagues with LA (31). Discordant cases were significantly associated with weak positives, as seen in the comparison to LA. This is a common observation, underscoring the stochastic nature of PCR amplification of low-starting-copy-number targets in the context of primer/polymerase competition.
Five laboratories using PGMY-CHUV participated in the 2009 proficiency panel study within the WHO HPV LabNet after technology transfer from our laboratory. Two were nonproficient because of contaminations, one was proficient at 88% (sensitivity issues) but used only one PCR with buffer A (containing 1.5 mM MgCl2), which is known to be less sensitive analytically than buffer B (containing 3 mM MgCl2), and two were fully proficient (15). These results were not different from those obtained with LA, where two laboratories out of four were proficient (15). Recent results of the 2010 proficiency panel showed that four laboratories out of six using PGMY-CHUV were fully proficient this time, one was proficient at more than 90%, and another was not proficient, suffering from a lack of sensitivity overall (13). Again, these results were not different from those obtained with LA, where 8 laboratories out of 17 were fully proficient (13). The successful participation of several laboratories in proficiency testing confirmed that PGMY-CHUV is suitable for epidemiological studies with adequate sensitivity and specificity, and that it can be transferred to other laboratories. We feel that these proficiency testing results reflect the ease of PGMY-CHUV implementation by laboratories provided they are using PCR on a regular basis in research as well as in routine clinical testing. Compared to commercial assays, PGMY-CHUV requires strict, additional quality control, as expected for an in-house assay.
PGMY-CHUV PCR was never associated with defects that had an impact on the routine use of the assay in more than 10 years in our laboratory. In contrast, PGMY-CHUV RBH was affected twice by probes being cross-contaminated during synthesis. Contamination appeared more recently with high automation levels and the utilization of reusable devices. For this reason, we now order desalted probes rather than high-performance liquid chromatography (HPLC)-purified ones and split the probe order time-wise or among different manufacturers' production sites. Cross-contaminated probes were detected at delivery by our membrane array quality control, underscoring its value and necessity. The quality assurance program used with PGMY-CHUV is described in chapter 5 of the WHO HPV Laboratory Manual (33). Additional aspects of quality assurance also are covered in chapters 3 and 6 of that manual.
Excluding DNA extraction (on the order of $2.20 per sample with a commercial kit), the estimated cost of performing a single genotyping under the most sensitive algorithm, consisting of duplicate PCR and a single RBH per sample, was $4.30 per sample. The least expensive, but slightly less sensitive, yet proficient genotyping algorithm was estimated at $2.40 per sample. It consisted of processing 82 samples in parallel with a single PCR condition (buffer B containing 3 mM MgCl2) and one RBH per sample. This algorithm appears to be well adapted to epidemiological studies. DNA sequencing was not included in the cost estimates, because it is not essential for the routine use of PGMY-CHUV owing to the high sensitivity of RBH (98.61%) (see the second table in File S4 in the supplemental material). Knowing that there were only 32 RBH-negative cases without mismatches among 6,703 genotype-sample combinations (see the third table in File S4 in the supplemental material), probe sensitivity could be improved even further at more than 99.5% with updated probes currently being evaluated. They target HPV subtypes affected by mismatches within probes of the present version of the array (HPV genotypes 45, 51, 54, 56, 58, 68, and 82 in particular). Their sequences are available upon request.
In conclusion, we present an in-house HPV genotyping assay which is proficient for HPV vaccine monitoring. With its proven transferability to other laboratories, its low cost, and robustness, PGMY-CHUV is a valuable method for HPV genotyping, especially in low-resource settings.
Supplementary Material
ACKNOWLEDGMENTS
The development of the assay was supported by the Institute of Microbiology of the Centre Hospitalier Universitaire Vaudois, University of Lausanne, and by a grant of the Ligue Vaudoise Contre le Cancer to R.S.
We thank Patty Gravitt and Janet Kornegay for providing sequences of PGMY primers prior to their publication in 1999. We gratefully acknowledge the expert technical help of the staff of the diagnostic virology and molecular diagnostic laboratories at our institute. Technology transfer within the WHO HPV LabNet was supported by the WHO via a project funded by the Bill and Melinda Gates Foundation that was coordinated by Tiequn Zhou (WHO). Elizabeth Unger and Joakim Dillner provided helpful advice during the course of this evaluation. Stefan Gerber, Phillip Shaw, and Elizabeth Unger critically reviewed the manuscript.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W. M., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Berkhof J., et al. 2006. Human papillomavirus type-specific 18-month risk of high-grade cervical intraepithelial neoplasia in women with a normal or borderline/mildly dyskaryotic smear. Cancer Epidemiol. Biomarkers Prev. 15:1268–1273 [DOI] [PubMed] [Google Scholar]
- 3. Castle P. E., Gravitt P. E., Solomon D., Wheeler C. M., Schiffman M. 2008. A comparison of linear array and line blot assay for detection of HPV and cervical precancer and cancer in the ASCUS LSIL triage study. J. Clin. Microbiol. 46:109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castle P. E., et al. 2008. A comparison of two PCR-based human papillomavirus genotyping methods. J. Clin. Microbiol. 46:3437–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castle P. E., Sadorra M., Garcia F., Holladay E. B., Kornegay J. 2006. A Pilot study of a commercialized human papillomavirus (HPV) genotyping assay: comparison of HPV risk group to cytology and histology. J. Clin. Microbiol. 44:3915–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clavel C., et al. 2001. Human papillomavirus testing in primary screening for the detection of high-grade cervical lesions: a study of 7932 women. Br. J. Cancer 84:1616–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clifford G. M., Smith J. S., Plummer M., Munoz N., Franceschi S. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer 88:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cogliano V., et al. 2005. Carcinogenicity of human papillomaviruses. Lancet Oncol. 6:204. [DOI] [PubMed] [Google Scholar]
- 9. Coupe V. M., et al. 2009. HPV16/18 vaccination to prevent cervical cancer in The Netherlands: model-based cost-effectiveness. Int. J. Cancer 124:970–978 [DOI] [PubMed] [Google Scholar]
- 10. Coutlée F., et al. 2007. Confirmatory real-time PCR assay for HPV-52 infection in anogenital specimens screened for HPV infection with the Linear array HPV genotyping test. J. Clin. Microbiol. 45:3821–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coutlée F., et al. 2006. Enhanced detection and typing of human papillomavirus (HPV) DNA in anogenital samples with PGMY primers and the linear array HPV genotyping test. J. Clin. Microbiol. 44:1998–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuzick J., et al. 1999. HPV testing in primary screening of older women. Br. J. Cancer 81:554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dillner J. 2011. Technical report of the 2010 WHO HPV LabNet proficiency study on HPV testing and typing. WHO HPV LabNet Global Reference Laboratory, Malmö, Sweden [Google Scholar]
- 14. Dillner J., Arbyn M., Dillner L. 2007. Translational mini-review series on vaccines: monitoring of human papillomavirus vaccination. Clin. Exp. Immunol. 148:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eklund C., Zhou T., Dillner J. 2010. A global proficiency study of human papillomavirus genotyping. J. Clin. Microbiol. 48:4147–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferguson M., Wilkinson D. E., Zhou T. 2009. WHO meeting on the standardization of HPV assays and the role of the WHO HPV Laboratory Network in supporting vaccine introduction held on 24–25 January 2008, Geneva, Switzerland. Vaccine 27:337–347 [DOI] [PubMed] [Google Scholar]
- 17. Gravitt P. E., et al. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herrero R., et al. 2005. Epidemiologic profile of type-specific human papillomavirus infection and cervical neoplasia in Guanacaste, Costa Rica. J. Infect. Dis. 191:1796–1807 [DOI] [PubMed] [Google Scholar]
- 19. Hiller T., Poppelreuther S., Stubenrauch F., Iftner T. 2006. Comparative analysis of 19 genital human papillomavirus types with regard to p53 degradation, immortalization, phylogeny, and epidemiologic risk classification. Cancer Epidemiol. Biomarkers Prev. 15:1262–1267 [DOI] [PubMed] [Google Scholar]
- 20. Hwang H. S., Park M., Lee S. Y., Kwon K. H., Pang M. G. 2004. Distribution and prevalence of human papillomavirus genotypes in routine pap smear of 2,470 Korean women determined by DNA chip. Cancer Epidemiol. Biomarkers Prev. 13:2153–2156 [PubMed] [Google Scholar]
- 21. Kjaer S. K., et al. 2009. A pooled analysis of continued prophylactic efficacy of quadrivalent human papillomavirus (types 6/11/16/18) vaccine against high-grade cervical and external genital lesions. Cancer Prev. Res. 2:868–878 [DOI] [PubMed] [Google Scholar]
- 22. Kleter B., et al. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 37:2508–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meijer C. J., et al. 2009. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int. J. Cancer 124:516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muñoz N., et al. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527 [DOI] [PubMed] [Google Scholar]
- 25. Munoz N., Castellsague X., de Gonzalez A. B., Gissmann L. 2006. Chapter 1: HPV in the etiology of human cancer. Vaccine 24(Suppl. 3):S1–S10 [DOI] [PubMed] [Google Scholar]
- 26. Naucler P., et al. 2007. HPV type-specific risks of high-grade CIN during 4 years of follow-up: a population-based prospective study. Br. J. Cancer 97:129–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paavonen J., et al. 2009. Efficacy of human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by oncogenic HPV types (PATRICIA): final analysis of a double-blind, randomised study in young women. Lancet 374:301–314 [DOI] [PubMed] [Google Scholar]
- 28. Safaeian M., et al. 2007. Comparison of the SPF10/LiPA system to the HC2 assay for detection of carcinogenic HPV genotypes among 5683 young women in Guanacaste, Costa Rica. J. Clin. Microbiol. 45:1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schiffman M., et al. 2005. A study of the impact of adding HPV types to cervical cancer screening and triage tests. J. Natl. Cancer Inst. 97:147–150 [DOI] [PubMed] [Google Scholar]
- 30. Stanley M., Villa L. L. 2008. Monitoring HPV vaccination. Vaccine 26(Suppl. 1):A24–A27 [DOI] [PubMed] [Google Scholar]
- 31. Steinau M., Swan D. C., Unger E. R. 2008. Type-specific reproducibility of the Roche linear array HPV genotyping test. J. Clin. Virol. 42:412–414 [DOI] [PubMed] [Google Scholar]
- 32. Szarewski A., et al. 2008. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol. Biomarkers Prev. 17:3033–3042 [DOI] [PubMed] [Google Scholar]
- 33. Unger E. R., Dillner J., Zhou T. (ed.). 2009. Human papillomavirus laboratory manual, 1st ed WHO, Geneva, Switzerland [Google Scholar]
- 34. van den Brule A. J., et al. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Hamont D., van Ham M. A., Bakkers J. M., Massuger L. F., Melchers W. J. 2006. Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the Roche linear array HPV genotyping test. J. Clin. Microbiol. 44:3122–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wheeler C. M., et al. 1996. Short-term fluctuations in the detection of cervical human papillomavirus DNA. Obstet. Gynecol. 88:261–268 [DOI] [PubMed] [Google Scholar]
- 37. Wheeler C. M., et al. 2009. The impact of quadrivalent human papillomavirus (HPV types 6, 11, 16, and 18) L1 virus-like particle vaccine on infection and disease due to oncogenic nonvaccine HPV types in sexually active women aged 16–26 years. J. Infect. Dis. 199:936–944 [DOI] [PubMed] [Google Scholar]
- 38. Wilkinson D. E., et al. 2010. Establishment of the 1(st) world health organization international standards for human papillomavirus type 16 DNA and type 18 DNA. Int. J. Cancer 126:2969–2983 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.