Abstract
A simple, rapid, sensitive, qualitative, colorimetric loop-mediated isothermal amplification (LAMP) with hydroxynaphthol blue dye (HNB) was established to detect high-risk human papillomavirus (HPV) genotypes 16, 18, 45, 52, and 58. All initial validation studies with the control DNA proved to be type specific. The colorimetric type-specific LAMP assay could achieve a sensitivity of 10 to 100 copies at 63°C for 65 min, comparable to that of real-time PCR. In order to evaluate the reliability of HPV type-specific LAMP, the assay was further evaluated with HPV DNAs from a panel of 294 clinical specimens whose HPV status was previously determined with a novel one-step typing method with multiplex PCR. The tested panel comprised 108 HPV DNA-negative samples and 186 HPV-DNA-positive samples of 14 genotypes. The results showed that the sensitivity of HPV type-specific LAMP for HPV types 16, 18, 45, 52, and 58 was 100%, 100%, 100%, 100%, and 100%, respectively, and the specificity was 100%, 98.5%, 100%, 98.8%, and 99.2%, respectively, compared with a novel one-step typing method with multiplex PCR. No cross-reactivity with other HPV genotypes was observed. In conclusion, this qualitative and colorimetric LAMP assay has potential usefulness for the rapid screening of HPV genotype 16, 18, 45, 52, and 58 infections, especially in resource-limited hospitals or rural clinics of provincial and municipal regions in China.
INTRODUCTION
It is well established that virtually all cervical cancers are caused by persistent high-risk oncogenic human papillomavirus (HPV) infections in the cervix. Thus, cervical screening could be improved by testing for the DNA of high-risk types of HPV as a primary screening tool (3, 9, 24). So far, at least 14 high-risk HPV genotypes have been shown to cause cervical cancer; those most important for cervical cancer are HPV types 16, 18, 45, 31, 33, 52, 58, and 35 (6, 7, 26). HPV16 is by far the predominant type, causing more than 50% of cancers; HPV18 follows, causing 14%; and HPV45 causes approximately 2.8%. These genotypes are highly prevalent in all regions of the world and account for approximately 70% of all cancers of the cervix (2, 13, 25). HPV types 52 and 58 are rare in Western countries; however, they are prevalent in Asian populations, especially in China (5, 14, 15, 28, 29).
Technologies available for HPV genotyping vary by method and platform and may offer type-specific characterization for a multitude of different high- and low-risk HPV types. For example, The Qiagen Hybrid Capture 2 HPV DNA (HC2) test and the Roche Amplicor HPV test (Amplicor) detect the DNA of 13 high-risk HPV types. The Linear Array HPV genotyping test allows for the discrimination of 37 HPV genotypes. The PapilloCheck HPV screening test (Greiner Bio-One GmbH, Frickenhausen, Germany) is a PCR-based DNA microarray system for the detection and identification of 24 HPV genotypes (4, 7, 13, 25). However, these methods might not be suitable in primary clinical settings in developing countries or for field use, because of the sophisticated instrumentation required, elaborate and complicated assay procedures, and expensive reagents. There is therefore a growing demand for simple and economical molecular tests. Loop-mediated isothermal amplification (LAMP) is a nucleic acid amplification method developed by Notomi et al. (18) and has emerged as a powerful gene amplification tool due to its simplicity, speed, specificity, and cost effectiveness. The use of this technique with hydroxynaphthol blue (HNB) dye was first developed in our laboratory and is being used increasingly for rapid and visual detection and typing of emerging viruses (1, 16, 27).
In this study, a simple and visual type-specific LAMP assay for the detection of high-risk HPV genotypes 16, 18, 45, 52, and 58 is described, in which the reaction was carried out in a single tube by mixing primers and DNA polymerase together with the tested samples at 63°C for 65 min. The LAMP assay was further evaluated with HPV DNAs from 294 clinical cervical scrape samples, and the results demonstrate that this assay is sensitive and specific. One of the most attractive features of this LAMP assay is that the results can be observed and determined by HNB dye-mediated visualization using the naked eye and without opening the tubes after amplification.
MATERIALS AND METHODS
Clinical samples.
A panel of 294 cervical scrape samples, including samples positive for HPV6 (n = 11), HPV11 (n = 9), HPV16 (n = 41), HPV18 (n = 12), HPV31 (n = 9), HPV33 (n = 10), HPV39 (n = 7), HPV45 (n = 9), HPV51 (n = 3), HPV52 (n = 37), HPV56 (n = 3), HPV58 (n = 30), HPV59 (n = 3), and HPV66 (n = 2) and HPV-negative samples (n = 108), were collected from Beijing University First Hospital between January 2009 and July 2011 and were tested by a novel one-step typing method with multiplex PCR (17) with the published set of specific primers according to the protocols as described in the paper.
Preparation of DNA samples.
DNA was extracted from the specimen using a QIAamp DNA minikit (Qiagen, Chatsworth, CA) according to the manufacturer's instructions. After extraction, DNA was eluted in 100 μl distilled water and was stored at −80°C.
Preparations of type-specific HPV DNA templates for control.
To determine the specificity of the type-specific LAMP method, 12 samples with single infections of HPV types 16, 18, 31, 33, 39, 45, 51, 52, 56, 58, 59, and 66, tested previously by a novel one-step typing method with multiplex PCR (17), were prepared and stored at −80°C.
HPV sequence alignment and genotype-specific primer design.
The E6, E7, and L1 gene sequences of HPV genotypes 16, 18, 45, 52, and 58 were obtained from GenBank database and the Los Alamos National Laboratory HPV Database (http://hpv-web.lanl.gov/) and were aligned by using ClustalX software. HPV genotype-specific primers were designed with a software program for LAMP primer design on the basis of the multiple-sequence alignments (http://primerexplorer.jp/e/). A BLAST search of the GenBank nucleotide database was also performed for the primers' sequences in order to verify genotype specificity. Each type-specific primer set, comprising two outer primers (F3 and B3), two inner primers (FIP and BIP), and two loop primers (LF and LB), was tested by the type-specific HPV DNA control template, and the best set for each genotype was selected. High-pressure liquid chromatography (HPLC)-purified oligonucleotide primer sets of each genotype were then synthesized by Sangon Biotech (Shanghai) Co., Ltd. The names and sequences of each primer set are shown in Table 1.
Table 1.
Name and sequence of each primer
| Name | Sequence (5′–3′) |
|---|---|
| HPV16E7F3 | AGACAACTGATCTCTACTGTT |
| HPV16E7B3 | CTTCCAAAGTACGAATGTCTAC |
| HPV16E7FIP | TTCTGCTTGTCCAGCTGGACGCAATTAAATGACAGCTCAGAG |
| HPV16E7BIP | CCGGACAGAGCCCATTACAATGTGTGTGCTTTGTACGCA |
| HPV16E7LF | CATCTATTTCATCCTCCTC |
| HPV16E7LB | TGCAAGTGTGACTCTACGCT |
| HPV18L1F3 | CGCGTCCTTTATCACAGG |
| HPV18L1B3 | TGGAATCCCCATAAGGATC |
| HPV18L1FIP | GGCACCATATCCAGTATCTACCATAATTGCCCCCCTTTAGAACT |
| HPV18L1BIP | TGCAAGATACTAAATGTGAGGTACCGCAGACATTTGTAAATAATCAGGAT |
| HPV18L1LF | TCACCATCTTCCAAAACTG |
| HPV18L1LB | ATTGGATATTTGTCAGTCT |
| HPV45L1F3 | ACTAAGTTTAAGCACTATAGTAGAC |
| HPV45L1B3 | CCTTTTGACAGGTAACAGC |
| HPV45L1FIP | ATGACATAACCTCTGCAGTTAAAGTTGTGGAGGAATATGATTTACAGTT |
| HPV45L1BIP | AATTGGAATTTTGGTGTCCCTCCACTGATTGCACAAAACGATA |
| HPV45L1LF | AGTGCACAACTGAAAA |
| HPV45L1LB | ACCACCTACTACAAGTTTAGTGGA |
| HPV52E6F3 | TTGAGGATCCAGCAACAC |
| HPV52E6B3 | GCGTAGGCACATAATACACA |
| HPV52E6FIP | GCACACACTGCAGCCTTATTTCTTTTGACCCTGCACGAATTGTG |
| HPV52E6BIP | AAGAGCTACAACGAAGAGAGGTCGCCATATGGATTATTGTCTC |
| HPV52E6LF | CACCGATTCTTCCAGCACC |
| HPV52E6LB | GTTTCTATTTACAGATTTACG |
| HPV58L1F3 | ACAGGGAATGCTTATCTATGG |
| HPV58L1B3 | TGTACCAAAGTCCATGCA |
| HPV58L1FIP | GCATTATTGTTACAGGCAACACCTTGTTTAATTGGCTGTAAACCTC |
| HPV58L1BIP | GCTGCTACTGATTGTCCTCCATTCCAAACCCTGTATCTACCAT |
| HPV58L1LF | ACCCCAATGCTCACCAGTG |
| HPV58L1LB | TTCTATTATTGAGGATGG |
HPV type-specific LAMP.
LAMP was performed in a one-step reaction in a 25-μl mixture containing 2.5 μl Bst DNA polymerase buffer (10×), 2.5 μl deoxynucleoside triphosphates (dNTPs) (10 mM; New England BioLabs, Ipswich, MA), 1 μl Bst DNA polymerase (8 U/μl; New England BioLabs, Ipswich, MA), 1 μl betaine (250 mM), 1 μl MgSO4 (150 mM), 1 μl HNB (3 mM; Lemongreen, Shanghai, China),1 μl of each primer (F3 and B3, 5 pmol/μl; BIP and FIP, 40 pmol/μl; and LF and LB, 20 pmol/μl), and 2 μl DNA. The mixtures were incubated at 63°C for 65 min. A Loopamp real-time LA-320 turbidimeter (Eiken Chemical Co., Ltd., Tokyo, Japan) was used to monitor the accumulation of magnesium pyrophosphate spectrophotometrically at 650 nm. The cutoff value for positive samples was determined when the turbidity increased above the threshold value, which was fixed at 0.2 over time. At the same time, a positive amplification was also indicated by a color change from violet to sky blue.
Specificity and sensitivity of type-specific LAMP assays.
To determine the specificities of the type-specific LAMP assays, 12 types of HPV DNA template controls (HPV types 16, 18, 31, 33, 39, 45, 51, 52, 56, 58, 59, and 66) were prepared, and HPV type-specific LAMP was performed as described above. In addition, 5 types of cloned HPV DNAs (HPV types 16, 18, 45, 52, and 58) were integrated into a pMD18-T plasmid, serial dilutions of the pMD18-T plasmid to cover the range of 107 to 100 copies/tube were prepared, and LAMP was performed as described above to determine the detection limit of each HPV type-specific LAMP.
RESULTS
Specificity of HPV type-specific LAMP.
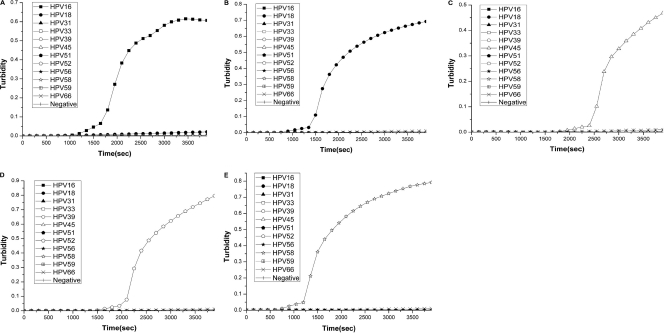
The specificity of type-specific primers for HPV types 16, 18, 45, 52, and 58 was evaluated. HPV type-specific LAMP was performed with each primer on template control DNA of 12 different HPV types (HPV types 16, 18, 31, 33, 39, 45, 51, 52, 56, 58, 59, and 66). HPV type-specific LAMP primers amplified only their respective types of HPV DNA; no LAMP products were detected in reactions carried out with other types of HPV DNAs by real-time turbidimeter (Fig. 1) or colorimetric determination (data not shown).
Fig. 1.
To determine the LAMP method's specificity, DNAs extracted from samples positive for HPV16, HPV18, HPV31, HPV33, HPV39, HPV45, HPV51, HPV52, HPV56, HPV58, HPV59, and HPV66 and from HPV-negative samples were amplified using HPV16 (A), HPV18 (B), HPV45 (C), HPV52 (D), and HPV58 (E) type-specific LAMP. The detection of LAMP products was assessed by turbidity assay using a Loopamp LA-320.
Sensitivity of HPV type-specific LAMP.
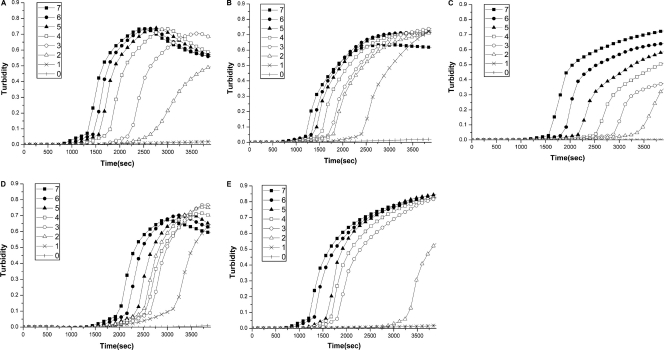
The sensitivity of the type-specific LAMP method for HPV genotypes 16, 18, 45, 52, and 58 was determined. Serial dilutions of the pMD18-T plasmid to cover the range of 107 to 100 copies/tube were used to determine the detection limits of HPV type-specific LAMP. The sensitivity of type-specific LAMP for HPV genotype 16, 18, 45, 52, and 58 was 100, 10, 100, 10, and 100 copies/tube, respectively, by real-time turbidimeter (Fig. 2) and colorimetric determination (Fig. 3A). The reaction was repeated three times for each template concentration, and the error bar was obtained by adding two standard deviations to the means at an optical density of 650 nm (OD650) (Fig. 3B).
Fig. 2.
To determine the sensitivity of the type-specific LAMP assay, serial dilutions of a cloned pMD18-T plasmid containing HPV16, HPV18, HPV45, HPV52, or HPV58 DNA were amplified by HPV16 (A), HPV18 (B), HPV45 (C), HPV52 (D), or HPV58 (E) type-specific LAMP, respectively. The detection of LAMP products was assessed by turbidity assay using a Loopamp LA-320. The numbers in the keys are the exponents of the log dilution from 107 to 100.
Fig. 3.
Sensitivity analysis of colorimetric type-specific LAMP assay using serial dilutions of a cloned pMD18-T plasmid containing HPV16, HPV18, HPV45, HPV52, or HPV58 DNA, respectively. (A) Tubes contained HPV16 (a), HPV18 (b), HPV45 (c), HPV52 (d), and HPV58 (e). Dilutions were 107 copies (tube 1), 106 copies (tube 2), 105 copies (tube 3), 104 copies (tube 4), 103 copies (tube 5), 102 copies (tube 6), 101 copies (tube 7), and 100 copies (tube 8). (B) The average result for each concentration of HPV DNA was determined from three independent colorimetric tests and is represented as the means ± 2 times the standard deviation. The absorbance (OD650) cutoff value (0.3) is calculated by adding two standard deviations to the mean of the negative reaction. When the absorbance (OD650) value is over 0.3, it is defined as a positive reaction.
Evaluation of HPV type-specific LAMP with clinical samples.
The HPV type-specific LAMP assay was further evaluated with a panel of 294 clinical cervical scrape samples, including samples positive for HPV6 (n = 11), HPV11 (n = 9), HPV16 (n = 41), HPV18 (n = 12), HPV31 (n = 9), HPV33 (n = 10), HPV39 (n = 7), HPV45 (n = 9), HPV51 (n = 3), HPV52 (n = 37), HPV56 (n = 3), HPV58 (n = 30), HPV59 (n = 3), and HPV66 (n = 2) and HPV-negative samples (n = 108), collected from Beijing University First Hospital between January 2009 and July 2011 and tested previously by a novel one-step typing method with multiplex PCR (17) using the published set of specific primers according to the protocols as described in the paper. The average reaction time of the LAMP assay was 30 min. The results of HPV type-specific LAMP were compared with those of HPV type-specific multiplex PCR, as shown in Table 2. The results demonstrated that the sensitivities of HPV type-specific LAMP for HPV types 16, 18, 45, 52, and 58 were 100%, 100%, 100%, 100%, and 100%, respectively, and the specificities were 100%, 98.5%, 100%, 98.8%, and 99.2%, respectively, compared with the results of a novel one-step typing method with multiplex PCR (17). No cross-reactivity with other HPV genotypes was observed.
Table 2.
Comparison of detection of HPV from clinical specimens by type-specific LAMP assay and a novel one-step typing method with multiplex PCR
| Result | No. of samples with result |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HPV16 |
HPV18 |
HPV45 |
HPV52 |
HPV58 |
||||||
| Multiplex PCRa | LAMP | Multiplex PCR | LAMP | Multiplex PCR | LAMP | Multiplex PCR | LAMP | Multiplex PCR | LAMP | |
| Positive | 41 | 41 | 12 | 16 | 9 | 9 | 37 | 40 | 30 | 32 |
| Negative | 253 | 253 | 282 | 278 | 285 | 285 | 257 | 254 | 264 | 262 |
| Total | 294 | 294 | 294 | 294 | 294 | 294 | 294 | 294 | 294 | 294 |
Specimens were tested previously by a novel one-step typing method with multiplex PCR (17) with the published set of specific primers according to the protocols as described in the paper.
DISCUSSION
High-risk types of human papillomavirus (HPV) are the causative agents of cervical cancer (3, 9, 24), and cervical screening could be improved by testing for the DNA of high-risk types of HPV as a primary screening tool (9, 24). Several longitudinal studies have shown that being positive for the DNA of high-risk types of HPV is a predictor of cervical dysplasia in women without cytological abnormality (9).
HPV types 16 and 18 are the most prevalent genotypes worldwide (6, 24, 25). HPV45 is highly prevalent in all regions of the world and frequently identified in women with high-grade cervical intraepithelial neoplasia (CIN 2+) and invasive cervical carcinoma (ICC), which account for 2 to 8% of cervical cancers (6, 9, 25). HPV genotypes 52 and 58 are prevalent in Asian populations, especially in China (5, 14, 15, 28, 29).
LAMP was first described in 2000 (18) and proved to be rapid, simple, and highly sensitive and specific, suggesting that it might be used for rapid diagnosis. LAMP is being used increasingly for rapid detection and typing of emerging viruses (10, 12, 20, 21). The use of reverse transcription (RT)-LAMP with HNB dye for visual detection of pandemic influenza A H1N1 virus 2009 was developed recently in our laboratory (16).
In this study, LAMP with HNB dye was shown to be a sensitive and easy assay for the detection of high-risk HPV types 16, 18, 45, 52, and 58. The sensitivity of amplification of LAMP for the detection of HPV genotypes 16, 18, 45, 52, and 58 was 100, 10, 100, 10, and 100 copies/tube, respectively, by two independent detection methods, real-time turbidimeter and HNB dye-based colorimetric determination. The detection limit for HPV type-specific LAMP was comparable to that of real-time PCR (13, 23). In addition, HPV type-specific LAMP amplified only the respective type of HPV DNA, with no cross-reactivity. The HNB dye-based assay has the same sensitivity as the turbidity assay. The cost of each type-specific LAMP is less than $1 per test, versus $8 per test using the multiplex PCR assay, coupled with the cost of the QIAxcel automated electrophoresis instrument (about $35,000). The most attractive feature of the HNB dye-based LAMP assay is the visual observation of the reaction results. Thus, no electrophoresis instrument or PCR machine is needed in practical application, and only an ordinary water bath is required to perform the LAMP reaction. The simplicity and cost effectiveness of the HNB dye-based assay make it more appropriate for rapid monitoring in the clinical setting, especially in resource-limited hospitals or rural clinics. The HNB dye-based assay has a remarkable advantage compared with other color-based assays (8, 11, 19) in that the HNB is mixed in prior to amplification. A need to open the assayed samples to add the dye is thereby omitted, thus reducing the risk of cross-contamination during amplification and detection of amplified DNA.
The reliability of clinical performance by HPV type-specific LAMP was evaluated using a panel of 294 clinical specimens whose HPV status was previously determined by a novel one-step typing method with multiplex PCR (17). The tested panel comprised 108 HPV DNA-negative samples and 186 HPV DNA-positive samples consisting of 41 HPV16-positive samples, 12 HPV18-positive samples, 9 HPV45-positive samples, 37 HPV52-positive samples, 30 HPV58-positive samples, and 57 positive samples containing 9 other HPV genotypes (HPV6, HPV11, HPV31, HPV33, HPV39, HPV51, HPV56, HPV59, and HPV66). Overall, the results were almost consistent with those from multiplex PCR (17). The average reaction time of the HPV type-specific LAMP assay was 30 min, suggesting that most of the positive samples contained high copy numbers of viral DNA. HPV DNA was not detected by multiplex PCR (17) in samples that were negative by each of the type-specific LAMP tests. Four of 282 HPV18-negative samples, 3 of 257 HPV52-negative samples, and 2 of 264 HPV58-negative samples by multiplex PCR (17) were positive by the respective type-specific LAMP. Samples with discrepant results in the two tests were retested by E6 nested multiplex PCR with the published set of common and specific primers according to the protocols as described previously (22) and later by sequencing of nested PCR products, which confirmed the accuracy of the results of type-specific LAMP. Therefore, the sensitivity of type-specific LAMP for HPV18, HPV52, and HPV58 was greater than that of multiplex PCR (17). As shown in Table 2, the results showed sensitivities of HPV type-specific LAMP for HPV types 16, 18, 45, 52, and 58 of 100%, 100%, 100%, 100%, and 100%, respectively, and specificities of 100%, 98.5%, 100%, 98.8%, and 99.2%, respectively, compared with the results of the novel one-step typing method with multiplex PCR (17), demonstrating the high sensitivity and specificity of HPV type-specific LAMP in the analysis of clinical samples.
There was a linear correlation between the genome quantity and reaction time to reach the threshold by the LAMP method, making quantitative HPV type-specific LAMP detection of HPV DNAs in clinical samples possible. The determination of viral load can be accomplished either by turbidity measurement using a Loopamp LA-320 turbidimeter or by measurement of the OD650 using a spectrophotometer (HNB dye-mediated color change from violet to sky blue), provided a concentration-dependent curve of LAMP assay results for each HPV genotype is normalized.
In conclusion, this qualitative colorimetric LAMP assay with HNB has potential usefulness for the rapid screening of HPV type 16, 18, 45, 52, and 58 infections, especially in resource-limited hospitals or rural clinics of provincial and municipal regions in China.
ACKNOWLEDGMENTS
We are grateful to Eiken Chemical Co., Ltd., Tokyo, Japan, and Beijing University First Hospital for providing the Loopamp turbidimeter and cervical scrape samples, respectively.
This work was supported by the China Mega-Project for Infectious Disease (grants 2009ZX10004-101 and -202 and 2008ZX10004-001, -002, and -004) and a research grant from the State Key Laboratory for Genetic Engineering and Molecular Virology.
The opinions expressed by the authors contributing to this article do not necessarily reflect the opinions of the Chinese Center for Disease Control and Prevention or the institutions with which the authors are affiliated.
Footnotes
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Cardoso T. C., et al. 2010. Visual detection of turkey coronavirus RNA in tissues and feces by reverse-transcription loop-mediated isothermal amplification (RT-LAMP) with hydroxynaphthol blue dye. Mol. Cell. Probes 24:415–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Castle P. E., et al. 2008. A comparison of two PCR-based human papillomavirus genotyping methods. J. Clin. Microbiol. 46:3437–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cuzick J., et al. 2010. Performance of the Abbott RealTime high-risk HPV test in women with abnormal cervical cytology smears. J. Med. Virol. 82:1186–1191 [DOI] [PubMed] [Google Scholar]
- 4. Dalstein V., et al. 2009. Analytical evaluation of the PapilloCheck test, a new commercial DNA chip for detection and genotyping of human papillomavirus. J. Virol. Methods 156:77–83 [DOI] [PubMed] [Google Scholar]
- 5. Ding T., et al. 2010. Distribution of human papillomavirus 58 and 52 E6/E7 variants in cervical neoplasia in Chinese women. Gynecol. Oncol. 119:436–443 [DOI] [PubMed] [Google Scholar]
- 6. Dobec M., Bannwart F., Kaeppeli F., Cassinotti P. 2009. Automation of the linear array HPV genotyping test and its application for routine typing of human papillomaviruses in cervical specimens of women without cytological abnormalities in Switzerland. J. Clin. Virol. 45:23–27 [DOI] [PubMed] [Google Scholar]
- 7. Dockter J., et al. 2009. Clinical performance of the APTIMA HPV assay for the detection of high-risk HPV and high-grade cervical lesions. J. Clin. Virol. 45:55–61 [DOI] [PubMed] [Google Scholar]
- 8. Goto M., Honda E., Ogura A., Nomoto A., Hanaki K. 2009. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46:167–172 [DOI] [PubMed] [Google Scholar]
- 9. Halfon P., et al. 2010. Comparison of the clinical performance of carcinogenic HPV typing of the Linear Array and Papillocheck HPV-screening assay. J. Clin. Virol. 47:38–42 [DOI] [PubMed] [Google Scholar]
- 10. Haridas D. V., Pillai D., Manojkumar B., Nair C. M., Sherief P. M. 2010. Optimisation of reverse transcriptase loop-mediated isothermal amplification assay for rapid detection of Macrobrachium rosenbergii noda virus and extra small virus in Macrobrachium rosenbergii. J. Virol. Methods 167:61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hill J., et al. 2008. Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J. Clin. Microbiol. 46:2800–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuan C. P., Wu M. T., Lu Y. L., Huang H. C. 2010. Rapid detection of squash leaf curl virus by loop-mediated isothermal amplification. J. Virol. Methods 169:61–65 [DOI] [PubMed] [Google Scholar]
- 13. Leo E., Venturoli S., Cricca M., Musiani M., Zerbini M. 2009. High-throughput two-step LNA real time PCR assay for the quantitative detection and genotyping of HPV prognostic-risk groups. J. Clin. Virol. 45:304–310 [DOI] [PubMed] [Google Scholar]
- 14. Li C., et al. 2010. A population-based study on the risks of cervical lesion and human papillomavirus infection among women in Beijing, People's Republic of China. Cancer Epidemiol Biomarkers Prev. 19:2655–2664 [DOI] [PubMed] [Google Scholar]
- 15. Lin C. Y., et al. 2007. Quality assurance of genotyping array for detection and typing of human papillomavirus. J. Virol. Methods 140:1–9 [DOI] [PubMed] [Google Scholar]
- 16. Ma X. J., et al. 2010. Visual detection of pandemic influenza A H1N1 virus 2009 by reverse-transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. J. Virol. Methods 167:214–217 [DOI] [PubMed] [Google Scholar]
- 17. Nishiwaki M., et al. 2008. Genotyping of human papillomaviruses by a novel one-step typing method with multiplex PCR and clinical applications. J. Clin. Microbiol. 46:1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Notomi T., et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parida M., et al. 2005. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 43:2895–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parida M. M., et al. 2007. Rapid and real-time detection of chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J. Clin. Microbiol. 45:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saitou Y., et al. 2010. A method for simultaneous detection and identification of Brazilian dog- and vampire bat-related rabies virus by reverse transcription loop-mediated isothermal amplification assay. J. Virol. Methods 168:13–17 [DOI] [PubMed] [Google Scholar]
- 22. Sotlar K., et al. 2004. Detection and typing of human papillomavirus by E6 nested multiplex PCR. J. Clin. Microbiol. 42:3176–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takacs T., et al. 2008. Molecular beacon-based real-time PCR method for detection of 15 high-risk and 5 low-risk HPV types. J. Virol. Methods 149:153–162 [DOI] [PubMed] [Google Scholar]
- 24. Tang N., et al. 2009. High-risk HPV detection and concurrent HPV 16 and 18 typing with Abbott RealTime high risk HPV test. J. Clin. Virol. 45:25–28 [DOI] [PubMed] [Google Scholar]
- 25. Thai H., et al. 2009. An HPV 16, 18, and 45 genotyping test based on Hybrid Capture technology. J. Clin. Virol. 45:93–97 [DOI] [PubMed] [Google Scholar]
- 26. Travasso C., Anand M., Samarth M., Deshpande A., Kumar-Sinha C. 2008. Human papillomavirus genotyping by multiplex pyrosequencing in cervical cancer patients from India. J. Biosci. 33:73–80 [DOI] [PubMed] [Google Scholar]
- 27. Wastling S. L., Picozzi K., Kakembo A. S., Welburn S. C. 2010. LAMP for human African trypanosomiasis: a comparative study of detection formats. PLoS Negl. Trop. Dis. 4:e865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu D., Cai L., Huang M., Zheng Y., Yu J. 2010. Prevalence of genital human papillomavirus infection and genotypes among women from Fujian province, PR China. Eur. J. Obstet. Gynecol. Reprod. Biol. 151:86–90 [DOI] [PubMed] [Google Scholar]
- 29. Ye J., et al. 2010. Prevalence and risk profile of cervical human papillomavirus infection in Zhejiang Province, southeast China: a population-based study. Virol. J. 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]