Abstract
We developed a real-time PCR to quantify 16S rRNA gene levels in plasma from HIV-infected patients as a marker of microbial translocation. The assay uses shrimp nuclease (SNuc) to eliminate DNA contamination, giving high sensitivity and low variability. The 16S rRNA gene levels measured in plasma from HIV patients correlated significantly with lipopolysaccharide levels.
TEXT
Microbial translocation is one of the factors driving chronic immune activation during HIV infection (2). A number of biomarkers, including lipopolysaccharides (LPS), soluble CD14 (sCD14), and bacterial DNA, have been used to quantify microbial translocation (reviewed in reference 15). The LPS limulus amoebocyte lysate (LAL) assay (11) is the current benchmark and exhibits high sensitivity, but it is limited by reagent contamination and the activity of plasma inhibitors (interassay variability, approximately 25%) (18). Also, LPS is present only in Gram-negative bacteria; thus, the LAL assay does not detect potential Gram-positive bacteria. The more reproducible sCD14 enzyme-linked immunosorbent assay (ELISA) (20) is an unreliable surrogate marker for microbial translocation, because increases are also triggered by proinflammatory cytokines such as alpha interferon (IFN-α) and IFN-β (3, 7). An alternative PCR method is the quantification of bacterial DNA encoding the ribosomal 16S rRNA gene, which is a gene with well-conserved regions unique to and shared by most bacteria (9).
Most conventional 16S rRNA gene PCR assays have been designed for phenotyping bacterial strains by the use of a longer amplicon that includes two or more hypervariable regions (12, 14). In contrast, we developed a new and highly sensitive (199-bp fragment) real-time PCR method, with a focus on accurate quantification, using an amplicon spanning only one hypervariable (V5) region (19). We did not test whether this smaller amplicon has sufficient diversity for phenotyping. Primers, TaqMan probe sequences, master mix (MM) composition, and cycle conditions are given in Table 1. An Escherichia coli 16S rRNA gene amplicon cloned into pCR II TOPO (pCRII-16S) was used as a plasmid standard.
Table 1.
Technical components of 16S rRNA gene PCR: primers, probe, master mix and reaction cyclesa
| Master mix (MM) component and source or targetb | Vol (μl) |
|---|---|
| Brilliant II Fast QPCR mix (Roche) | 12.5 |
| 16S F [20 μM] (Integrated Sciences) | 0.375 |
| 16S R [20 μM] (Integrated Sciences) | 0.375 |
| 16S probe [20 μM] (Integrated Sciences) | 0.25 |
| DEPC-treated water (MP Biomedicals) | 6.0 |
| SNuc (Thermo Scientific and USB/ | |
| Millenium Science) (0.4 U/μl) | 0.5 |
| Total | 20 |
| Target DNA | 5.0 |
Cycling conditions for SNuc treatment of the master mix, 37°C for 10 min followed by 72°C for 20 min. Cycling conditions for PCR amplification after addition of target DNA, 95°C for 2 min followed by 40 cycles of 95°C for 2 min, 95°C for 5 s, and 60°C for 30 s. All runs were performed on a Stratagene MX3005P Real-time PCR machine.
The primers and the TaqMan probe and corresponding positions (numbering from E. coli [GenBank accession no. J01859.1]) (6) were as follows: for 16S F, 5′-AACAGGATTAGATACCCTGGTAG-3′ (nucleotide [nt] 780 to 802); for 16S R, 5′-GGTTCTKCGCGTTGCWTC-3′, where W = A and T and K = G and T (nt 962 to 979); and for the 16S probe, 5′-FAM-AAC7AC5TGCTCCACCGCT-BHQ1-3′ (nt 948 to 937). A, locked nucleic acid modification 5 (LNA mod 5); C, LNA mod 7; FAM, 6-carboxyfluorescein; BHQ1, Black Hole Quencher I. Target DNA, DNA extracted from 200 μl of EDTA-plasma or 0.1× TE buffer or pCRII-16S plasmid standards at the indicated concentrations. DEPC, diethyl pyrocarbonate; SNuc, shrimp nuclease.
The quantification of 16S rRNA gene levels by real-time PCR is very problematic, as any trace contamination of the PCR mixture with, e.g., exogenous DNA in bacterially expressed Taq polymerase (1) gives rise to false-positive signals. Contamination can be managed using UV irradiation or restriction digestion of the MM, but this often results in a significant loss of PCR sensitivity (8). Alternatively, a background threshold value based on negative controls can be used (10).
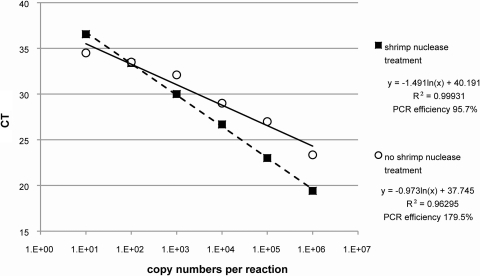
Our approach utilizes shrimp nuclease (SNuc) that has been expressed and purified from Pichia pastoris (yeast). SNuc is a selective endonuclease for double-stranded DNA that leaves primers, probes, and RNA intact. In the absence of SNuc, we detected the 16S rRNA gene in the no-template control (NTC); the detection limit of the pCRII-16S plasmids was 103 copies/reaction (Fig. 1). SNuc treatment of the MM efficiently removed the double-stranded DNA and lowered the limit of detection to 10 pCRII-16S plasmid copies, allowing consistent discrimination between true “negative” samples and low levels of the 16S rRNA gene in patient samples.
Fig. 1.
Comparison of results of the 16S rRNA gene assay performed with and without shrimp nuclease treatment. Standard curves were generated by amplification of pCRII-16S plasmid standards with and without shrimp nuclease treatment. CT, threshold cycle.
No contamination was introduced during DNA mock-extraction (DNeasy blood and tissue kit; Qiagen) performed using 0.1× Tris-EDTA (TE) buffer, which was subsequently used as the NTC. Levels of DNA from healthy control plasma ranged from 18.3 to 63.5 ± 10 copies/reaction. The origin of the bacterial DNA remains to be established, but its presence in blood has been reported before (16, 17).
Serial dilutions (10-fold) of pCRII-16S in four independent experiments demonstrated an overall detection limit of 10 copies/reaction, a correlation coefficient (r2) of ≥0.98, a mean slope of −3.39, and a PCR efficiency of 97.2% (Table 2). Samples for simulation of the low, middle, and upper ends of the quantitative range were prepared by spiking either 2 × 102, 2 × 103, or 2 × 105 pCRII-16S plasmid copies/reaction into 5 μl of plasma DNA from one healthy donor. Inter- and intra-assay precision values represented ≤1 threshold cycle (CT), and corresponding intra- and inter-assay coefficient of variation (CV) values were ≤3.2% for the low, middle, and upper ends of the quantitative range (Table 2). The assay precision for serial dilutions of pCRII-16S alone was also highly reproducible, with a CV of ≤3.3% for all dilutions (Table 2). These values are markedly lower than those determined for the real-time PCR assay by Jiang et al. (13) (5% for the intra-assay variation and 10% for the interassay variation) and the LAL assay (25% CV for the intra-assay and <5% CV for the interassay), demonstrating that our 16S real-time PCR assay is both reliable and highly sensitive.
Table 2.
Inter- and intra-assay precision of the 16S real-time PCR using shrimp nuclease pretreatment
| PCR target and category | Precision (CT ± SD)a |
Coefficient of variation (% CV)b |
||
|---|---|---|---|---|
| Interassay | Intra-assay | Interassay | Intra-assay | |
| DNA purified from HIV-negative plasma spiked with pCRII-16S rRNA gene plasmid standardsc | ||||
| Low (2 × 102 copies) | 31.3 ± 0.9 | 31.5 ± 1.0 | 2.78 | 3.15 |
| Med (2 × 103 copies) | 28.5 ± 0.4 | 28.5 ± 0.5 | 1.48 | 1.77 |
| High (2 × 105 copies) | 21.7 ± 0.5 | 21.8 ± 0.6 | 2.46 | 2.74 |
| Plasmid standards (no. of copies)d | ||||
| 101 | 35.4 ± 0.9 | 35.0 ± 1.1 | 3.26 | 2.48 |
| 102 | 32.8 ± 0.3 | 32.7 ± 0.4 | 1.25 | 0.83 |
| 103 | 29.3 ± 0.1 | 29.3 ± 0.1 | 0.26 | 0.24 |
| 104 | 25.8 ± 0.2 | 25.8 ± 0.2 | 0.75 | 0.83 |
| 105 | 22.2 ± 0.3 | 22.2 ± 0.2 | 0.95 | 1.15 |
| 106 | 18.7 ± 0.1 | 18.7 ± 0.1 | 0.33 | 0.04 |
Interassay and intra-assay precision values (representing the standard deviation of the CT values obtained from replicate experiments) were calculated from three independent real-time PCR runs performed on 3 different days for triplicate measurements.
The coefficient of variation (CV) values represent the ratio of the standard deviation of the distribution to its arithmetic mean.
DNA from HIV-negative plasma was spiked with 2 × 102, 2 × 103, and 2 × 105 copies of the pCRII-16S rRNA gene plasmid standards, simulating the lower (Low), middle (Med), and upper (High) ends of the quantitative range, respectively.
Copy numbers are given per reaction. The detection limit of all runs was 10 copies per reaction. The mean PCR efficiency was 97.24%, and the mean correlation coefficient (r2) was ≥0.98.
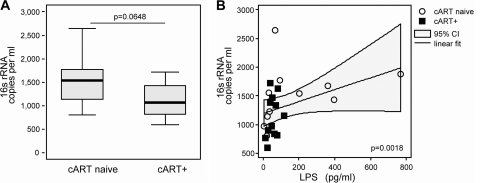
To compare measurements of 16S rRNA gene levels with LPS levels in human plasma, we recruited 23 HIV-positive patients who were receiving effective combination antiretroviral therapy (cART+) (median HIV RNA < 50 [range, <50 to 1,700] copies/ml; median CD4+ count = 558 [range, 209 to 1,199] cells/ml [n = 12]) or were therapy naive (cART−) (median HIV RNA = 21,500 [range, 3,700 to 100,000] copies/ml; median CD4+ count = 442 [range, 368 to 815] cells/ml [n = 11]) (HiACT cohort [G. F. Lichtfuss, W. Cheng, Y. Farsakoglu, G. Paukovics, R. Rajasuriar, P. Velayudham, A. C. Hearps, P. U. Cameron, S. R. Lewin, S. M. Crowe, and A. Jaworowski, submitted for publication]). Plasma LPS levels were measured using the LAL assay (Lonza, Walkersville, MD) (20). 16S rRNA gene levels were higher in cART− than in cART+ patients, with medians of 1.5 × 103 and 1.0 × 103 copies/ml plasma, respectively, although those results were not statistically significant (P = 0.0648) (Fig. 2A). This trend is in agreement with previous studies (13). We were unable to confirm the previously reported correlation with CD4+ counts (2), probably due to similar ranges of CD4+ counts in our groups (P = 0.62). LPS plasma levels in cART− and cART+ patients were similar, with median values of 66.96 pg/ml (range, 4.3 to 765.9 pg/ml) and 50.5 pg/ml (range, 14.1 to 119.2 pg/ml), respectively (P = 0.538). LPS levels significantly correlated with 16S rRNA gene copy numbers (Spearman's rho = 0.6156 [P = 0.0018]) (Fig. 2B). The correlation of LPS levels and 16S rRNA gene levels was also confirmed in a larger study (4).
Fig. 2.
Measurement of levels of bacterial products in HIV-infected patients. (A) 16S rRNA gene copies per milliliter plasma from HIV-infected cART-naive patients (left) and from HIV-infected patients receiving cART (right) (Wilcoxon rank-sum test [P = 0.0648]. The centered lines indicate the medians, the box corners the upper and lower quartiles, and the whiskers the minimums and maximums. ART, antiretroviral therapy. (B) Correlation between copy numbers of 16S rRNA gene and LPS levels in plasma from HIV-infected patients who were treatment naive or were receiving cART (Spearman's rho = 0.6156 [P = 0.0018]).
To establish the 16S rRNA gene as a biomarker for microbial translocation during HIV infection, careful analysis of potential confounders such as age, comorbidities, and lifestyle is required.
In conclusion, we have developed a sensitive, specific, and quantitative real-time PCR assay for the determination of 16S rRNA gene levels in human plasma by improving specificity through pretreatment with SNuc. The strong correlation between 16S rRNA gene levels and LPS levels in plasma from HIV-infected patients proves that the 16S rRNA gene assay would be a valuable adjunct to the quantification of LPS. This assay would be useful for studies of microbial translocation during HIV infection but could also be applied to other diseases, such as inflammatory bowel disease (5), in which pathogenesis has been associated with microbial translocation.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia (grants 400302 and 510488) and by Career Development Award 359233 to R.J.C. S.R.L. is an NHMRC practitioner fellow and receives funding from the Alfred Foundation.
Footnotes
Published ahead of print on 3 August 2011.
REFERENCES
- 1. Böttger E. C. 1990. Frequent contamination of Taq polymerase with DNA. Clin. Chem. 36:1258–1259 [PubMed] [Google Scholar]
- 2. Brenchley J. M., et al. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12:1365–1371 [DOI] [PubMed] [Google Scholar]
- 3. Brettschneider J., et al. 2002. The macrophage activity marker sCD14 is increased in patients with multiple sclerosis and upregulated by interferon beta-1b. J. Neuroimmunol. 133:193–197 [DOI] [PubMed] [Google Scholar]
- 4. Byakwaga H., et al. Intensification of antiretroviral therapy with raltegravir or addition of hyperimmune bovine colostrum in HIV-infected patients with suboptimal CD4+ T-cell response: a randomized controlled trial. J. Infect. Dis., in press [DOI] [PubMed] [Google Scholar]
- 5. Caradonna L., et al. 2000. Enteric bacteria, lipopolysaccharides and related cytokines in inflammatory bowel disease: biological and clinical significance. J. Endotoxin Res. 6:205–214 [PubMed] [Google Scholar]
- 6. Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. 1979. The complete nucleotide sequence of the ribosomal 16-S RNA from Excherichia coli. Experimental details and cistron heterogeneities. Eur. J. Biochem. 100:399–410 [DOI] [PubMed] [Google Scholar]
- 7. Carotenuto P., et al. 2005. Antiviral treatment with alpha interferon up-regulates CD14 on liver macrophages and its soluble form in patients with chronic hepatitis B. Antimicrob. Agents Chemother. 49:590–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corless C. E., et al. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drancourt M., et al. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferri E., et al. 2010. Plasma levels of bacterial DNA in HIV infection: the limits of quantitative polymerase chain reaction. J. Infect. Dis. 202:176–177 [DOI] [PubMed] [Google Scholar]
- 11. Hurley J. C. 1995. Endotoxemia: methods of detection and clinical correlates. Clin. Microbiol. Rev. 8:268–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janda J. M., Abbott S. L. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang W., et al. 2009. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J. Infect. Dis. 199:1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kroes I., Lepp P. W., Relman D. A. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. U. S. A. 96:14547–14552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lichtfuss G. F., et al. 2011. Biomarkers of immune dysfunction following combination antiretroviral therapy for HIV infection. Biomark. Med. 5:171–186 [DOI] [PubMed] [Google Scholar]
- 16. Moriyama K., et al. 2008. Polymerase chain reaction detection of bacterial 16S rRNA gene in human blood. Microbiol. Immunol. 52:375–382 [DOI] [PubMed] [Google Scholar]
- 17. Nikkari S., McLaughlin I. J., Bi W., Dodge D. E., Relman D. A. 2001. Does blood of healthy subjects contain bacterial ribosomal DNA? J. Clin. Microbiol. 39:1956–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nixon D. E., Landay A. L. 2010. Biomarkers of immune dysfunction in HIV. Curr. Opin. HIV AIDS 5:498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qin J., et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajasuriar R., et al. 2010. Biological determinants of immune reconstitution in HIV-infected patients receiving antiretroviral therapy: the role of interleukin 7 and interleukin 7 receptor α and microbial translocation. J. Infect. Dis. 202:1254–1264 [DOI] [PubMed] [Google Scholar]