Abstract
Haemophilus influenzae type b (Hib) is one of the leading causes of meningitis in developing countries. To establish and evaluate a novel loop-mediated isothermal amplification (LAMP) assay for Hib, we designed a LAMP primer set targeting the Hib-specific capsulation locus. LAMP detected 10 copies of purified DNA in a 60-min reaction. This indicated that the detection limit of LAMP was >100-fold lower than the detection limits of both a PCR for the detection of bexA and a nested PCR for Hib (Hib PCR). No H. influenzae, other than Hib or control bacteria, was detected. Linear determination ranged from 10 to 1,000,000 microorganisms per reaction mixture using real-time turbidimetry. We evaluated the Hib LAMP assay using a set of 52 randomly selected cerebrospinal fluid (CSF) specimens obtained from children with suspected meningitis. For comparison, the CSF specimens were tested using a conventional Hib PCR assay. Hib was detected in 30 samples using LAMP and in 22 samples using the Hib PCR assay. The Hib PCR showed a clinical sensitivity of 73.3% and a clinical specificity of 100% relative to the Hib LAMP assay. These results suggest that further development and evaluation of the Hib LAMP will enhance the global diagnostic capability for Hib detection.
INTRODUCTION
Haemophilus influenzae type b (Hib) causes meningitis, epiglottitis, bacteremia, and pneumonia predominantly in infants and young children (18). Over the past 2 decades, the introduction of Hib conjugate vaccines into routine immunization schedules has dramatically reduced the incidence of Hib-associated disease in many countries (2). Despite the introduction of these vaccines, surveillance for invasive Hib disease continues to be an important tool for monitoring vaccine impact and the potential reemergence of invasive Hib disease (7, 19, 23). To assess Hib carriage, laboratory facilities for the reliable cultivation of Hib and identification of the capsular polysaccharide by immunological techniques are necessary. Such facilities are found in well-equipped clinical microbiology laboratories, but serotyping of the capsular antigen may produce inconsistent results (11). In developing countries, accurate diagnosis of Hib remains a challenge due to the limited availability of routine microbiology laboratory services and injudicious use of antimicrobial agents.
Molecular assays are inherently valuable due to their enhanced analytical and clinical sensitivity and specificity and because the likelihood of detection is not diminished with nonviable organisms (4, 10, 11, 15). The development of a PCR for detection of bexA (bexA PCR) (3) and a nested PCR for Hib (Hib PCR) (5) has been an important milestone in the evolution of the laboratory diagnosis of Hib. Unfortunately, PCR-based assays are relatively expensive and complex to perform in resource-limited laboratory settings that are common in developing countries. The challenges for detection and diagnosis of Hib mirror those found in the detection of other invasive bacterial pathogens such as Streptococcus pneumoniae and Neisseria meningitidis.
In 2000, a novel nucleic acid detection method called loop-mediated isothermal amplification (LAMP) was reported (16). This method utilizes a unique priming mechanism that yields specific DNA products in a shorter period of time than PCR. From the PubMed, more than 500 articles related the LAMP method, including evaluation for the detection of bacteria, viruses, and parasites (14), have now been published (6). The availability of the LAMP method offers the opportunity to develop a novel assay for detection of Hib that is more reliable and easier to perform than bacterial culture, antigen detection, and PCR-based assays. However, to date, no Hib-specific LAMP assay has been reported. In the present study, we have established a novel Hib LAMP assay and compared its performance to that of Hib PCR using cerebrospinal fluid (CSF) specimens collected from patients with suspected meningitis.
MATERIALS AND METHODS
Bacterial strains.
Overall, 46 H. influenzae strains (including serotypes a to f, nontypeable, and biotype aegyptius) plus 21 strains representing other Haemophilus species and non-Haemophilus genera were evaluated. Among the 21 other Haemophilus species were H. parainfluenzae (IID991), H. parahaemolyticus (GTC1529), and H. haemolyticus (HK45), and the non-Haemophilus genera were Streptococcus mitis (ATCC 9811), S. oralis (ATCC 10557), S. gordonii (ATCC 10558), S. mutans (XC47), S. sanguis (ATCC 10556), S. salivarius (HHT), S. pneumoniae (ATCC 6305, R6, GTC261, IID553, and IID554), Escherichia coli (DH5α), Actinobacillus actinomycetemcomitans (Y-4), Porphyromonas gingivalis (381 and ATCC 49417), Actinomyces naeslundii (WVU627), Prevotella intermedia (ATCC 25611), and P. nigrescens (ATCC 25261). For the present study, 8 standard and 18 reference H. influenzae strains (9 Hib, 9 other serotypes, and 8 nontypeable strains, including one biotype aegyptius) were evaluated (Table) 1. Eight standard H. influenzae strains were IID983 (serotype a), IID984 (serotype b), IID985 (serotype c), IID986 (serotype d), IID987 (serotype e), IID 988 (serotype f), IID989 (nontypeable), and IID993 (nontypeable, biotype aegyptius). The 18 reference strains included 1 serotype a (HK390), 8 serotype b (HK176, HK177, HK179, HK180, HK195, HK196, HK827, and HK838), 1 serotype c (HK635), 2 serotype f (HK638 and HK2109), and 6 nontypeable strains (HK856, HK2112, HK2115, HK2117, HK2119, and HK2121). Capsule production for the 18 reference strains was previously confirmed by agglutination using burro antisera against serotypes a to f, provided by Rachel Schneerson (National Institutes of Health, Bethesda, MD). In addition, 20 clinical H. influenzae strains from nasopharyngeal swab were evaluated (Table 1).
Table 1.
Characteristics of 46 H. influenzae strains
| H. influenzae (no. of strains)a | No. of strains | Origin of isolateb | Capsule type(s)c | Hib agglutination testd |
Hib LAMP testinge | |
|---|---|---|---|---|---|---|
| Slide | Latex | |||||
| Standard strains (8)* | 5 | — | a, c, d, e, f | – | – | – |
| 1 | — | b | + | + | + | |
| 2 | — | NT | – | – | – | |
| Reference strains (18)† | 2 | RT | a, f | – | – | – |
| 1 | CSF | b | – | + | + | |
| 7 | CSF | b | + | + | + | |
| 2 | CSF | c, f | – | – | – | |
| 5 | RT | NT | – | – | – | |
| 1 | RT | NT | + | – | – | |
| Clinical strains (20)‡ | 8 | N | b | + | + | + |
| 1 | N | b | – | + | + | |
| 11 | N | NT | – | – | – | |
*, Obtained from the Institute of Medical Science, University of Tokyo, Tokyo, Japan; †, provided by Mogens Kilian, Institute of Medical Microbiology and Immunology, Aarhus University, Aarhus, Denmark; ‡, obtained from the Department of Microbiology, Gifu University School of Medicine, Gifu, Japan.
RT, respiratory tract specimen; CSF, cerebrospinal fluid; N, nasopharyngeal swab.
Capsule types were confirmed by PCR (5). NT, nontypeable.
+, Positive; –, negative. Slide agglutination serotyping was performed with H. influenzae antisera specific for type b. Latex agglutination serotyping was performed for the detection of H. influenzae serotype b.
+, Amplification after a 35-min incubation; –, no amplification after a 60-min incubation.
Serotyping by agglutination test.
To confirm capsule production by H. influenzae serotype b, a Hib-specific antiserum slide agglutination test (Denkaseiken, Tokyo, Japan) and a Hib latex agglutination test were performed (Slidex Meningite kit 5; bioMérieux, Lyon, France). Although it is of practical value only when used on CSF according to the manufacturer's instructions, we were able to preliminarily confirm the accuracy of detecting Hib polysaccharide even when used on suspension of bacterial cells (Table 1).
Preparation of chromosomal DNA.
Genomic DNA was purified from the 67 strains listed above using a QIAamp DNA minikit (Qiagen, Valencia, CA) according to the manufacturer's protocol. For the detection limit study, genomic DNA from Hib IID984 was obtained as described above, and the concentration was determined using an Ultrospec 3300 Pro spectrophotometer (Amersham Pharmacia Biotech, Cambridge, United Kingdom). The number of genome copies in the LAMP mixture was calculated based on a molecular size of 1.83 Mbp (H. influenzae RD KW20, GenBank accession no. NC000907). To ascertain the detection limit of the Hib LAMP assay, serial 10-fold dilutions of genomic DNA amplified, and the results were compared to those obtained using conventional PCR.
For the detection limit study, triplicate Hib LAMP testing was performed over a 3-day period using 10-fold dilutions of genomic DNA. Two technicians independently tested the same samples to confirm the reproducibility of LAMP results. The supernatant of a pooled Hib-negative CSF specimen (1) was used for a spiking assay, serial 10-fold dilutions of genomic Hib DNA were amplified, and the results were compared between Hib LAMP and conventional PCR.
Hib LAMP primer design.
Five Hib LAMP primers were designed based on published sequences of the Hib capsulation locus region II (GenBank accession no. X78559) using the LAMP primer support software program (Net Laboratory, Kanagawa, Japan) (22). The Hib LAMP primers included two outer primers (F3 and B3), a forward inner primer (FIP), a backward inner primer (BIP), and a loop primer forward (LF; Fig. 1).
Fig. 1.
(A) Nucleotide sequences of H. influenzae type b capsulation locus region II used to design the LAMP primer. The sequences used for LAMP primers are indicated by arrows. (B) Structure and sequence of the primers used in the LAMP reaction.
Hib LAMP reaction.
The reaction mixture (25 μl) contained 1.6 μM concentrations (each) of FIP and BIP, 0.2 μM concentrations (each) of F3 and B3, 0.4 μM LF, 8 U of Bst DNA polymerase large fragment (New England Biolabs, Ipswich, MA), 1.4 mM deoxynucleoside triphosphates, 0.8 M betaine (Sigma, St. Louis, MO), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, 0.1% Tween 20, and template DNA (2 μl). The mixture was incubated at 63°C for 35 or 60 min and then heated at 80°C for 2 min to terminate the reaction.
Analysis of Hib LAMP products.
The LAMP reaction results in the generation of turbidity proportional to the amount of amplified DNA (13). Amplified products were resolved by electrophoresis in 3% agarose gels, followed by visualization by ethidium bromide staining. For further confirmation, a Loopamp real-time turbidimeter (LA-200; Teramecs, Kyoto, Japan) monitored the amount of turbidity of the reaction tube in real-time by reading the optical density at 650 nm (OD650) every 6 s. The presence of a greater quantity of initial template DNA shortened the threshold time to detection of Hib. Using the application software for the turbidimeter, the amplification time required to exceed the turbidity level of 0.1 (Tt) was obtained according to the manufacturers' protocols (13).
To confirm the fidelity of amplification, a portion of the amplified products was digested with the restriction enzyme Hpy188I (New England Biolabs), and their sizes were analyzed by electrophoresis in 3% agarose gels, followed by ethidium bromide staining. To verify the structure of the amplified LAMP products, the amplified products were sequenced by using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI Prism 377 DNA sequencer (Applied Biosystems) in accordance with the manufacturer's instructions. The primers used to sequence the target region (between F1 and B1) were the primers F2 (5′-TTGCGTTTGTTGAATTCTGG-3′) and B2 (5′-CGAAGAATGAGAAGTTTTGTGG-3′) (Fig. 1).
Hib PCR assay.
For capsular typing of H. influenzae strains, we used Hib PCR amplification with previously published primers (5). The PCR mixture (10 μl) consisted of 0.2 mM concentrations of each deoxyribonucleoside triphosphate, 10 mM Tris-HCl buffer (pH 8.3), 50 mM KCl, 2 mM MgCl2, 1 U of ExTaq DNA polymerase (Takara Bio, Tokyo, Japan), 0.5 μM concentrations of each primer, and 1 μl of template DNA. Nested PCR was performed using a thermal cycler (MJ Research, Waltham, MA) for 25 cycles, each consisting of 1 min of denaturation at 94°C, 1 min of annealing at 60°C, and 1 min of extension at 72°C. After the final cycle, all reactions were incubated for an additional 10 min at 72°C. The products were visualized by resolution on a 3% agarose gel, followed by staining with ethidium bromide.
Clinical CSF specimens.
To conduct a pilot evaluation of the Hib LAMP, 52 CSF specimens (22 bexA PCR positive and 30 bexA PCR negative) were randomly selected from CSF collected during a 2-year prospective study of bacterial meningitis in Hanoi (1) for evaluation by Hib PCR (5) and Hib LAMP. In the present study, children with suspected meningitis less than 5 years of age were prospectively enrolled at study hospitals (1). Clinicians classified patients as suspected, probable, or confirmed bacterial meningitis based on clinical signs and symptoms, CSF parameters (e.g., white blood cell [WBC] count and glucose and protein levels) and bacterial testing (i.e., culture and latex agglutination testing).
CSF specimens were pretreated at 95°C for 2 min and centrifuged (13,000 × g, 5 min). The supernatant was saved for PCR and LAMP analysis. PCR primers (bexAF [5′-TAT CAC ACA AAT AGC GGT TGG-3′] and bexAR [5′-GGC CAA GAG ATA CTC ATA GAA CGT T-3′]) were designed to amplify a 181-bp bexA fragment and performed as described previously (3). In addition, a 5-μl aliquot from each CSF specimen was subjected to Hib PCR (25-μl reaction volume) and Hib LAMP as described above.
Statistical analysis.
The clinical sensitivity and specificity of conventional Hib PCR diagnosis were compared to those of Hib LAMP. The linear regression analysis was performed between a plot of the amplification time required to exceed the turbidity level of 0.1 (Tt) versus the log of the initial template DNA.
RESULTS
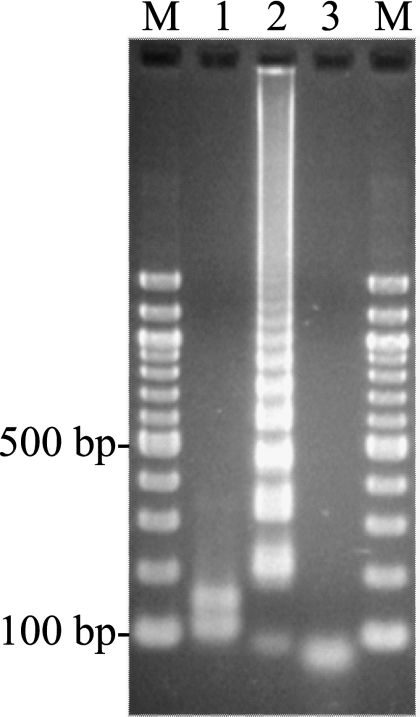
The LAMP assay performed at 63°C for 35 min successfully amplified the 247-bp target sequence of the Hib capsulation locus region II (Tables 1 and 2). The product was visible on an agarose gel (Fig. 2). The amplified product had a ladder-like pattern on the gel that is characteristic of the LAMP reaction and indicates the production of stem-loop DNAs with inverted repeats of the target sequence (16).
Table 2.
Detection limit of the Hib LAMP and Hib PCR assays for H. influenzae type b IID984
| Assaya | Detection with Hib genome copy numberb: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | 102 | 10 | 1 | 0 | |
| PCR | + | + | + | + | – | – | – | – |
| LAMP (35 min) | + | + | + | + | + | + | ± | – |
| LAMP (60 min) | + | + | + | + | + | + | ±* | – |
PCR results were obtained by electrophoretic analysis. LAMP results were determined visually.
+, Amplification occurred; –, amplification did not occur; ±, amplification occurred once within three trials; ±*, amplification occurred twice within three trials.
Fig. 2.
Electrophoretic analysis of the Hib LAMP-amplified products. Lane M, 100-bp ladder (New England Biolabs) used as a size marker; lane 1, LAMP product from lane 2 after digestion with Hpy188I (New England Biolabs) (the digested fragments were 111 and 152 bp); lane 2, 106 copies of the genomic DNA of H. influenzae type b IID984; lane 3, no template.
Analytical specificity of the Hib LAMP.
To evaluate the species-specificity of the LAMP primers, we tested 46 H. influenzae strains (Table 1) and 21 non-H. influenzae strains. For each assay mixture, a standard genomic DNA concentration (106 copies) was used for each strain. In the Hib LAMP reaction, amplification of Hib DNA was observed after 35 min (Table 1). In contrast, genomic DNA of the non-Hib strains was not amplified even after 60 min of incubation (Table 1). Neither nontypeable H. influenzae nor H. influenzae serotypes a, c, d, e, and f yielded a positive Hib LAMP reaction. In addition, 18 standard non-Haemophilus strains were negative by Hib LAMP.
Amplification specificity was confirmed by Hpy188I digestion to ensure that the product contained sequences corresponding to the selected target gene sequence of the Hib capsulation locus region II. The products of Hpy188I digestion were 111 and 152 bp in size, a finding consistent with the predicted sizes (Fig. 2). Amplified products were further analyzed by sequencing, and the sequences were compared to those of the targeted region (bases 5528 to 5559 in the original sequence) of the Hib capsulation locus region II (between F1 and B1; Fig. 1). The sequences obtained were identical to the expected nucleotide sequences (data not shown).
Detection limit of the Hib LAMP reaction.
The serial 10-fold dilutions of genomic Hib DNA were reliably amplified at 10 genome copies per reaction, whereas amplification occurred twice within three trials until one copy per reaction (Table 2). The bexA and Hib PCRs demonstrated detection limits of 2.5 × 103 (data not shown) and 103 (Table 2) genome copies, respectively. Thus, the detection limit of the Hib LAMP assay was over 100 times greater than that of bexA and Hib PCRs. No amplification was apparent in the Hib LAMP reaction when the sample tube lacked target DNA. These results were observed in triplicate during 3 days of experiments, and identical results were obtained within two persons. In addition, the detection limit of Hib LAMP assay using CSF spiking specimens was 10 genome copies, while the Hib PCR demonstrated a detection limit of 103 genome copies (data not shown).
Hib LAMP analysis of clinical isolates.
The results obtained by the Hib LAMP were concordant with Hib PCR and latex agglutination test results for among standard (n = 8), reference (n = 18), and clinical (n = 20) strains (Table 1). We confirmed the identities of 8 Hib and 10 other H. influenzae strains among the reference strains (Table 1). Similarly, the identities of 9 Hib and 11 other H. influenzae strains were confirmed among the group of clinical strains (Table 1). Serotyping by slide agglutination correctly identified 87.5% (7 of 8) of the reference Hib strains with 1 false-negative and 1 false-positive result observed among 18 reference strains (Table 1). One false-negative result was observed among the 20 clinical strains (Table 1).
Real-time turbidity measurement.
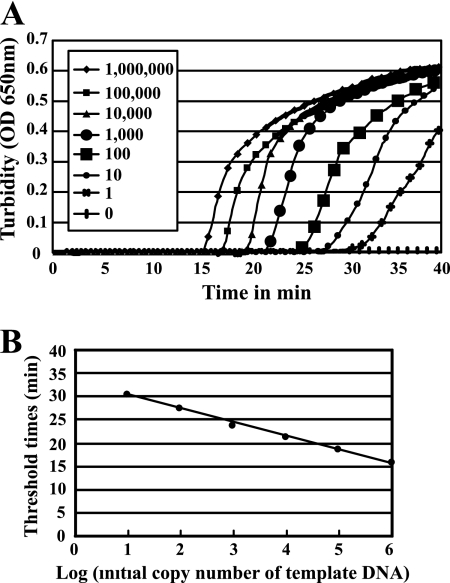
Real-time turbidity measurements of the Hib LAMP reaction solutions contained from one to 106 copies of Hib template DNA (Fig. 3) revealed that one copy of Hib DNA could be detected in 60 min (Fig. 3A). The detection limit of the Hib LAMP assay was identical among the Loopamp real-time turbidimeter, direct visual inspection, or gel electrophoresis findings (Table 2, Fig. 3A). The Tt versus the log of the initial template DNA (Fig. 3B) showed a linear relationship (correlation coefficient, r2 = 0.979). The linear determination range was from 10 to 1,000,000 microorganisms per reaction mixture.
Fig. 3.
(A) Detection limit and real-time reaction of the H. influenzae type b LAMP assay, as monitored by real-time measurements of turbidity. Shown from left to right are the curves of decreasing bacterial concentrations, from 1,000,000 to one genome copy. (B) The relationship between the threshold times (Tt) of each sample and the log of the amount of initial template DNA. The linear determination range was from 10 to 1,000,000 microorganisms per reaction mixture.
Hib LAMP and Hib PCR analysis of CSF specimens.
The Hib PCR was applied to 52 CSF specimens. Hib PCR results were identical with that of the conventional bexA PCR. Among 30 Hib LAMP-positive specimens, 22 (73.3%) were Hib PCR positive, while eight specimens were Hib PCR negative (Table 3). Twenty-two Hib LAMP-negative specimens (100%) were also determined to be negative using the Hib PCR assay (Table 3).
Table 3.
Comparison of Hib PCR and Hib LAMP results from testing 52 CSF specimens
| PCR result | No. of specimens |
||
|---|---|---|---|
| LAMP positive | LAMP negative | Total | |
| Positive | 22 | 0 | 22 |
| Negative | 8 | 22 | 30 |
| Total | 30 | 22 | 52 |
Twenty-one CSF specimens were H. influenzae culture positive. Among the 21 H. influenzae culture-positive specimens, 20 (95%) were both Hib LAMP and Hib PCR positive, while only one was both Hib LAMP and Hib PCR negative. Gram-negative rods consistent with Hib were identified on Gram stain in five CSF specimens and, of these, two CSF specimens were Hib culture, Hib PCR, and Hib LAMP positive, and three CSF specimens (one Klebsiella pneumoniae culture positive, one Escherichia coli culture positive, and one Pseudomonas aeruginosa culture positive) were Hib culture, Hib PCR, and Hib LAMP negative.
Among the 51 CSF specimens with WBC information, 35 (69%) showed WBC counts above the normal range. Among 29 Hib LAMP-positive specimens with WBC information, 26 (90%) had elevated WBC counts; in contrast, 21 (100%) Hib PCR-positive specimens with WBC information showed elevated WBC counts.
To ensure that the Hib LAMP products corresponded to the selected gene target, we confirmed their identities by sequencing and digestion with Hpy188I (New England Biolabs [data not shown]). Compared to Hib LAMP, the Hib PCR assay demonstrated a clinical sensitivity and a specificity of 73.3% (22/30) and 100% (22/22), respectively.
DISCUSSION
The Hib LAMP assay established in the present study accurately discriminated standard reference Hib strains from non-Hib strains of known identity. The Hib LAMP established here demonstrated an analytical specificity equivalent to Hib PCR. Notably, the Hib LAMP was found to have an detection limit more than 100 times lower than previously described bexA and Hib PCR methods (3, 5). The linear determination range of the Hib LAMP by real-time turbidimetry was from 10 to 106 microorganisms. Even though we used CSF spiked samples, a detection limit in the Hib LAMP reaction was still lower than that observed with Hib PCR method. The lower detection limit and higher reaction robustness (6) of the LAMP assay conceivably contributed to its higher detection rate (30 positive) than that of Hib PCR (22 positive) using 52 clinical CSF specimens from children with suspected meningitis.
LAMP assays for detection of meningitis bacteria in oral swab specimens have been established (20, 21). However, the efficacy for the diagnosis of bacterial meningitis has not been tested (9). In fact, to our knowledge, this is the first trial of LAMP detection of bacterial meningitis using clinical CSF specimens.
In the present study, the two false-negative results from slide agglutination serotyping tests may have been caused by low levels of capsule production. A recent study revealed that 68% of H. influenzae isolates identified as Hib by the serotyping did not contain the corresponding capsule type genes (11). That study also suggested that the most common and significant laboratory error was misidentification of nontypeable strains as serotype b. However, in our study, the Hib LAMP successfully distinguished Hib from other H. influenzae strains. The Hib LAMP assay used here was more analytically sensitive and specific than slide agglutination serotyping. The higher analytical specificity and lower detection limit of the Hib LAMP assay is in agreement with our previously reported findings for S. pneumoniae LAMP (20).
As the LAMP reaction progresses, the by-product pyrophosphate ions bind to magnesium ions and form a white precipitate of magnesium pyrophosphate. The resulting turbidity can be judged simply by the naked eye. This characteristic feature of the LAMP reaction can be used to detect the reaction endpoint, simply by gauging the presence of precipitate. Real-time turbidity measurements of the LAMP reaction using a simple and inexpensive apparatus (Loopamp real-time turbidimeter) (13) permit the quantitative analysis of minute amounts of nucleic acids with high precision and over a wide range. In the present study, we assessed the amount of Hib template DNA in real-time by reading the OD650 every 6 s in a Loopamp real-time turbidimeter. The curve of real-time turbidity measurements had high linearity (Fig. 3B) as in the case of real-time PCR (12). This observation indicates that the concentration of any template DNA can be determined by comparing the Tt value with the Tt values of template DNA samples of known concentrations. Thus, using a real-time turbidity monitoring system, the concentrations of Hib DNA in clinical samples can be quantified.
In the present study, 95% (n = 20) of the H. influenzae culture-positive specimens were both Hib PCR and Hib LAMP positive. More than 90% of the Hib PCR-positive (100% [21/21]) and Hib LAMP-positive (90% [26/29]) specimens were CSF with abnormally elevated WBC counts.
The Hib LAMP assay was clinically more sensitive than the previously described Hib PCR method (30 CSF were positive by Hib LAMP versus 22 positive by Hib PCR). The higher clinical sensitivity of the Hib LAMP found in the present study is consistent with results of a reverse transcription-LAMP assay developed for the detection of Japanese encephalitis virus in CSF (17). The discordant results in our study (eight CSF specimens were negative by Hib PCR and positive by Hib LAMP) are likely due to the lower detection limit of Hib genome achieved by the Hib LAMP in the tested CSF specimens. In addition, previous studies demonstrated that the LAMP reaction is more tolerant of the presence of potentially perturbing biological substances than PCR (8). The robust performance of the Hib LAMP assay (6) in the present study suggests that LAMP-based detection of Hib and other invasive bacterial pathogens is feasible in a wide variety of clinical settings. Also, compared to standard Hib culture plus antigen detection (two false-negative results by slide agglutination test versus no false-negative result by LAMP, Table 1) and PCR-based assays (detection limit, 103 genome copies by Hib PCR and 10 genome copies by Hib LAMP), the Hib LAMP is more analytically sensitive. In addition, compared to PCR, LAMP reactions are less susceptible to enzymatic inhibitors (8).
This feature makes the LAMP assay particularly suitable for resource-limited settings that may be found in developing country laboratories. Our laboratory's experience with the Hib LAMP, as well as our experience using LAMP for the detection of other pathogens such as S. pneumoniae, suggests that the cost of the Hib LAMP assay per specimen tested is remarkably lower than PCR and can be performed in a laboratory with limited technology. Further evaluation of the Hib LAMP in prospective studies is now under way to confirm the Hib LAMP test characteristics, including clinical sensitivity, clinical specificity, predictive values, and likelihood ratios compared to bacterial culture, antigen detection, and PCR.
In summary, we have successfully established a LAMP-based Hib DNA amplification method and confirmed that its analytical specificity is high and its detection limit is low. Compared to PCR-based detection methods, this assay allows the detection of Hib from both spiked CSF specimens and clinical CSF specimens with higher analytical and clinical sensitivity. Because the LAMP reaction is easy to set up and does not require special equipment, it has obvious advantages in clinical settings and in population-based studies with limited access to well-equipped laboratories.
ACKNOWLEDGMENTS
This investigation was supported by the Uemura Fund, Nihon University School of Dentistry, a Grant-in-Aid for Scientific Research (C) (no. 10201004), and a grant to promote multidisciplinary research projects from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. D.W.K. was supported by grant RTI05-01-01 from the Regional Technology Innovation Program of the Ministry of Knowledge and Economy of South Korea. This study was supported, in part, through funding from the governments of Kuwait, Sweden, and the Republic of Korea.
We are grateful to Yoshiaki Kawamura, Gifu University Graduate School of Medicine, for helpful advice. We are especially grateful to Morgens Kilian (Aarhus University), Yoshihisa Yamashita (Kyushu University), and Masao Maeno and Hirotaka Torigoe (Nihon University) for their support in this study.
Footnotes
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Anh D. D., et al. 2006. Haemophilus influenzae type b meningitis among children in Hanoi, Vietnam: epidemiologic patterns and estimates of H. influenzae type b disease burden. Am. J. Trop. Med. Hyg. 74:509–515 [PubMed] [Google Scholar]
- 2. Barbour M. L. 1996. Conjugate vaccines and the carriage of Haemophilus influenzae type b. Emerg. Infect. Dis. 2:176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corless C. E., et al. 2001. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39:1553–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falla T. J., et al. 1995. Characterization of capsular genes in Haemophilus influenzae isolates from H. influenzae type b vaccine recipients. J. Infect. Dis. 171:1075–1076 [DOI] [PubMed] [Google Scholar]
- 5. Falla T. J., et al. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Francois P., et al. 2011. Robustness of loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 62:41–48 [DOI] [PubMed] [Google Scholar]
- 7. Hviid A. 2006. Postlicensure epidemiology of childhood vaccination: the Danish experience. Expert Rev. Vaccines 5:641–649 [DOI] [PubMed] [Google Scholar]
- 8. Kaneko H., Kawana T., Fukushima E., Suzutani T. 2007. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70:499–501 [DOI] [PubMed] [Google Scholar]
- 9. Kim K. S. 2010. Acute bacterial meningitis in infants and children. Lancet Infect. Dis. 10:32–42 [DOI] [PubMed] [Google Scholar]
- 10. Kroll J. S., Ely S., Moxon E. R. 1991. Capsular typing of Haemophilus influenzae with a DNA probe. Mol. Cell Probes 5:375–379 [DOI] [PubMed] [Google Scholar]
- 11. LaClaire L. L., et al. 2003. Identification of Haemophilus influenzae serotypes by standard slide agglutination serotyping and PCR-based capsule typing. J. Clin. Microbiol. 41:393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marty A., et al. 2004. Detection of Haemophilus influenzae type b by real-time PCR. J. Clin. Microbiol. 42:3813–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mori Y., Kitao M., Tomita N., Notomi T. 2004. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 59:145–157 [DOI] [PubMed] [Google Scholar]
- 14. Mori Y., Notomi T. 2009. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muhlemann K., Balz M., Aebi S., Schopfer K. 1996. Molecular characteristics of Haemophilus influenzae causing invasive disease during the period of vaccination in Switzerland: analysis of strains isolated between 1986 and 1993. J. Clin. Microbiol. 34:560–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Notomi T., et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parida M. M., et al. 2006. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid and real-time detection of Japanese encephalitis virus. J. Clin. Microbiol. 44:4172–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peltola H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13:302–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prasad K., Karlupia N. 2007. Prevention of bacterial meningitis: an overview of Cochrane systematic reviews. Respir. Med. 101:2037–2043 [DOI] [PubMed] [Google Scholar]
- 20. Seki M., et al. 2005. Loop-mediated isothermal amplification method targeting the lytA gene for detection of Streptococcus pneumoniae. J. Clin. Microbiol. 43:1581–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torigoe H., Seki M., Yamashita Y., Sugaya A., Maeno M. 2007. Detection of Haemophilus influenzae by loop-mediated isothermal amplification (LAMP) of the outer membrane protein P6 gene. Jpn. J. Infect. Dis. 60:55–58 [PubMed] [Google Scholar]
- 22. Van Eldere J., et al. 1995. Region II of the Haemophilus influenzae type b capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol. Microbiol. 15:107–118 [DOI] [PubMed] [Google Scholar]
- 23. von Gottberg A., et al. 2006. Impact of conjugate Haemophilus influenzae type b (Hib) vaccine introduction in South Africa. Bull. World Health Organ. 84:811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]