Abstract
The single-nucleotide variation 823C to T (His275Tyr), responsible for oseltamivir drug resistance has been detected in some isolates of the influenza A/H1N1/2009 virus. Early detection of the presence of this oseltamivir-resistant strain allows prompt consideration of alternative treatment options. An isolated-probe–asymmetric amplification PCR (Roche LightCycler v2.0) and high-resolution melting (HRM) method using unlabeled probes and amplified products (Idaho LightScanner 32) was designed and optimized to detect and estimate the proportion of H275Y mutants in influenza A/H1N1/2009 virus samples. The lower limit of quantification within the linear range of PCR assay detection was 200 copies/reaction. The melting peaks of the H275Y-specific unlabeled probe for the wild-type A/H1N1/2009 and H275Y mutant viruses were clearly distinguishable at 65.5°C and 69.0°C, respectively, at various ratios of wild-type/mutant virus population standards. The 95% detection limit for the 10% mutant sample pool was 1,200 copies/reaction (95% confidence interval, 669.7 to 3,032.6 copies/reaction). This HRM assay was tested with 116 archived clinical specimens. The quantitative HRM results obtained with samples containing mixed mutant–wild-type virus populations, at threshold cycle (CT) values of <29, compared well to those obtained with a pyrosequencing method performed by an independent laboratory. The quantitative feature of this assay allows the proportions of mutant and wild-type viral populations to be determined, which may assist in the conventional clinical management of infected patients and potentially more preemptive clinical management. This validated quantitative HRM method, with its low running cost, is well positioned as a rapid, high-throughput screening tool for oseltamivir resistance mutations in influenza A/H1N1/2009 virus-infected patients, with the potential to be adapted to other influenza virus species.
INTRODUCTION
In recent years, increasing resistance to antiviral therapy among seasonal influenza virus strains has been documented, with the majority of the seasonal A/H1N1 and A/H3N2 influenza virus strains being resistant to oseltamivir (Tamiflu) and adamantanes, respectively (3). With the currently circulating pandemic A/H1N1/2009 strain, the oseltamivir drug resistance amino acid mutation His275Tyr or H275Y (resulting from a cytosine-to-thymidine nucleotide substitution at position 823), has been found mostly in patients with a history of oseltamivir treatment (1, 16, 23), though occasional sporadic transmission of this drug resistance virus has been reported in certain populations (17, 21). Early identification of an oseltamivir-resistant H275Y mutant strain allows the timely institution of alternative treatment of A/H1N1/2009-infected patients, especially among the immunocompromised, to improve clinical outcomes.
Several molecular methods are currently available for the detection of the H275Y drug resistance mutation in influenza A/H1N1/2009 virus. These include qualitative assays using restriction fragment length polymorphism (18) and qualitative and quantitative real-time reverse transcription (RT)-PCR assay methods (2, 5, 12, 19), some of which can also detect mixtures of wild-type and drug-resistant virus populations.
Two of these quantitative assays rely on minor differences in threshold cycle (CT) values among the different influenza virus strains, derived from a second derivative approach based on the amplification curves, to estimate the H275Y mutant population in a mixed sample (5, 19). This makes them susceptible to imprecision arising from reagent preparation and the matrix effects of different samples. The Centers for Disease Control and Prevention (CDC) has developed a pyrosequencing protocol which accurately quantifies the proportion of H275Y mutants in the virus population (9). Nonetheless, this method is too expensive for routine, large-scale use in most diagnostic laboratories.
This study describes an efficient and economical isolated-probe–asymmetric amplification PCR (IPAA-PCR) and high-resolution melting (HRM) approach using unlabeled probes and amplified products to detect and estimate the proportion of H275Y mutant influenza A/H1N1/2009 virus in mixed-virus-population samples.
MATERIALS AND METHODS
Clinical specimens.
A total of 116 archived influenza A/H1N1/2009 virus-positive clinical specimens collected from 74 intensive care unit (ICU) patients and received for routine influenza testing at the National University Hospital, Singapore, were selected for this study. These clinical specimens included nasal/nasopharyngeal and throat swabs suspended in universal transport medium, endotracheal tube aspirates, and sputum samples. During this recruitment period, only severely ill patients were given oseltamivir therapy and many of these patients were eventually moved to the ICU for further management. All of the samples had been stored in their native forms without additives at −80°C since January to August 2010, following the initial testing without any additional freeze-thawing. The protocols for RNA extraction and detection of influenza A/H1N1/2009 virus, using a validated in-house dual-gene real-time RT-PCR assay, have been previously described (15).
Primers and unlabeled LunaProbe design.
A pair of primers (AITBiotech, Singapore) flanking the H275Y single-nucleotide variant (SNV) with an amplicon size of 155 bp was selected from the most conserved region of the neuraminidase gene by using the influenza A/Bethesda/NIH106-D14/2009 virus neuraminidase gene (GenBank accession no. GU571155) as the reference. The unlabeled LunaProbe (Suprenom, Singapore) was designed to match the mutant sequence containing the SNV. The primer and probe sequences were as follows: forward 424F23, 5′-CAAGTGATGGACAGGCCTCATAC-3′; reverse 558R21, 5′-ATGCCAGTTATCCCTGCACAC-3′; unlabeled probe 498L30, 5′-GGAGCATTCCTCATAGTAATAATTAGGGGC(C3 Spacer)-3′.
RNA transcript control.
Wild-type and H275Y mutant amplicons were cloned into the pCR2.1-TOPO vector and in vitro transcribed into RNA transcripts using RiboMAX large-scale RNA production system T7 (Promega, Madison, WI) according to the manufacturer's instructions. Both transcripts were quantified using the NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE) at an absorption wavelength of 260 nm. The RNA transcripts were then diluted with pooled influenza virus-negative RNA extracts from clinical specimens as tested by the dual-gene real-time influenza A/H1N1/2009 virus RT-PCR assay (15), in order to prepare standards of the desired concentrations. All standards were aliquoted into individual vials and kept at −20°C for short-term storage (up to 2 weeks) and at −80°C for longer-term storage.
IPAA-PCR assay.
The IPAA-PCR assay was carried out with either the LightScanner 32 (Idaho Technology Inc., Salt Lake City, UT) or the LightCycler v2.0 system (Roche Molecular Diagnostics, Pleasanton, CA) using a sample-seeking temperature of 30°C and SuperScript III Platinum One-Step quantitative RT-PCR reagents (Invitrogen, Carlsbad, CA). The 8-μl reaction mixture used in the IPAA-PCR assay, prepared by adding 2 μl of RNA sample to a 6-μl reaction mixture volume consisting of 4 μl of 2× Reaction Mix, 0.2× LC Green Plus dye (Idaho Technology Inc., Salt Lake City, UT), 0.3 μmol/liter forward primer, 0.25 μmol/liter reverse primer, and 0.2 μl of SuperScript III RT/Platinum Taq Mix, was added to the capillary and centrifuged at 1,500 rpm for 15 s. A 2-μl volume of a forward primer-probe mixture consisting of each component at 10 μmol/liter was then deposited into the capillary and not centrifuged.
The IPAA-PCR was initiated with an RT step of 55°C for 8 min and a 2.5-min denaturation step of 95°C, followed by 50 amplification cycles consisting of 10 s at 95°C, 20 s at 60°C, and 25 s at 68°C. The ramping rate of the PCR machine was set at 20°C/s. Note that this amplification step can be performed using either the LightCycler or the LightScanner with this protocol and should give equivalent results.
Upon completion of the 50 amplification cycles, the capillary was immediately reverse centrifuged at 1,500 rpm for 15 s to mix the reaction mixture with the predeposited primer-probe mixture near the capillary cap. The mixture was then centrifuged, and the amplification was extended for an additional 15 cycles using the thermal cycling profile described above.
HRM.
After the additional PCR cycles, the capillaries were analyzed using the LightScanner 32 and premelting at 95°C for 15 s. They were then rapidly cooled to 40°C and held there for 30 s. Subsequently, HRM data were continuously acquired at a ramping rate of 0.1°C/s from 58°C to 75°C, with an initial holding temperature of 55°C.
To screen for the presence of mutant A/H1N1/2009 virus, a wild-type and a mutant RNA transcript control were included in each run. If a specimen is found to contain a mixture of mutant and wild-type strains, it is subjected to reanalysis using the protocol described above to estimate the proportion of the H275Y mutant population. This is achieved by the inclusion of a series of 0 to 100% mixed mutant–wild-type standards prepared from clinical influenza virus RNA extracts at 10% intervals. These standards were diluted with pooled negative RNA extracts to achieve a total RNA concentration of 104 copies/reaction.
Analysis of HRM data was performed using the unlabeled-probe genotyping mode by setting the normalized range at 61.75 to 73°C. The data from each melting analysis were studied using both −dF/dT versus temperature/melting peak plots and relative fluorescence signal changes versus temperature/melting curve plots. The mutant proportion of the population was estimated by the position of the melting peak in relation to the mixed mutant–wild-type standards. For example, if a peak fell between the 20% and 30% mixed mutant–wild-type standards, it would signify a 20 to 30% mutant population.
Performance evaluation.
The lower limit of detection (LLOD) and lower limit of quantification (LLOQ) of the mutant strain by the IPAA-PCR assay were determined by duplicate RNA transcripts of concentrations of 2 to 2 × 107 copies/reaction. The analytical sensitivity of the IPAA-PCR assay for a 10% mutant population was further tested with dilutions of 1 to 3.5 log RNA copies/reaction in 8 replicates by 3 different operators on 3 different days. The 95% detection limit was determined by polynomial probit analysis using SPSS version 17.0 (SPSS Inc., Chicago, IL).
The 116 clinical specimens tested were subjected to HRM analysis for the presence of the H275Y mutation. These specimens were also directly sequenced by the previously described Sanger method (22). Specimens containing mixed mutant–wild-type populations were reanalyzed to estimate the H275Y mutant percentage of the population using the quantitative HRM analysis and verified by a pyrosequencing method based on the CDC protocol in another laboratory (9).
RESULTS
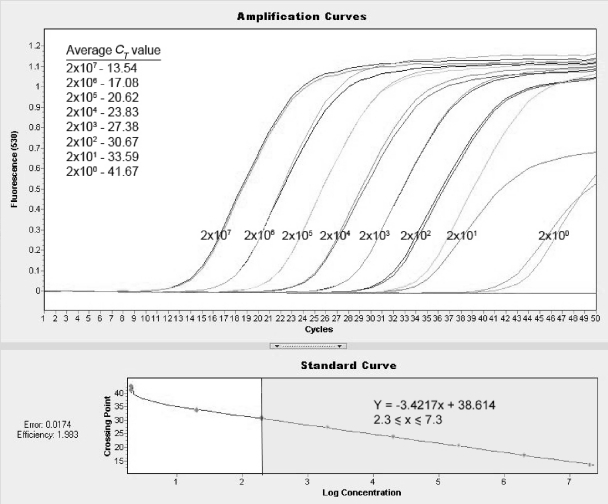
The IPAA-PCR assay was able to amplify all duplicate RNA transcript standards at 2 × 101 to 2 × 107 copies/reaction at a CT of <40 cycles, and the LLOD was thus determined to be 20 copies/reaction. The LLOQ, defined as the lowest concentration whereby a difference in the CT value of approximately 3.33/log is maintained, was 200 copies/reaction (Fig. 1). The 95% detection limit in the 10% mutant sample pool was 1,200 copies/reaction (95% confidence interval, 669.7 to 3,032.6 copies/reaction). The PCR efficiency calculated using the LC v2.0 software was 1.983.
Fig. 1.
Amplification curves and standard curve of duplicate mutant RNA transcript standards at 2 × 100 to 2 × 107 copies/reaction. The linear range was found to lie between 2 × 102 and 2 × 107 copies/reaction. The linear equation was derived from the standard curve at a value of 2.3 ≤ log concentration/x ≤ 7.3 (shaded), with a gradient of −3.4217. The PCR efficiency calculated using the LC v2.0 software was 1.983.
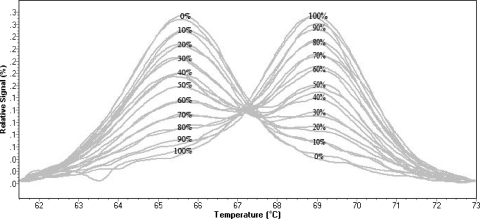
The melting peaks of the SNV unlabeled probe for the mismatched base C (wild type) and the perfectly matched base T (H275Y mutant) were clearly distinguishable at 65.5°C and 69.0°C, respectively, at all mixed mutant–wild-type standards (Fig. 2). However, the LightScanner analyzer automatically assigned a melting temperature only for a minimum mutant population percentage of 20%.
Fig. 2.
Normalized melting peaks, analyzed on the LightScanner 32, at 61.75 to 73°C of mutant–wild-type standards in duplicate from 0 to 100% mutants in 10% increments. Wild-type and mutant melting peaks appeared at 65.5°C and 69°C, respectively, for all mixed mutant–wild-type standards.
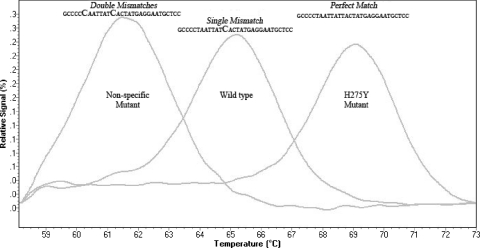
The HRM approach detected 1 nonspecific mutant sample, 3 H275Y mutant samples, 7 mixtures, 104 wild-type samples, and 1 sample that could not be amplified. The nonspecific mutant was later found to be a silent mutation located at nucleotide position 816 (T to C) by Sanger sequencing. The HRM profile of the silent mutation is shown in Fig. 3. The comparative results of the percentages of the H275Y mutant in mixed samples, determined by the HRM and pyrosequencing methods, are shown in Table 1. Here, the quantitative results of the HRM method were comparable to those of the pyrosequencing method for samples with CT values below 29. The Sanger sequencing method was the least sensitive.
Fig. 3.
Normalized melting peaks between 58.12°C and 73°C showing a nonspecific mutant (T816C), the wild type, and the H275Y mutant at 61.5°C, 65.25°C, and 69°C, respectively. The nonspecific mutant (two mismatches) is clearly differentiated from the wild type (single mismatch) and the H275Y mutant (perfect match).
Table 1.
Comparison of mutant–wild-type mixture detection and quantification using in-house Sanger sequencing, HRM, and pyrosequencing methods
| Sample no. | Sanger sequencing | HRM (% mutant [range]) | Pyrosequencing (% mutant) | CT value |
|---|---|---|---|---|
| 1 | Mixture | Mixture (10–20) | Mixture (21.2) | 26.2 |
| 2 | Mixture | Mixture (80–90) | Mixture (67.8) | 27.0 |
| 3 | Mixture | Mixture (60–70) | Mixture (68.9) | 25.9 |
| 4 | Mutant | Mixture (90–100) | Mixture (86.2) | 21.8 |
| 5 | Not amplified | Mixture (50–60) | Wild type | 34.6 |
| 6 | Mixture | Mixture (80–90) | Mixture (70.8) | 29.0 |
| 7 | Not amplified | Mixture (70–80) | Mixture (13.7) | 33.6 |
DISCUSSION
The HRM assay described above was developed with the aim of rapidly determining the presence and proportions of drug-resistant mutant and wild-type influenza A/H1N1/2009 viruses in clinical samples to aid patient management in a timely manner.
The use of unlabeled probe in this assay has several benefits. The smaller size of the probe, in this instance, 30 bp, limits the probability of detecting a nontarget mutation by allowing larger temperature differences to result from relatively few base mismatches within a shorter sequence. The large melting temperature difference of approximately 3.5°C between the wild type and the H275Y mutant, due to the use of Lunaprobe, buffers the minor variations between runs arising from reagent preparation and matrix effects from the samples. It also renders the assay more sensitive to nontarget mutations.
It has recently been reported that the presence of a synonymous change, Tyr274Tyr, at nucleotide position 822 (T to C) may negatively affect the analytical sensitivity of many current H275Y assays (24). These assays include real-time, reverse transcriptase PCR, and reverse transcriptase PCR–rolling-circle amplification methods that rely on perfect matching of the probe sequences to their target sequences for detection. Consequently, the synonymous change may render poor binding of the probes to their target, giving rise to false-negative results (24).
This assay is highly unlikely to be affected by such a nontarget change. In this assay, the unlabeled probe was designed to fully complement the H275Y mutant sequence. This design meant that the wild-type sequence would behave like a single base mismatch, lowering the melting temperature of the Lunaprobe. Any novel variation along the probe will act as an additional mismatch. When this occurs, the presence of a double mismatch will further widen the temperature differentiation from that of the wild type or the H275Y mutant, allowing confident identification of nontarget variants occurred. This was illustrated by the detection of the silent mutation at nucleotide position 816 (T to C) in our study. Similarly, this assay is expected to show a different melting temperature for a synonymous change compared to the wild-type and H275Y mutant sequences.
Although unlabeled-probe HRM is routinely used to detect small gene deletions and single-nucleotide polymorphisms and for genotyping in human genetic analysis (4, 13, 25–27), this approach is less commonly applied to virus gene analysis. A possible reason for this could be the challenge of balancing the inverse relationship between asymmetric ratios, a high value of which improves the unlabeled-probe signal, and low-level sensitivity, which is crucial for accurate detection of low-viral-load samples (7). Another major concern about unlabeled-probe HRM analysis is the appearance of aberrant melting peaks that complicate interpretation. These peaks are generated by the extension of the unlabeled probe during asymmetric amplification (7). To minimize these limitations, we isolated the excess forward primers and probes during the initial amplification by depositing them in the capillary without centrifuging them. Additionally, the 3′ end of the unlabeled probe was blocked using a C3 Spacer modification to prevent further enzymatic extension by the polymerase during the HRM analysis, as previously recommended (6). In this study, the additional 15 cycles of amplification were sufficient for asymmetric amplification by the excess forward primers.
The discrepancy between the HRM and pyrosequencing method results obtained with samples 5 and 7 was due to viral titers that fell below the LLOQ of the HRM assay. Furthermore, the low viral titer hinders the accurate sampling of representative mutant–wild-type mixtures from the clinical specimens due to stochastic effects; i.e., minority species may be identified with a disproportionate frequency, purely by chance. Two samples were not amplified by the Sanger sequencing method as a result of assay sensitivity limitation.
The reproducibility and accuracy of the relative quantification of this assay are highly dependent on the use of exact quantities of mutant–wild-type mixtures in the quantitative standards of every experiment. By using the LLOD for the 10% mutant population (1,200 copies/reaction), the minimum CT value required for HRM quantification, estimated from the standard curve of the IPAA-PCR assay, was 28. For routine use, plasmid DNA standards may be preferred to RNA standards, as they are more resistant to degradation. New standard controls were introduced in each individual experiment run in both qualitative and quantitative assay, to eliminate the matrix differences between the PCR master mixtures. All RNA dilutions for assay performance testing were done using negative RNA extracts from routine screening to simulate the matrix of the real RNA specimens. This is an important point, as we have found that using water as a diluent alters the melting temperature of the probe and template (A-T and A-C; data not shown). The gradient of the standard curve (−3.42 CT/log concentration) of the IPAA-PCR assay suggests the usability of this method for viral load quantification.
Although the oseltamivir resistance mutation in the influenza A/H1N1/2009 virus is currently relatively rare, this was also the case for the seasonal H1N1 and H3N2 viruses previously, which have since developed more widespread oseltamivir resistance (14, 20), and this possibility cannot be ruled out for A/H1N1/2009. Hence, it is prudent to develop rapid and accurate means of determining such drug resistance in the still relatively new pandemic A/H1N1/2009 virus. Also, the economical running cost of this HRM method makes it an ideal candidate for high-throughput oseltamivir resistance mutation screening. In addition, the use of a single-closed-tube format from RT to melting analysis saves manual handling and minimizes the amplicon contamination risk. This assay can be modified and adapted to other strains of influenza virus, including the prevalent H3N2 subtype. It also lends confidence in the application of this unlabeled-probe HRM technique in other virological assays.
It is likely that incomplete viral suppression with oseltamivir allows the development of drug resistance within these influenza A/H1N1/2009 virus populations (1, 16, 23), as has been seen in human infections with other influenza virus subtypes, notably avian H5N1 (8). Hence, close monitoring of oseltamivir resistance among all influenza virus subtypes, including the A/H1N1/2009 strain, is important to allow a timely switch to an more effective alternative agent, e.g., zanamivir (10, 11).
In conclusion, the results obtained with this HRM technique, as demonstrated in this study, are comparable to those obtained by pyrosequencing, and in the absence of this platform, this HRM assay offers an alternative, cost-effective, rapid screening method for close monitoring for the emergence of oseltamivir-resistant H275Y mutant A/H1N1/2009 influenza viruses in this postpandemic period.
ACKNOWLEDGMENTS
We thank Lily Chiu and Thomas Ho Kiat Tay of the Molecular Diagnosis Center of the National University Hospital for their kind assistance during the assay validation process.
Funding support for this study was provided by a National University Health System interdepartmental grant from the Departments of Medicine and Laboratory Medicine to J.W.-T.T., P.A.T., and E.S.-C.K.
Footnotes
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Baz M., et al. 2009. Emergence of oseltamivir-resistant pandemic H1N1 virus during prophylaxis. N. Engl. J. Med. 361:2296–2297 [DOI] [PubMed] [Google Scholar]
- 2. Bennett S., Gunson R. N., MacLean A., Miller R., Carman W. F. 2011. The validation of a real-time RT-PCR assay which detects influenza A and types simultaneously for influenza A H1N1 (2009) and oseltamivir-resistant (H275Y) influenza A H1N1 (2009). J. Virol. Methods 171:86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention 2008. Update: influenza activity—United States, September 28-November 29, 2008. MMWR Morb. Mortal. Wkly. Rep. 57:1329–1332 [PubMed] [Google Scholar]
- 4. Chen W. J., et al. 2009. Rapid diagnosis of spinal muscular atrophy using high-resolution melting analysis. BMC Med. Genet. 10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chidlow G. R., et al. 2010. The detection of oseltamivir-resistant pandemic influenza A/H1N1 2009 viruses using a real-time RT-PCR assay. J. Virol. Methods 169:47–51 [DOI] [PubMed] [Google Scholar]
- 6. Dames S., Margraf R. L., Pattison D. C., Wittwer C. T., Voelkerding K. V. 2007. Characterization of aberrant melting peaks in unlabeled probe assays. J. Mol. Diagn. 9:290–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dames S., Pattison D. C., Bromley L. K., Wittwer C. T., Voelkerding K. V. 2007. Unlabeled probes for the detection and typing of herpes simplex virus. Clin. Chem. 53:1847–1854 [DOI] [PubMed] [Google Scholar]
- 8. de Jong M. D., et al. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667–2672 [DOI] [PubMed] [Google Scholar]
- 9. Deyde V. M., et al. 2009. Detection of molecular markers of antiviral resistance in influenza A (H5N1) viruses using a pyrosequencing method. Antimicrob. Agents Chemother. 53:1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dulek D. E., et al. 2010. Use of intravenous zanamivir after development of oseltamivir resistance in a critically Ill immunosuppressed child infected with 2009 pandemic influenza A (H1N1) virus. Clin. Infect. Dis. 50:1493–1496 [DOI] [PubMed] [Google Scholar]
- 11. Gaur A. H., et al. 2010. Intravenous zanamivir for oseltamivir-resistant 2009 H1N1 influenza. N. Engl. J. Med. 362:88–89 [DOI] [PubMed] [Google Scholar]
- 12. Hindiyeh M., et al. 2010. Rapid detection of influenza A pandemic (H1N1) 2009 virus neuraminidase resistance mutation H275Y by real-time reverse transcriptase PCR. J. Clin. Microbiol. 48:1884–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kramer D., et al. 2009. A fast, sensitive and accurate high resolution melting (HRM) technology-based assay to screen for common K-ras mutations. Cell. Oncol. 31:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lackenby A., Thompson C. I., Democratis J. 2008. The potential impact of neuraminidase inhibitor resistant influenza. Curr. Opin. Infect. Dis. 21:626–638 [DOI] [PubMed] [Google Scholar]
- 15. Lee H. K., et al. 2010. Diagnostic testing for pandemic influenza in Singapore: a novel dual-gene quantitative real-time RT-PCR for the detection of influenza A/H1N1/2009. J. Mol. Diagn. 12:636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Memoli M. J., Hrabal R. J., Hassantoufighi A., Eichelberger M. C., Taubenberger J. K. 2010. Rapid selection of oseltamivir- and peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin. Infect. Dis. 50:1252–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moore C., et al. 2011. Evidence of person-to-person transmission of oseltamivir-resistant pandemic influenza A(H1N1) 2009 virus in a hematology unit. J. Infect. Dis. 203:18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nukiwa N., et al. 2010. Simplified screening method for detecting oseltamivir resistant pandemic influenza A (H1N1) 2009 virus by a RT-PCR/restriction fragment length polymorphism assay. J. Virol. Methods 170:165–168 [DOI] [PubMed] [Google Scholar]
- 19. Renaud C., Kuypers J., Corey L. 2010. Diagnostic accuracy of an allele-specific reverse transcriptase-PCR assay targeting the H275Y oseltamivir resistant mutation in 2009 pandemic influenza A/H1N1 virus. J. Clin. Virol. 49:21–25 [DOI] [PubMed] [Google Scholar]
- 20. Sheu T. G., et al. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 52:3284–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. South Carolina Department of Health and Environmental Control 10 July 2009, posting date Three reports of oseltamivir resistant novel influenza A (H1N1) viruses. South Carolina Department of Health and Environmental Control, Columbia, SC: http://www.scdhec.gov/health/disease/han/docs/HAN-20090710-01.pdf [Google Scholar]
- 22. Suhaila M., et al. Mixtures of oseltamivir-sensitive and resistant pandemic influenza A/H1N1/2009 viruses in immunocompromised hospitalized children. Pediatr. Infect. Dis. J. 30:625–627 [DOI] [PubMed] [Google Scholar]
- 23. Tramontana A. R., et al. 2010. Oseltamivir resistance in adult oncology and hematology patients infected with pandemic (H1N1) 2009 virus, Australia. Emerg. Infect. Dis. 16:1068–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trevino C., Bihon S., Pinsky B. A. 2011. A Synonymous change in the influenza A virus neuraminidase gene interferes with PCR-based subtyping and oseltamivir resistance mutation detection. J. Clin. Microbiol. 49:3101–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van der Stoep N., et al. 2009. Diagnostic guidelines for high-resolution melting curve (HRM) analysis: an interlaboratory validation of BRCA1 mutation scanning using the 96-well LightScanner. Hum. Mutat. 30:899–909 [DOI] [PubMed] [Google Scholar]
- 26. Zhou L., Myers A. N., Vandersteen J. G., Wang L., Wittwer C. T. 2004. Closed-tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye. Clin. Chem. 50:1328–1335 [DOI] [PubMed] [Google Scholar]
- 27. Zhou L., Wang L., Palais R., Pryor R., Wittwer C. T. 2005. High-resolution DNA melting analysis for simultaneous mutation scanning and genotyping in solution. Clin. Chem. 51:1770–1777 [DOI] [PubMed] [Google Scholar]