Abstract
The HIV-1 BED incidence assay was developed at the Centers for Disease Control and Prevention and since 2005 has been available as a commercial kit for use in HIV-1 incidence surveillance. A BED-specific proficiency testing (PT) program was initiated in 2006 that included a panel of eight coded specimens (six unique and two duplicates) to participating laboratories. The number of participating laboratories increased from 12 to 38 from 2006 to 2009. Overall, 96.1% of the laboratories reported results, and 95.4% of those reporting achieved a 100% score. The observed mean normalized optical density (OD-n) values of all participants correlated well with the expected OD-n values for all specimens (R2 = 0.98) used for seven PT rounds. BED testing demonstrated high reproducibility among all laboratories, with an agreement of 99.3% (574/578) between initial and confirmatory classification and regression statistics of R2 = 0.96, slope = 1.022, and intercept = 0.0066. Reproducibility among duplicate specimens was very high during each PT round, with mean deviation of 1.8%. Analysis of controls and calibrator specimen for all 343 runs showed a coefficient of variation of ca. 20% for raw ODs in the dynamic range, which was reduced to <10% when the OD was normalized (OD-n). Most laboratories that failed the PT assessment had transcriptional errors, kit reagent problems, or specimen handling errors. Thus, the BED-specific PT program enabled us to track performance of different laboratories conducting the BED assay while identifying areas for improvements. This program will also serve as a template for future PT programs for new incidence assays as they become available.
INTRODUCTION
The HIV-1 BED Incidence EIA (BED assay) is an IgG capture enzyme immunoassay to detect and distinguish recent (<6 months) from long-term HIV-1 infection. This assay was developed to measure HIV-1 incidence for population surveillance (7, 23) and has been applied in worldwide settings (8, 10, 11, 15, 16, 19, 26). Although there are several reports of in-house assays (1, 24, 30) or modified diagnostic assays (4, 13, 14, 17, 18, 25, 28) to detect recent infections, the BED assay is the only assay developed specifically to detect recent HIV-1 infections for incidence estimation and commercially available as a kit. The number of laboratories using the BED assay has increased steadily. As with all laboratory testing, a comprehensive quality assurance (QA) program is a vital component of incidence testing. Quality of testing can be assured by multiple steps implemented at different levels that include training, monitoring, and corrective actions as part of the continuous quality improvement. The BED assay includes four kit control specimens, including one calibrator (CAL) specimen (7). Control specimens validate the runs, while CAL is used to normalize the optical density values which minimize inter-run variations. Because of the quantitative nature of the BED assay, it is critically important that the testing is performed with highest quality. Proficiency testing (PT) programs have been shown to be very effective in improving the quality of laboratory testing, including HIV testing (2, 3, 5, 6, 9, 12). The goals of PT for HIV incidence testing are to assess the performance of participating laboratories and to identify issues for corrective action and technical assistance. The Centers for Disease Control and Prevention (CDC) initiated a voluntary and cost-free PT program in 2006 with 12 participating laboratories and has expanded to more laboratories over time. We report here the results of this PT program, which provides information about precision and reproducibility of the BED assay among participating laboratories over seven rounds.
MATERIALS AND METHODS
Specimens.
The specimens chosen for each PT round were from a collection of 25 undiluted HIV-1-positive bulk volume donor plasma samples purchased from SeraCare Life Sciences, Inc. (Milford, MA). The donor plasma were heat inactivated at 56°C for 30 min, fully characterized using the HIV-1 BED Incidence Assay, and then divided into aliquots and stored at −70°C. Twenty-five specimens covered a range of OD-n values from low to high and were classified as a recent (OD-n = <0.8) or a long-term (OD-n > 0.8) infection based on their BED results.
PT panels.
After preparation of PT panels for each round, the specimens were retested for verification in CDC laboratory before distribution. The number of specimens with recent or long-term classification varied with each PT round. Occasionally laboratories requested the panel as dried plasma spots (DPS) to match their ongoing testing with dried blood spot specimens. DPS panels were prepared simply by spotting 50 μl of plasma onto Whatman 903 filter paper and allowing it to dry completely before packaging them into plastic bags with desiccant (22).
For each round, the panel specimens were labeled with the appropriate anonymous identifiers (e.g., A1 to A8) and then collated into a set of eight specimens for each participant, including six unique and two duplicate specimens. The PT panel was distributed to the participating laboratories twice a year. The panels were packaged within a specimen box and shipped on dry ice to the participant, along with a cover letter and a Microsoft Excel data report form. The cover letter and data report form were also e-mailed to each participant with the expectation that results be returned in an e-mail attachment within the 2-month reporting deadline. The report form requested typical assay information, including test date, technician name, and kit lot and expiration date, as well as all quality control (QC) and specimen data with final interpretation.
For each round, the PT program coordinator collected information from each participant, including current contacts and shipping address, and compiled all of the reported data.
BED assay.
The BED assay was performed by the participating laboratory as described earlier (7, 23). Initial testing for all specimens was done in singlet, followed by confirmatory testing of specimens with an OD-n of <1.2, in triplicate. The controls were run on each plate in triplicate, and median values were used and reported.
Data analysis and corrective actions.
The reported data were summarized in a database that includes participant QC and specimen data. The reported results were compared to the panel's expected results for normalized OD (OD-n) and to the final classification as recent or long-term infection. Additional parameters measured included interlaboratory comparison, interassay reproducibility of specimens retested by confirmatory testing, and intra-assay reproducibility of duplicate specimens.
In order to pass a PT round, the participant must correctly classify all specimens as either a recent or a long-term infection, as classified by the CDC. In addition, the program coordinator ensured that the reported QC values were within the established ranges for both OD and OD-n, as published in the BED EIA HIV-1 Incidence Test kit inserts (Calypte Biomedical Corp., Portland, OR; Sedia Biosciences, Inc., Portland, OR). If an error was discovered, the participant was notified within a few days to file a corrective action form in order to determine the underlying cause. If requested, the coordinator assisted the participant with this nonpunitive discovery process, where the goal was to ensure the highest quality of laboratory performance with incidence testing.
Within 2 weeks of the reporting deadline, the program coordinator provided all participants with a summary report. The participants were assigned a lab code such that the results remained anonymous. The report summarized overall findings, allowing each participant to compare their results to the expected results.
For the purpose of this study, the data from all seven rounds were analyzed in aggregate. Mean OD-n values of participants were compared to the expected OD-n values for each specimen. Intralaboratory reproducibility was measured by agreement between initial and confirmatory testing results and by the consistency of results for coded, duplicate specimens.
RESULTS
Participation and responses.
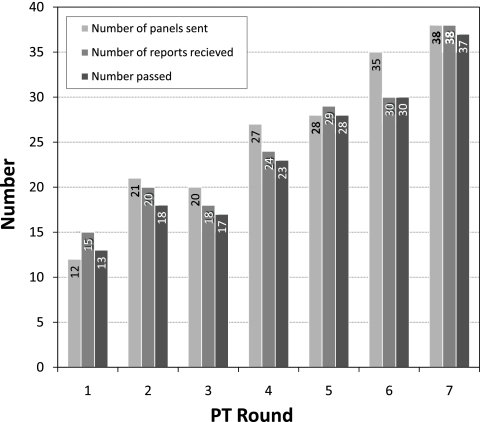
Participation in the PT program increased progressively from 12 laboratories in 2006 to 38 in 2009 (Fig. 1), reflecting increased use of the BED assay worldwide. The number of reports received varied from 85.5% (round 6) to 125% (round 1). Occasionally, the same panel was tested and reported by more than one operator from the same laboratory, resulting in more reports received than the number of panels sent (rounds 1 and 5). The number of laboratories that scored 100% (“pass”) ranged from 13 to 37, reflecting 86.7 to 100% of the reports with all correct results. Overall, 174 (96.1%) reports were received from all seven rounds of PT testing comprising 181 panels. Of those reporting, 166 (95.4%) received a 100% score. Overall, there were eight reports that did not pass with a <100% score.
Fig. 1.
Participation and performance of laboratories in the BED-specific PT program over seven rounds.
PT data analysis.
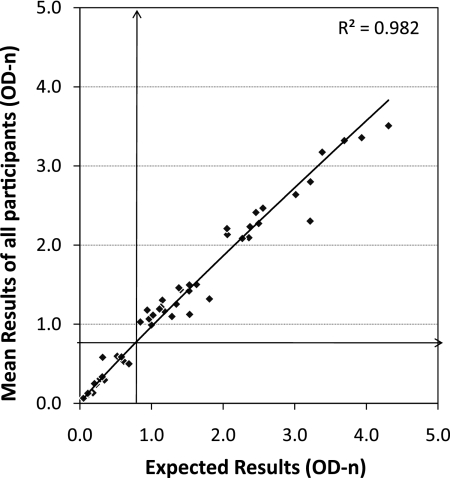
Overall, there were 56 specimens distributed over seven PT rounds (eight specimens per round). The mean OD-n result of a specimen, calculated from all submitted results for each round, was very close to the expected OD-n results (Fig. 2), with a regression coefficient of R2 = 0.97, demonstrating outstanding performance of the BED assay in multiple laboratories and over multiple kit lots. When we compared the mean OD-n values of participants with the expected OD-n values, as a group all of the specimens were correctly classified as either recent (lower left quadrant) or long-term (upper right), with no misclassification of any specimens (lower-right and upper-left areas).
Fig. 2.
Overall concordance of mean participant results with expected CDC results for 56 specimens examined over seven rounds. The regression line with the best fit is shown (R2 = 0.982). Vertical and horizontal arrows indicate 0.8 cutoffs.
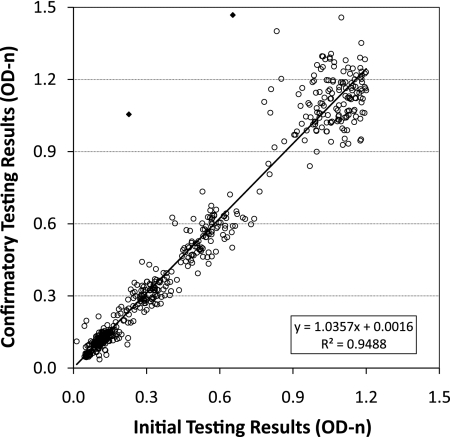
Intralaboratory reproducibility was examined by two different approaches. First, overall concordance between initial BED results and confirmatory BED results (for 579 results with an OD-n of <1.2, as recommended by the algorithm) was assessed by using an x-y scatter plot (Fig. 3). The agreement between initial and confirmatory testing was outstanding with an R2 = 0.95, a slope of 1.03, and an intercept close to zero, demonstrating that the assay is highly reproducible. Only 2 (0.3%) of 579 results showed significant deviation (closed diamonds, Fig. 2).
Fig. 3.
Reproducibility of the BED assay showing concordance between initial and confirmatory test results. The best-fit regression line is shown with statistics in the lower right portion of the figure. Two specimen results with very divergent initial and confirmatory results are indicated by diamonds (◆).
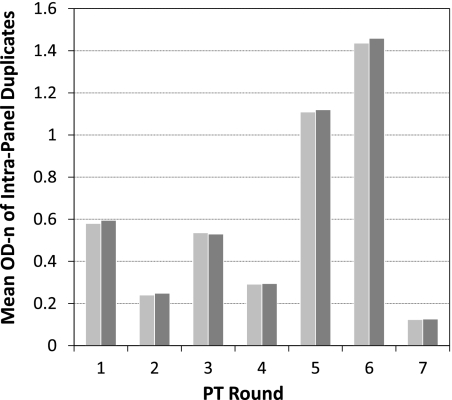
Each PT round included coded duplicate specimens. This allowed us to compare reproducibility. The overall mean of duplicate specimens of all participating laboratories is shown in Fig. 4. The mean OD-n results of the duplicates were very similar for each round (mean deviation, 1.8%; range, 0.95 to 3.75%), again demonstrating the high precision of the assay.
Fig. 4.
Reproducibility of coded, duplicate specimens sent during each round of PT program. The mean OD-n of all participants is shown for duplicate specimens for all seven rounds.
The seven PT rounds resulted in 343 plates contributing to as many values for each of the four controls routinely used in the assay. The mean OD of all runs indicated a coefficient of variation (CV) of ca. 20% for specimens in the dynamic range (0.5 to 1.5 OD), which was reduced to <10% when the OD was normalized by using CAL specimen, as recommended (7, 23) (Table 1). The CV for the negative control (NC) specimen remained ca. 50%, which is expected at the low end.
Table 1.
Mean OD and OD-n values for calibrator and control specimens for 343 laboratory reports obtained during seven rounds of the PT program, along with standard deviations and the percent coefficients of variationa
| OD | Parameter | NC | CAL | LPC | HPC |
|---|---|---|---|---|---|
| Raw OD450 | Mean | 0.049 | 0.901 | 0.552 | 1.394 |
| SD | 0.026 | 0.170 | 0.112 | 0.262 | |
| %CV | 53.5 | 18.9 | 20.3 | 18.8 | |
| OD-n | Mean | 0.054 | 1.000 | 0.614 | 1.549 |
| SD | 0.026 | 0.0 | 0.044 | 0.103 | |
| %CV | 47.2 | 0.0 | 7.2 | 6.6 |
%CV, percent coefficient of variation; NC, negative control; CAL, calibrator; LPC, low-positive control; HPC, high-positive control.
Cause of laboratory error and corrective actions.
We further investigated the cause of PT failure for a small number of reports (n = 8). Common problems identified are listed in Table 2; these problems include transcription error and no explanation for the failure as the most common occurrence (three each). Transcription errors typically occurred when the technician transferred data from the laboratory form to the PT report form. There were three occasions when a specimen was misclassified but no explanation was available. The three unresolved misclassifications were due to either the lab not having another test kit to repeat the testing or the participant not returning a corrective action form. Two laboratories each had kit reagent problems and specimen processing errors. Examples of specimen handling errors included specimen mix-up and testing an older PT panel by mistake. Kit problems included the use of an expired kit, and one laboratory was not notified by the kit manufacturer of a reagent substitution, which resulted in invalid QC. Additional identified problems included invalid QC and use of an incorrect cutoff (one each). In most cases, we worked with individual laboratories to identify problems and improve laboratory practices, where possible. The total number of issues listed is higher than the number of failing reports (n = 8) because multiple issues were identified for some reporting laboratories.
Table 2.
Summary of issues that were identified for eight participants with a <100% score in the PT programa
| Issue | No. of occurrences |
|---|---|
| Transcription error | 3 |
| Misclassified specimen (no explanation) | 3 |
| Kit problems | 2 |
| Specimen processing error | 2 |
| Invalid QC | 1 |
| Use of incorrect cutoff | 1 |
For some participants, more than one issue was identified.
DISCUSSION
We reported the development of the BED assay in 2002 (23), and the assay was commercially available in 2005. Since then, the number of laboratories using the BED assay for incidence surveillance has increased over time. Unlike HIV diagnostic enzyme immunoassays (EIAs), which are qualitative in nature, incidence tests are quantitative antibody assays that require higher precision to ensure that the classification of recent HIV infection is based on validated criteria and is not influenced by variability among laboratories. Since 2006, we have successfully implemented the PT program for the BED assay. Our PT program has helped to monitor the performance of participating laboratories while assessing the reproducibility of the BED assay in the field. Since inception of the program, the participation has increased from 12 laboratories to 38. We observed a high return rate of results (overall, 96.1%), which is most likely attributable to preshipment confirmation of participation. Occasional nonparticipation in the program was due to a lack of kits in the inventory and staff turnover. Because HIV surveillance is a periodic event, BED kits are not regularly purchased and kept in the inventory. Staff turnover is a common problem; therefore, it is important to train additional staff to maintain competency.
Overall, the results have been accurate, with 95.4% of those participating in PT program matching with the expected results (100% score). Our comprehensive analyses of data from seven rounds indicated that the precision and reproducibility of the BED assay were outstanding among multiple laboratories, operators, and kit lots. This was clearly demonstrated by comparison of the mean participant data for each round with the expected results (Fig. 2). Duplicate specimens included in each round of PT showed that reproducibility was high when the mean OD-n values of all participants were compared for duplicate specimens (Fig. 4). Each lot of the BED assay is checked for quality and consistency by the CDC before it is distributed widely to other laboratories by manufacturers, which is reflected in its field performance. In addition, the assay is less prone to variability due to its capture format compared to the earlier, less-sensitive EIA version of the incidence assay.
Since the assay is commercially available, there are more laboratories using the BED assay than those participating in the PT program. This program is voluntary but cost-free; therefore, participation in this program is a nonpunitive opportunity for laboratories to monitor their performance and implement improvement measures. Enrollment in the PT program is expected to increase as more laboratories begin to implement incidence testing. With growth come several challenges, including the high cost of shipping plasma specimens on dry ice. We are looking at alternative approaches, including dried blood, serum, or plasma spots (22) or dried tube specimens (20), which would significantly reduce shipping costs because these specimens can be shipped at room temperature. In addition, we are exploring the online submission of results, which should greatly improve data acquisition and analysis.
In conclusion, our PT program is part of a comprehensive quality assurance program for HIV-1 incidence testing and provides an opportunity for laboratories to monitor performance of the BED assay and take corrective actions. Several groups have reported the development of different incidence assays (1, 21, 25, 27, 29, 30). As these assays become implemented more widely, it will be important to ensure the quality of testing by multiple approaches, including the PT program. The PT program implemented by us should serve as a good template to monitor and improve quality of incidence testing as new assays become available.
ACKNOWLEDGMENTS
We thank all participants who contributed data summarized in this report and Connie Sexton for her review and comments.
Footnotes
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Barin F., et al. 2005. Development and validation of an immunoassay for identification of recent human immunodeficiency virus type 1 infections and its use on dried serum spots. J. Clin. Microbiol. 43:4441–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cha Y. J., Cho H. I. 2002. External quality assurance in diagnostic immunology: a twenty-year experience in Korea. Southeast Asian J. Trop. Med. Public Health 33(Suppl. 2):104–111 [PubMed] [Google Scholar]
- 3. Chalermchan W., Pitak S., Sungkawasee S. 2007. Evaluation of Thailand national external quality assessment on HIV testing. Int. J. Health Care Quality Assurance 20:130–140 [DOI] [PubMed] [Google Scholar]
- 4. Constantine N. T., et al. 2003. Improved classification of recent HIV-1 infection by employing a two-stage sensitive/less-sensitive test strategy. J. Acquir. Immune Defic. Syndr. 32:94–103 [DOI] [PubMed] [Google Scholar]
- 5. Demmler G. J., Istas A., Easley K. A., Kovacs A. 2000. Results of a quality assurance program for detection of cytomegalovirus infection in the pediatric pulmonary and cardiovascular complications of vertically transmitted human immunodeficiency virus infection study. J. Clin. Microbiol. 38:3942–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dhingra-Kumar N., Sharma A. K., Madan N. 1997. Analysis of quality assurance programmes for HIV screening in blood transfusion centres in Delhi. Bull. World Health Organ. 75:223–228 [PMC free article] [PubMed] [Google Scholar]
- 7. Dobbs T., Kennedy S., Pau C. P., McDougal J. S., Parekh B. S. 2004. Performance characteristics of the immunoglobulin G-capture BED-enzyme immunoassay, an assay to detect recent human immunodeficiency virus type 1 seroconversion. J. Clin. Microbiol. 42:2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hall H. I., et al. 2008. Estimation of HIV incidence in the United States. JAMA 300:520–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hannon W. H., Lewis D. S., Jones W. K., Powell M. K. 1989. A quality assurance program for human immunodeficiency virus seropositivity screening of dried-blood spot specimens. Infect. Control Hosp. Epidemiol. 10:8–13 [DOI] [PubMed] [Google Scholar]
- 10. Hargrove J. W., et al. 2008. Improved HIV-1 incidence estimates using the BED capture enzyme immunoassay. AIDS 22:511–518 [DOI] [PubMed] [Google Scholar]
- 11. Hu D. J., et al. 2003. HIV type 1 incidence estimates by detection of recent infection from a cross-sectional sampling of injection drug users in Bangkok: use of the IgG capture BED enzyme immunoassay. AIDS Res. Hum. Retrovir. 19:727–730 [DOI] [PubMed] [Google Scholar]
- 12. Jackson J. B., et al. 1993. Establishment of a quality assurance program for human immunodeficiency virus type 1 DNA polymerase chain reaction assays by the AIDS Clinical Trials Group. J. Clin. Microbiol. 31:3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janssen R. S., et al. 1998. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 280:42–48 (Erratum, 281:1893, 1999.) [DOI] [PubMed] [Google Scholar]
- 14. Jenner J., Grazioplene M., Kazianis A., Phinney K., Werner B. 2003. Modification of a commercial HIV-1 enzyme immunoassay for identification of recent HIV-1 infections: use of differential antibody avidity, abstr. 653. Tenth Conference on Retroviruses and Opportunistic Infections, Boston, MA [Google Scholar]
- 15. Jiang Y., et al. 2007. HIV-1 incidence estimates using IgG-capture BED-enzyme immunoassay from surveillance sites of injection drug users in three cities of China. AIDS 21(Suppl. 8):S47–S51 [DOI] [PubMed] [Google Scholar]
- 16. Kim A., et al. 2010. Evaluating the BED capture enzyme immunoassay to estimate HIV incidence among adults in three countries in sub-Saharan Africa. AIDS Res. Hum. Retrovir. 28:1051–1061 [DOI] [PubMed] [Google Scholar]
- 17. Kothe D., et al. 2003. Performance characteristics of a new less sensitive HIV-1 enzyme immunoassay for use in estimating HIV seroincidence. J. Acquir. Defic. Syndr. 33:625–634 [DOI] [PubMed] [Google Scholar]
- 18. Masciotra S., et al. 2010. Antibody avidity-based assay for identifying recent HIV-1 infections based on Genetic Systems TM1/2 Plus O EIA, abstr. 937. 17th Conference on Retroviruses and Opportunistic Infections, San Francisco, CA [Google Scholar]
- 19. Nesheim S., et al. 2005. Temporal trends in HIV type 1 incidence among inner-city childbearing women in Atlanta: use of the IgG-capture BED-enzyme immunoassay. AIDS Res. Hum. Retrovir. 21:537–544 [DOI] [PubMed] [Google Scholar]
- 20. Parekh B., et al. 2010. Dried tube specimens: a simple and cost-effective method for preparation of HIV proficiency testing panels and quality control materials for use in resource-limited settings. J. Virol. Methods 163:295–300 [DOI] [PubMed] [Google Scholar]
- 21. Parekh B., et al. 2007. Development of a limiting-antigen avidity-based enzyme immunoassay using a chimeric recombinant gp41 protein to detect recent HIV-1 infection, abstr. 342. 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA [Google Scholar]
- 22. Parekh B. S. 2005. Use of dried blood spots (DBS) or dried serum/plasma spots (DSS/DPS) to detect recent HIV-1 seroconversion by the BED-CEIA. HIV Diagnostics: New Developments and Challenges, Orlando, FL [Google Scholar]
- 23. Parekh B. S., et al. 2002. Quantitative detection of increasing HIV type 1 antibodies after seroconversion: a simple assay for detecting recent HIV infection and estimating incidence. AIDS Res. Hum. Retrovir. 18:295–307 [DOI] [PubMed] [Google Scholar]
- 24. Parekh B. S., Pau C. P., Kennedy M. S., Dobbs T. L., McDougal J. S. 2001. Assessment of antibody assays for identifying and distinguishing recent from long-term HIV type 1 infection. AIDS Res. Hum. Retrovir. 17:137–146 [DOI] [PubMed] [Google Scholar]
- 25. Rawal B. D., et al. 2003. Development of a new less-sensitive enzyme immunoassay for detection of early HIV-1 infection. J. Acquir. Immune Defic. Syndr. 33:349–355 [DOI] [PubMed] [Google Scholar]
- 26. Saphonn V., et al. 2005. Trends of HIV seroincidence among HIV sentinel surveillance groups in Cambodia, 1999-2002. J. Acquir. Immune Defic. Syndr. 39:587–592 [PMC free article] [PubMed] [Google Scholar]
- 27. Soroka S. D., Granade T. C., Candal D., Parekh B. S. 2005. Modification of rapid human immunodeficiency virus (HIV) antibody assay protocols for detecting recent HIV seroconversion. Clin. Diagn. Lab. Immunol. 12:918–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suligoi B., et al. 2002. Precision and accuracy of a procedure for detecting recent human immunodeficiency virus infections by calculating the antibody avidity index by an automated immunoassay-based method. J. Clin. Microbiol. 40:4015–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wei X., et al. 2010. Development of two avidity based assays to detect recent HIV-1 seroconversion using a multi-subtype gp41 recombinant protein. AIDS Res. Hum. Retrovir. 26:61–71 [DOI] [PubMed] [Google Scholar]
- 30. Wilson K. M., et al. 2004. Incidence immunoassay for distinguishing recent from established HIV-1 infection in therapy-naive populations. AIDS 18:2253–2259 [DOI] [PubMed] [Google Scholar]